Abstract
Covalent attachment of the small ubiquitin-related modifier, SUMO, to substrate proteins plays a significant role in plants under stress conditions, which can alter target proteins' function, location, and protein-protein interactions. Despite this importance, information about SUMOylation in the major legume crop, soybean, remains obscure. In this study, we performed a bioinformatics analysis of the entire soybean genome and identified 40 genes belonged to six families involved in a cascade of enzymatic reactions in soybean SUMOylation system. The cis-acting elements analysis revealed that promoters of SUMO pathway genes contained different combinations of stress and development-related cis-regulatory elements. RNA-seq data analysis showed that SUMO pathway components exhibited versatile tissue-specific expression patterns, indicating coordinated functioning during plant growth and development. qRT-PCR analysis of 13 SUMO pathway members indicated that majority of the SUMO pathway members were transcriptionally up-regulated by NaCl, heat and ABA stimuli during the 24 h period of treatment. Furthermore, SUMOylation dynamics in soybean roots under abiotic stress treatment were analyzed by western blot, which were characterized by regulation of SUMOylated proteins. Collectively, this study defined the organization of the soybean SUMOylation system and implied an essential function for SUMOylation in soybean abiotic stress responses.
Keywords: soybean, SUMO pathway, expression profiling, abiotic stress, SUMOylation
Introduction
The conjugation of protein tags or modifiers to target proteins is one important protein post-translational modification for the maintenance of cellular homeostasis and biological activities in eukaryotes during rapid environmental changes, when physiological responses often occur both quickly and reversibly. In recent years, the small ubiquitin-related modifier (SUMO) proteins of approximately 100–115 amino acids, has emerged as an influential class of protein modifiers which has been shown to regulate various nuclear processes, including transcriptional control, subcellular trafficking, and regulation of the cell cycle (Miller et al., 2010, 2013; Park H. C. et al., 2011).
The modification process of SUMOylation is mechanistically similar to the conjugation of ubiquitin, the most prominent representative of small protein modifiers with conserved structure (Rodriguez et al., 2001; Matic et al., 2010). SUMO is synthesized as a pre-protein that needs to be processed by ubiquitin-like proteases (ULP) to expose a carboxyl-terminal double glycine (di-Gly) motif. Analogous to ubiquitination, SUMO conjugation in plants is facilitated through a multi-step enzymatic pathway including E1 enzymes for SUMO activation, E2 enzymes for conjugation and E3 enzymes for ligation (Castro et al., 2012). The end result is SUMO isopeptide conjugated via its C-terminal diGly motif to accessible lysine (Lys) within substrate proteins. Interestingly, SUMOylation is a reversible process, and de-SUMOylation is accomplished through ULP class of SUMO proteases which release free SUMOs from protein conjugates for further conjugation cycles (Colby et al., 2006; Mukhopadhyay and Dasso, 2007).
Unlike ubiquitylation, SUMOylation is not typically committing proteins to breakdown by the 26S proteasome, but can change target protein localization, repress or enhance activity of target transcription factor, regulate interactions between proteins, and mediate cross talk with other post-translational modifications (Bossis and Melchior, 2006). SUMO conjugation was reported to be essential in Arabidopsis, and an increasing number of papers have demonstrated that mutants that were not able to attach SUMO1/2 onto substrate proteins exhibit typical phenotypes such as growth reduction, impaired salinity, drought, heat and freezing tolerance, and altered response to phosphate starvation (Roden et al., 2004; Catala et al., 2007; Miura et al., 2007b, 2011; van den Burg et al., 2010). Previous studies with rice (Oryza sativa) and poplar (Populus strichocarpa) showed that SUMOylation can be significantly enhanced upon heat, cold, high salinity, and abscisic acid exposure (Chaikam and Karlson, 2010; Reed et al., 2010; Li et al., 2013), which was always accompanied by a decrease in the pool of free SUMO and correlated with the duration and intensity of the stress. Interestingly, SUMOylation levels decrease within hours or even minutes when the heat shocked plants returned to the normal temperature, suggesting that SUMOylation acts transiently and reversibly (Kurepa et al., 2003; van den Burg et al., 2010). Recently, SUMOylation was reported to display a striking memory that dampened succeeding conjugate increases if a second stimulus was provided too close to the first one (Kurepa et al., 2003; Miller et al., 2013).
Further genetic studies provided evidence that several members of SUMOylation system contributed to the regulation of development and adaptation to abiotic stress conditions. SUMO1 and SUMO2 in Arabidopsis appeared to be functionally redundant, but elimination of them was embryo lethal (Park H. J. et al., 2011). On the other hand, over-expression of either AtSUMO1 or AtSUMO2 gene was correlated with ABA-mediated growth inhibition (Tatham et al., 2001; Aguilar-Martinez et al., 2015). Besides, null E3 ligase SIZ1 alleles (siz1-1, siz1-2, and siz1-3) in Arabidopsis displayed several phenotypes including hypersensitive to extreme temperature and drought stress, as well as enhanced tolerance to salinity stress, indicating that SIZ1-mediated SUMOylation play an important role in response to abiotic stress (Miura et al., 2007b, 2011; Cheong et al., 2009; Chen et al., 2011). Moreover, Arabidopsis mutants lacking SUMO proteases OTS1 and OTS2 showed inhibition of root growth under salt treatment (Conti et al., 2008), while transgenic rice plants over-expressing OsOTS1 gene exhibited increased salt tolerance concomitant with reduced levels of SUMOylated proteins (Srivastava et al., 2016). It indicated that manipulation of conjugation or de-conjugation of SUMO from target proteins could be an effective method in plants to cope with stress conditions.
Soybean is one of the most important legume crops in the world, offering high-quality protein and vegetable oil for human and animal consumption, but the growth and development of soybean seedlings is always affected by adverse conditions (Nakashima et al., 2009; Schmutz et al., 2010). Given the potential importance of SUMOylation to stress defense, presented here is a comprehensive description of core components of SUMO system in soybean. We also investigated the responses of SUMOylation system under salt, high temperature and abscisic acid stress conditions both at transcriptional and translational levels. Our results may suggest a role for SUMOylation in soybean abiotic stress responses, and provide valuable information for further functional studies of SUMOylation cascade components in soybean.
Materials and methods
Identification and analysis of soybean SUMO pathway genes
Soybean SUMO pathway genes were identified by tblastn searches against the soybean genome at Phytozome v11.0 (https://phytozome.jgi.doe.gov/pz/portal.html), using the amino acid sequences of the known pathway genes from Arabidopsis as queries. The molecular weight and isoelectric point (pI) of each gene were calculated by ExPASy (http://www.expasy.org/tools/). Protein subcellular localization was predicted by the WoLF PSORT (http://www.genscript.com/wolf-psort.html) (Horton et al., 2007).
Multiple sequence alignments of SUMO pathway components in soybean, Arabidopsis, poplar, rice, maize and tomato were carried out using clustalW with default parameters (Thompson et al., 2002). The phylogenetic tree was constructed by MEGA (V7.0.20) using the neighbor-joining (NJ) method with the following parameters: Poisson correction, pair-wise deletion and 1000 bootstrap replicates (Tamura et al., 2007). Protein domains were predicted using Pfam (http://pfam.xfam.org) and visualized using BoxShade online software (http://www.ch.embnet.org/software/BOX_form.html) (Beitz, 2000). In addition, possible SUMOylation sites and SIM sequences were predicted using the medium settings of program GPS-SUMO v1.0.1 (Zhao et al., 2014).
Cis-acting elements analysis in promoters of the soybean SUMO pathway genes
PlantCARE online program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html; Lescot et al., 2002) was used to predict cis-acting elements in promoters of soybean SUMO pathway genes. Sequence of 1,500 bp upstream of the start codon downloaded from Phytozome 11.0 database was used for cis-acting elements scan.
RNA-Seq atlas analysis of SUMO pathway genes in different tissues
The normalized transcriptome data (Reads/Kb/Millon, RPKM) of 14 soybean samples from six different tissues harvested at different growth periods was downloaded from the SoyBase website (https://soybase.org/soyseq/). These samples include young leaf, flower, one cm pod, pod-shell 10-DAF (days after flowering), pod-shell 14-DAF, root, nodule and developing seeds (seed harvested at 10-DAF, 14-DAF, 21-DAF, 25-DAF, 28-DAF, 35-DAF, and 42-DAF) (Severin et al., 2010). The criteria of expressed gene was the RPKM value equal to or greater than two in the expression atlas (Belamkar et al., 2014). The RPKM normalized read count data of expressed gene was then log2-transformed and displayed in the form of heatmap using the OmicShare tools (http://www.omicshare.com/tools).
Plant growth and treatments
Soybean seeds (Glycine max cv Dongnong 50) were germinated on two layers of filter paper soaked in distilled water at 22°C in the dark. After 2 days, germinated seedlings were transferred to vermiculite and irrigated with half-strong modified Hoagland nutrient solution (pH 5.8) in a growth chamber under normal conditions (25/21°C day/night temperature, relative humidity of 60–80% and 16 h light period/day at intensity of 120 μmol photons m−2 s−1) for 2 weeks. When the second trifoliate leave unrolled, soybean seedlings were transferred to 1/2 Hoagland solution. Stress treatments were performed as follows: for salt treatment, the roots of soybean plants were immersed in nutrient solution containing 200 mM NaCl at room temperature; for heat treatment, hydroponic seedlings were transferred into a 42°C growth chamber; for ABA treatment, soybean seedlings were grown in nutrient solution added 100 μM ABA. Root tissues were harvested after 0, 0.5, 1, 6, 12, and 24 h exposure to salt, heat, and ABA treatment. Three independent sets of control and stress treatment samples were collected, and each replicate represented a pooled sample of three individual plants. After collection, all the samples were immediately frozen in liquid nitrogen and stored at −80°C until use.
RNA extraction and qRT-PCR analysis
The total RNA samples were isolated using Ultrapure RNA Kit (CWBIO, China). cDNA was synthesized using 2 μg RNA using the HiScript II Q Select RT SuperMix for qPCR Kit (Vazyme, China). Gene specific primers for quantitative real-time RT-PCR (qRT-PCR) analysis were designed using Primer 5.0 according to soybean cDNA sequences (Table S1). The soybean actin 11 was used as internal reference gene. qRT-PCR reaction was performed using ChamQ SYBR qPCR Master Mix (Vazyme, China) and was conducted on ABI 7300 Real-time Detection System (Applied Biosystems, USA). The PCR reaction was carried out with the following reaction conditions: 95°C for 20 s; followed by 40 cycles of 95°C for 15 s, 60°C for 20 s and 72°C for 20 s. Samples for qRT-PCR were run in 3 biological replicates with 3 technical replicates and the data were represented as the mean ± SD (n = 3) for Student's t-test analysis. The relative gene expression was calculated using the ΔΔCt algorithm (Livak and Schmittgen, 2001; Bustin et al., 2009).
Prokaryotic expression and purification of soybean SUMO proteins
GmSMO1, GmSMO2, and GmSUMO4 genes were amplified by PCR using soybean cDNA as template. After sequencing, the amplified products were added Sac I and Hind III restriction sites to clone into the pET32a expression vector. The resultant vectors were transformed into E. coli BL21 cells, which were induced for expression by adding 1 mM IPTG after achieving OD600 0.6 in LB broth. Cells were collected after 4 h of growth at 28°C, and centrifuged at 4000 × g for 30 min then resuspended in PBS buffer. After sonication, the soluble protein was used directly for SDS-PAGE or purification using Ni Sepharose 6 Fast Flow resin (GE Healthcare) according to protocols in the manual.
Protein extraction and western blot analysis
Total protein samples from three biological replicates were isolated from soybean root tissues using previously described method (Cai et al., 2017). Protein concentration was determined by Bradfrod method using BSA as the standard. 15 μg of purified GmSUMO protein and each protein sample of soybean roots was separated on 12% SDS−PAGE and transferred to nitrocellulose membrane according to standard protocols. The membrane was blocked using phosphate buffered saline (PBS) buffer containing 5% milk for 1 h at room temperature, then probed with AtSUMO1 primary antibody (AbcamInc., China, ab5316; a rabbit polyclonal antibody that reacts with ArabidopsisSUMO1/2) which was prepared in the PBS buffer containing 1% BSA (1:4000) at 4°C for overnight. After removing unbound antibodies by washing with a PBST buffer (PBS containing 0.05% Tween 20), the blot was incubated with goat antirabbit IgG secondary antibody (horseradish peroxidase conjugated, Thermo FisherScientific, USA) in the PBST buffer at a dilution of 1:20000. Chemiluminescence was carried out using the ECL Plus kit according to manufacturer's (Merck Millipore, USA) instructions.
Results
SUMO pathway genes in soybean
Using homology searches and domain confirmation, a total of 40 genes encoding SUMO pathway members were found in soybean, including six family groups. The detailed characteristics of each family member were shown in Table 1.
Table 1.
Characteristics of soybean SUMO pathway genes.
| Group | Gene Name | Locus Name | Chromosome | Gene length (bp) | Number of Exon | Basic features of Protein | Subcellular location | ||
|---|---|---|---|---|---|---|---|---|---|
| Length (aa) | MW (Da) | pI | |||||||
| SUMO | GmSUMO1 | Glyma.08G320500 | 8 | 300 | 3 | 99 | 11,203.5 | 4.71 | mito: 6, nucl: 5, cyto: 1, plas: 1 |
| GmSUMO2 | Glyma.18G165200 | 18 | 345 | 3 | 114 | 12,969.1 | 5.58 | nucl: 8, cyto: 2, chlo: 1, mito: 1, plas: 1 | |
| GmSUMO3 | Glyma.08G350600 | 8 | 354 | 3 | 117 | 13,210.6 | 5.68 | nucl: 10, cyto: 2, chlo: 1 | |
| GmSUMO4 | Glyma.08G111700 | 8 | 321 | 3 | 106 | 12,199.7 | 6.67 | nucl: 7, cyto: 5, extr: 2 | |
| GmSUMO5 | Glyma.08G111,800 | 8 | 297 | 3 | 98 | 11,115.8 | 9.14 | cyto: 5, nucl: 4, chlo: 2, mito: 2 | |
| GmSUMO6 | Glyma.05G154000 | 5 | 297 | 3 | 98 | 11,144 | 9.59 | nucl: 6, cyto: 5, mito: 2 | |
| GmSUMO1 | Glyma.08G320500 | 8 | 300 | 3 | 99 | 11,203.5 | 4.71 | mito: 6, nucl: 5, cyto: 1, plas: 1 | |
| GmSUMO2 | Glyma.18G165200 | 18 | 345 | 3 | 114 | 12,969.1 | 5.58 | nucl: 8, cyto: 2, chlo: 1, mito: 1, plas: 1 | |
| E1 | GmSAE1a | Glyma.08G011400 | 8 | 846 | 9 | 281 | 31,131.4 | 5.28 | extr: 11, cyto: 2 |
| GmSAE1b | Glyma.05G204000 | 5 | 996 | 10 | 331 | 36,839.8 | 5.56 | nucl: 6, cyto: 6, mito: 1 | |
| GmSAE2a | Glyma.13G201500 | 13 | 2,052 | 11 | 683 | 70,966 | 4.94 | chlo: 4, plas: 3, nucl: 2, E.R.: 2, cyto: 1, extr: 1 | |
| GmSAE2b | Glyma.12G236000 | 12 | 1,911 | 11 | 636 | 70,911.1 | 5.03 | chlo: 5, plas: 3, nucl: 2, cyto: 1, vacu: 1, E.R.: 1 | |
| E2 | GmSCEa | Glyma.11G053300 | 11 | 480 | 5 | 159 | 18,000.3 | 8.47 | nucl: 7, cyto: 3, mito: 2, plas: 2 |
| GmSCEb | Glyma.01G188900 | 1 | 480 | 5 | 159 | 18,000.3 | 8.47 | nucl: 7, cyto: 3, mito: 2, plas: 2 | |
| GmSCEc | Glyma.17G169700 | 17 | 483 | 5 | 160 | 18,067.4 | 8.47 | nucl: 10, plas: 2, cyto: 1 | |
| GmSCEd | Glyma.05G091100 | 5 | 483 | 5 | 160 | 18,067.4 | 8.47 | nucl: 10, plas: 2, cyto: 1 | |
| E3 | GmSIZ1a | Glyma.12G071300 | 12 | 2,643 | 16 | 880 | 95,834.9 | 4.89 | nucl: 13 |
| GmSIZ1b | Glyma.U020100 | – | 2,640 | 16 | 879 | 95,988.2 | 4.83 | nucl: 13 | |
| GmSIZ1c | Glyma.12G170900 | 12 | 2,649 | 16 | 882 | 96,853.1 | 4.75 | nucl: 12, cyto: 2 | |
| GmSIZ1d | Glyma.13G328100 | 13 | 2,643 | 17 | 880 | 96,878.1 | 4.78 | nucl: 12, cyto: 2 | |
| GmMMS21 | Glyma.13G273500 | 13 | 744 | 7 | 247 | 27,767.8 | 4.74 | chlo: 10.5, chlo_mito: 7, mito: 2.5 | |
| GmPIAL1 | Glyma.11G020900 | 11 | 2,631 | 16 | 876 | 95,516.7 | 7.54 | chlo: 10, pero: 2, nucl: 1 | |
| GmPIAL2 | Glyma.01G222500 | 1 | 2,631 | 16 | 876 | 95,665.8 | 7.7 | nucl: 7, chlo: 4, cysk: 2 | |
| SUMO Chain binding protein | GmCBa | Glyma.11G103800 | 11 | 618 | 4 | 205 | 23,015.4 | 9.21 | nucl: 5, mito: 4, chlo: 3, cyto: 1 |
| GmCBb | Glyma.12G028700 | 12 | 618 | 4 | 205 | 23,149.5 | 9.21 | chlo: 6, nucl: 4, cyto: 2, mito: 2 | |
| GmCBc | Glyma.20G004500 | 20 | 405 | 4 | 134 | 14,955.4 | 8.79 | Unknown | |
| GmCBd | Glyma.09G281300 | 9 | 621 | 4 | 206 | 23,152.8 | 6.5 | nucl: 11, cyto: 1, mito: 1 | |
| Ubiquitin like protease | GmPROa | Glyma.04G193100 | 4 | 945 | 7 | 314 | 36,882.6 | 10.2 | cyto: 7, chlo: 2, E.R.: 2, golg: 2 |
| GmOTSa | Glyma.18G137700 | 18 | 1,761 | 14 | 586 | 67,944 | 6.95 | nucl: 13 | |
| GmOTSb1 | Glyma.08G287800 | 8 | 1,131 | 10 | 376 | 43,485.7 | 7.76 | nucl: 13 | |
| GmOTSb2 | Glyma.08G287700 | 8 | 576 | 4 | 191 | 21,854.85 | 4.9 | extr: 7, chlo: 2, cyto: 2, vacu: 2 | |
| GmB2a | Glyma.06G095600 | 6 | 2,874 | 16 | 957 | 10,6715.9 | 4.19 | nucl: 8, cysk: 3, plas: 1.5, golg_plas: 1.5 | |
| GmB2b | Glyma.04G093800 | 4 | 2,802 | 16 | 933 | 104,220.9 | 4.24 | nucl: 8, cysk: 3, plas: 1.5, golg_plas: 1.5 | |
| GmB2c | Glyma.15G058100 | 15 | 1,869 | 10 | 622 | 70,841.3 | 5.46 | chlo: 8, nucl: 5 | |
| GmB2d | Glyma.13G256800 | 13 | 1,932 | 16 | 643 | 93,129.9 | 4.51 | nucl: 5, mito: 4, chlo: 3, cysk: 1 | |
| GmB2e | Glyma.02G214100 | 2 | 2,166 | 9 | 721 | 83,035.8 | 4.6 | nucl: 12, cyto: 2 | |
| GmESD4a | Glyma.15G148700 | 15 | 1,410 | 9 | 470 | 54,530.6 | 8.51 | nucl: 6, cyto: 5, chlo: 2 | |
| GmESD4b | Glyma.09G044400 | 9 | 1,407 | 9 | 468 | 54,626 | 8.88 | nucl: 8, cyto: 3, chlo: 2 | |
| GmESD4c | Glyma.17G027100 | 17 | 1,503 | 9 | 500 | 57,721.8 | 9 | chlo: 8, nucl: 3, pero: 2 | |
| GmESD4d | Glyma.07G246900 | 7 | 1,539 | 9 | 512 | 59,051.9 | 8.2 | chlo: 9, nucl: 3, mito: 1 | |
bp, base pair; aa, amino acids; Da, Dalton; pI, isoeletricponit Subcellular predictions; chlo, (chloroplast); cyto, (cytosol); nucl, (nucleus); E.R., (endoplasmic reticulum); mito, (mitochondria); plas, (plasma membrane); extr, (extracellular); cysk, (cytoskeleton); plas, (plasma membrane); vacu, (vacuolar membrane); cyto, (cytoplasmic); pero, (peroxisomal); golg, (golgi).
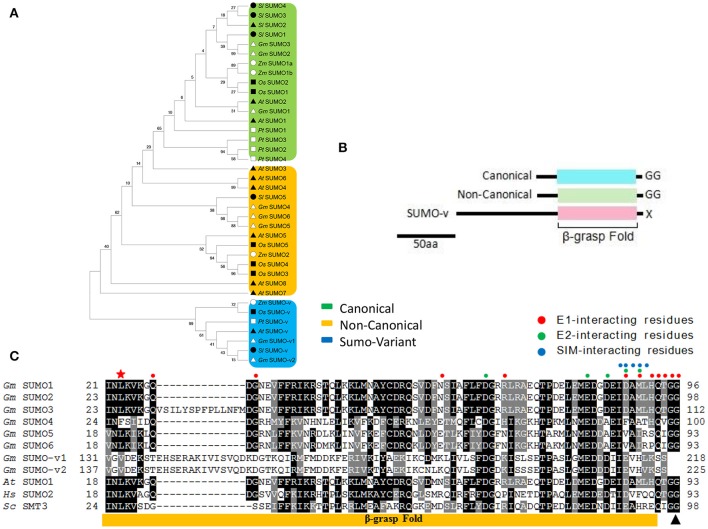
Our soybean list included six SUMO genes, designated as GmSUMO1 to GmSUMO6, all of which had three exons and encoded proteins harboring a SUMO-type β-grasp domain and predicted to localize in the nucleus (Table 1). Among these SUMO genes, GmSUMO2 and GmSUMO3 encoded proteins sharing 92 and 80% similarity with that from AtSUMO1, while GmSUMO1 encoded a protein with 85% identity to AtSUMO2. By contrast, GmSUMO4, GmSUMO5, and GmSUMO6 encoded products with little resemblance to either AtSUMO1/2 or GmSUMO1/2/3. To better understand the evolutionary relationship between these SUMO isoforms, we searched a number of other plant genomes for related sequences and constructed a phylogenetic tree (Figure 1A). Phylogenetic analysis revealed a highly conserved SUMO group including GmSUMO1/2/3, AtSUMO1/2, ZmSUMO1a/b and conserved representatives from other species. In contrast, a “non-canonical” group that included GmSUMO4/5/6, AtSUMO3/5, and ZmSUMO2 encoded proteins shared about 30% amino acid similarities to their canonical paralogs (Figure 1A). Regardless of their dissimilarities, it is noteworthy that non-canonical members also have the C-terminal di-Gly motif necessary for conjugation (Figure 1B) which indicated their potential abilities of covalent attachment to target proteins. In support, we identified some residues important for the non-covalent binding of SUMOs to E1, E2, and SUMO-interacting motifs (SIMs, ψψXψD/S/E, D/S/EψXψψ or ψψDLT, where ψ stands for the hydrophobic amino acids and X represents any residue) conserved across the biological kingdoms (Figure 1C). Besides these identified SUMO proteins above mentioned, two longer SUMO related proteins designated as SUMO-variants (V) were detected in soybean genome (Figure 1). GmSUMO-Vs contained the N-terminal extension of more than 200 residues upstream of the SUMO β-grasp domain (Table 1 and Figure 1B). Without an exposed diGly motif (Figures 1B,C), SUMO-v may not enter the well-studied E1/E2/E3 SUMOylation pathway, but probably have other functions to be explored in plants.
Figure 1.
The genomes of soybean and other plant species encode a family of SUMO-related proteins (A) Protein sequences from Arabidopsis thaliana, Zea mays, Oryza sativa, Solanum lycopersium, Populus trichocarpa and Glycine max (Table S2) were used to construct the phylogenetic tree by the neighbor-joining method in MEGA. They were classified into three groups: canonical SUMO, non-canonical SUMO, and SUMO-variant. (B) Domain structures of the plant SUMO family that include the canonical and non-canonical SUMO isoforms, and SUMO-variant containing a long, conserved N-terminal extension in front of the β-grasp domain. (C) Alignment of SUMO sequence reveals conserved and divergent residues. Included are soybean (Gm) SUMO-1, -2, -3, -4, -5, -6, and SUMO-v1, -v2, along with canonical SUMOs from Arabidopsis (AtSUMO1), human (HsSUMO2, NP_008868.3), and yeast (ScSmt3, KZV12750.1), only conserved region is shown in the figure. The yellow box locates the β-grasp-fold. The black triangle locates the processing site by ULP that exposes the diGly motif essential for conjugation in canonical SUMOs. Residues marked with red, green, and blue circle dots are important for the non-covalent binding of SUMOs to E1, E2, and SIMs (SUMO interacting motif), respectively. The asterisk denotes the conserved Lys required for forming SUMO-chains. Gray and black boxes identify similar and conserved amino acids, respectively. Dashes denote gaps.
The initial activation of SUMOylation is adenylation of mature SUMO on the C-terminus diGly by the heterodimeric SAE1/SAE2 E1. Soybean genome expressed two small subunit SAE1 isoforms (GmSAE1a/b) and two large subunit SAE2 isoforms (GmSAE2a/b) (Table 1). The ubiquitin-fold domain and cysteine domain important to E1 catalysis were shown in Figure S1. After being activated by E1, the bound SUMO is transferred to an active-site cysteine in the E2 by transesterification (Miura et al., 2007a). In contrast with the single SCE1 gene in Arabidopsis, the soybean genome contained four E2 genes named GmSCEa, GmSCEb, GmSCEc, and GmSCEd (Table 1). Conformed with the observations made earlier in the homologs AtUbc9 (Kraft et al., 2005) and OsSce1 (Nigam et al., 2008), all of these four genes were predicted to encode proteins localized predominantly to the nucleus along with SUMOs, however, whether GmSCEs function related to the nuclear pore complex is still not known and deserved further studies. Motif-scan analysis of GmSCEa-d revealed that they have highly conserved Ub-Conjugating Enzyme Catalytic (UBC) domain (spanning from 8 to 150aa residues; Figure S2A), which was common to UBC family members (Nigam et al., 2008). Phylogenetic analysis reflected that soybean SCE genes had high similarities with homologs in other species (Figure S2B) highlighted the conservation of E2 proteins. Collectively, we speculated that soybean homologs of SUMOylation system will be biochemically functional.
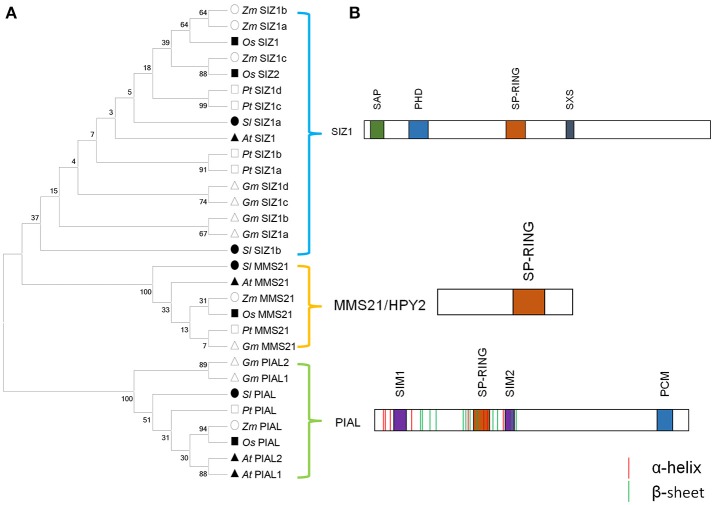
Although SCE itself can directly transfer SUMO to substrates, E3 ligases are often required to increase the rate of SUMOylation and increase the substrate specificity during SUMOylation process (Gareau and Lima, 2010). When searching the soybean genome using E3 genes in Arabidopsis, we got seven soybean E3 genes (Table 1), all of which contain the conserved Siz1/PIAS(SP)-RING domain which plays the role of binding to the SUMO-E2 intermediate. These E3 genes could be divided into three groups, including SIZ1, Methyl Methane sulfonate-Sensitivity protein-21/High Ploidy-2 (MMS21/HPY2), and Protein Inhibitor of Activated STAT (PIAS)-Like (PIAL) (Figure 2A). The soybean SIZ1 group contained four proteins (GmSIZ1a-d) that included the signature MIZ/SP-RING domain, as well as PINIT and SXS motifs that promoted selective SUMOylation (Figure 2B; Yunus and Lima, 2009), which was reported to be important for Arabidopsis SIZ1 activity (Cheong et al., 2009). Phylogenetic analysis of SIZ1 type SUMO E3 ligases from various plant species showed that GmSIZ1a-d were evolutionarily most closely to PtSIZ1a/b (Figure 2A).
Figure 2.
Soybean genome encodes three types of E3 ligase protein. (A) E3 protein sequences from Arabidopsis thaliana, Zea mays, Oryza sativa, Solanum lycopersium, Populus trichocarpa, and Glycine max (Table S2) were used to construct the phylogenetic tree. These E3 ligases can be classified into three groups. (B) Schematic representation of functional domains of SIZ1, HPY/MMS21 and PIAS-like type SUMO ligase.
Thirteen SUMO protease genes in soybean were classified into four groups (Table 1 and Figure 3), all of which have the signature peptidase_C48 domain characterized by a catalytic triad of His-Asp-Cys that leads to peptide bond cleavage (Colby et al., 2006; Mukhopadhyay and Dasso, 2007). For class A, there was only one member in soybean, confirmed the notion that most plants encoded a single gene of this class (Figure S3; Novatchkova et al., 2012). For GmESD4a-d, several SIM sequences were identified that could bind SUMO (Figure S4). For the OTS family in soybean, the predicted GmOTSa encoded protein contained intact peptidase_C48 domain, whereas two loci Glyma.08G287800 and Glyma.08G287700 in soybean genome encoded N-terminal and C-terminal of the peptidase_C48 domain respectively (Figure 3), and whether either of them could exhibit SUMO protease activity is not known and deserved further study.
Figure 3.
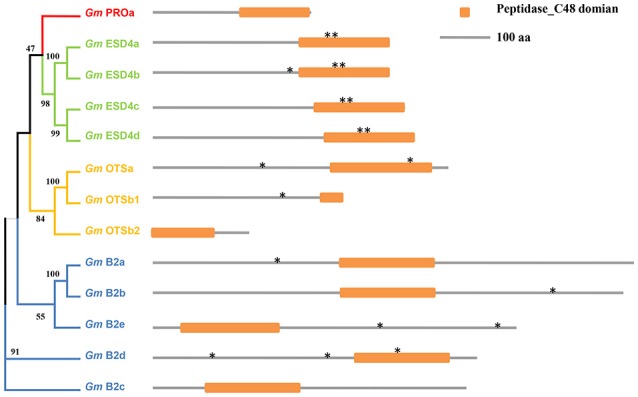
Schematic representation of functional domains of ULP family members in soybean genome. The phylogenetic analysis was carried out using MEGA (V7.0.20). Orange rectangles indicate the peptidase_C48 domain. Asterisks indicate the putative SIM motifs.
In addition, soybean genome contained another family of SUMO chain binding protein genes characterized by a tandem arrangement of four SIMs (Table 1 and Figure S5), which therefore have specific affinity for binding to SUMO chains. Besides, these proteins share a RING domain which was speculated to direct proteins with SUMO chains into the ubiquitin-proteasome degradation pathway (Plechanovová et al., 2011; Praefcke et al., 2012).
cis-acting regulatory elements in the promoters of SUMO pathway genes
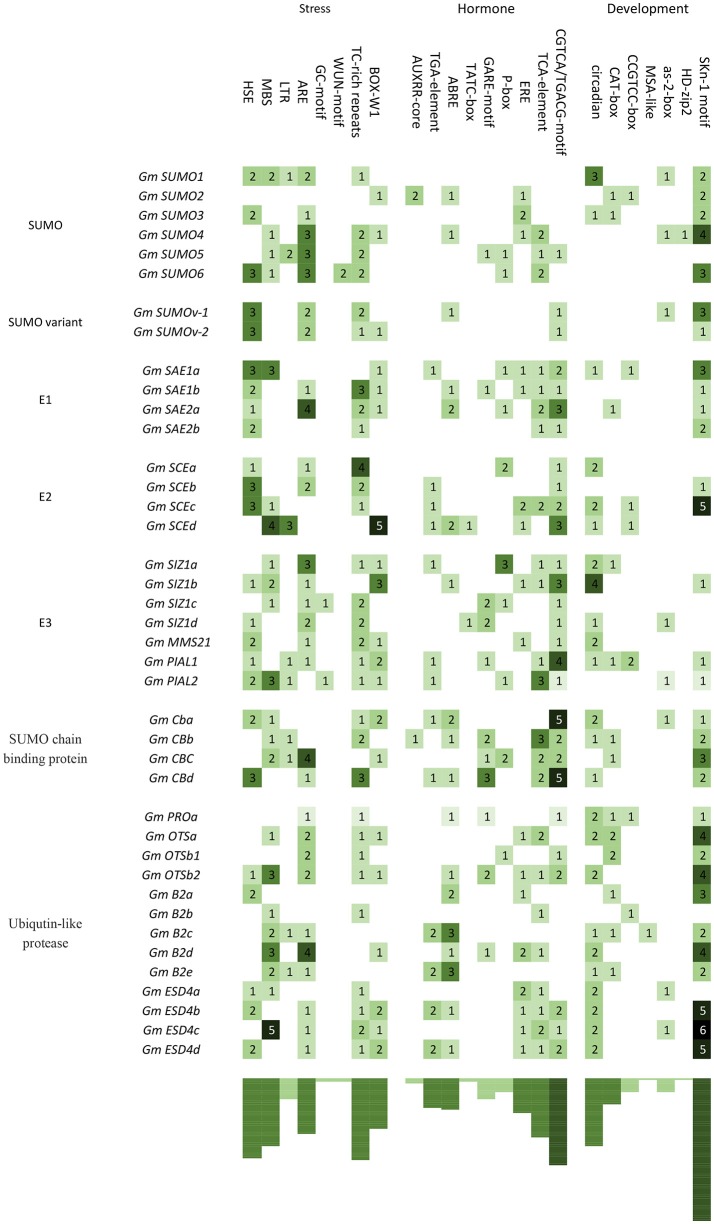
In order to get some information of stimulus induced as well as temporal and spatial specific expression patterns of these soybean SUMO pathway genes, 1,500 bp promoter regions upstream of all of these 40 soybean genes were extracted and used for cis-acting elements searches (Table S3). As shown in Figure 4, most genes involved in SUMOylation in soybean contained several environmental stress-related elements, including cis-elements that were related to plant responses to heat (HSE), drought (MBS), low temperature (LTR), anaerobic stress (ARE), and fungus (BOX-W1). Besides, several promoters contained cis-elements related to MeJA (CGTCA/TGACG-motif), salicylic acid (TCA-element), ABA (ABRE), GA (TATC-box, GARE-motif and P-box), auxin (AUXRR-core and TGA-element), and ethylene (ERE) (Figure 4). These results suggested that these genes were likely to be transcriptionally regulated upon environmental stresses and hormone signals. Moreover, seven types of cis-acting elements were involved in plant meristem (CAT-box and CCGTCC-box), endosperm (SKn-1 motif), leaf and shoot development (HD-zip2 and as-2-box), cell cycle (MSA-like) and circadian control (circadian), which covered most plant developmental stages (Figure 4). It suggested that SUMO pathway genes may be transcriptionally regulated during growth and development of soybean plants.
Figure 4.
cis-acting regulatory elements identified in soybean SUMO pathway genes. One Thousand and five hundred blood pressure upstream sequences of 40 SUMO pathway genes were analyzed. The number and abundance of each cis-element are shown in the figure. Promoter sequences, cis-element sequences and annotation of each cis-element are available in the Tables S3,S4.
Expression of SUMO pathway genes in different tissues of soybean
Gene functions are always closely related to where and how they are transcriptionally expressed. Transcript profiles from 14 tissues/organs were collected from soybean RNA-Seq data and downloaded from the Soybase (Table S5). A total of 30 SUMO pathway genes were expressed at a variety of developmental stages in soybean (Figure 5). Among these SUMO genes, GmSUMO1 was expressed at very high levels in almost all tissues, GmSUMO2 and GmSUMO3 showed high expression levels in aerial and underground tissues, but much lower expression levels in seed developing stages. It suggested that GmSUMO1 could be functional complement to the other two orthologs. In the E1 group, GmSAE1a/b and GmSAE2a/b genes exhibited relatively higher expression levels in aerial tissues, while low expression levels during seed development. Notably, all of the E2 genes showed high expression levels in all of these 14 tissues, suggesting this family of genes may support soybean normal growth and development. Interestingly, expression levels of E3 family members in some tissues were quite different. For example, expressions of GmSIZ1c and GmPIAL1 were hardly detected in nearly all tissues, while GmSIZ1a showed relatively high expression levels in most tissues, besides, GmSIZ1b showed higher expression levels in flower and root, but GmMMS21 expressed at relatively high levels in young leaf, one cm pod and nodule (Figure 5). Furthermore, our results showed that ULP family gene GmPROa expressed at low levels in almost all developmental stages, except the young leaf. Another interesting scenario was that GmB2d and GmESD4a/b/c were highly expressed in nodule, suggesting their potential roles in nodulation although experimental evidence is lacking for their involvement in nodule organogenesis or development. As shown in Figure 5, GmCBa and GmCBb exhibited similar expression patterns, suggesting potential redundant functions between the orthologs. Additionally, GmCBc was expressed at very low levels in almost all tissues, on the contrary, GmCBd gene showed strong expression in all of these investigated tissues. Given the results that most genes studied exhibited relatively higher expression levels in roots, the following transcriptional and translational expression profiles were performed using soybean root tissue.
Figure 5.
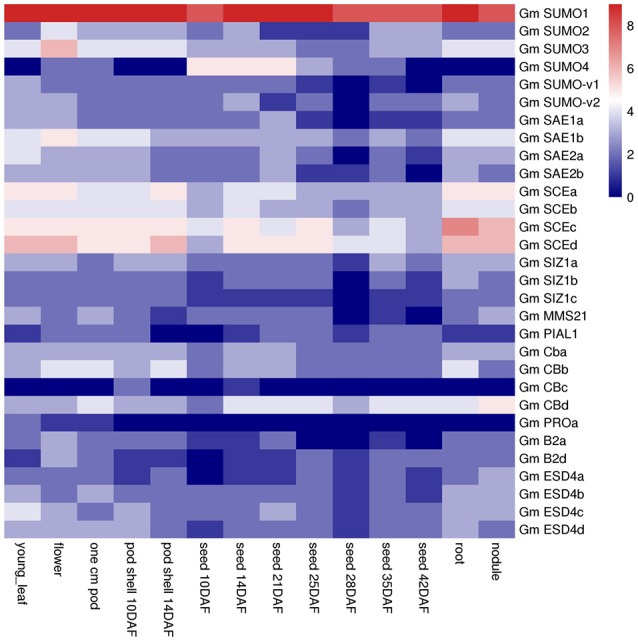
Expression pattern of soybean SUMO pathway genes in different tissues.The Reads/Kb/Million (RPKM) normalized values of expressed gene was log2-transformed. The abbreviation “DAF” in the tissue label indicates “Days after flowering.”
Transcriptional profiles of SUMO pathway genes in soybean roots under abiotic stress
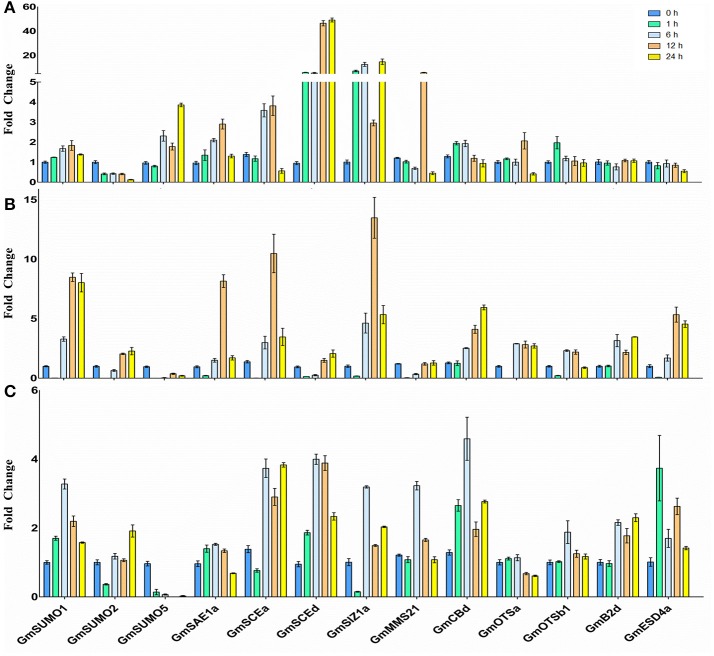
In order to help appreciate the potential roles and transcriptional regulations of the soybean SUMO system members under abiotic stress, we examined the expression changes of 13 SUMO pathway genes in soybean roots in response to NaCl (200 mM), heat (42°C) and ABA (100 μM) treatments.
Quantitative-PCR analysis revealed that the majority of the SUMO cascade component transcript levels were differentially regulated under salinity conditions (Figure 6A). GmSUMO2 transcripts decreased to very low levels after exposure to salt stress, whereas, GmSUMO5 transcripts showed up-regulated after treated for 6 h. The expression of GmSAE1a was slightly increased after 6 h of treatment, and was maintained nearly at initial levels after 24 h of salinity stress (Figure 6A). Among the E2 enzymes, GmSCE1a showed a transient increase, whereas, GmSCE1d transcript was significantly increased since treated for 1 h, and reached a maximum at 12–24 h which was induced about 50-fold. Among the E3 genes, GmSIZ1a began to increase after 1 h of treatment, whereas, GmMMS21 showed a transient increase and decreased when treated for 24 h, which exhibited similar expression patterns with GmOTSa gene. These increases in gene expression may contribute to the accumulation of SUMOylated proteins after exposure to salt stress. Besides, GmSUMO3, GmOTSb1, GmESD4a, GmB2d, and GmCBb showed no detectable changes in the 24 h period after salt treatment.
Figure 6.
Stress induces the SUMO pathway genes expression. Changes in mRNA levels of soybean SUMO pathway genes with stress treatment were determined by quantitative real-time PCR. (A) NaCl (200 mM) (B) Heat stress (42°C) (C) ABA (100 μM) Changes in transcript abundance were represented as fold change by calibrating the relative mRNA levels of each time point with the relative mRNA levels of the 0 h time point.
When treated with heat stress, GmSUMO1 and GmSUMO2 were found to be induced, whereas, GmSUMO5 showed a significant reduction under heat treatment (Figure 6B). The expression patterns of GmSAE1a under heat stress were similar with that under salinity treatment. Both of the E2 genes were up-regulated under heat stress, and GmSCE1a gene exhibited a faster response than GmSCE1d gene. Among these E3 genes, GmSIZ1a showed dramatic up-regulation 6–24 h after heat treatment; however, GmMMS21 was significantly down-regulated 1 h after treatment, and returned to the initial levels after treated for 12 h. In addition, similar expression patterns were observed among GmOTSa, GmESD4a, and GmB2d, which showed increased expression levels from 6 h to 24 h of treatment; while GmOTSb1 showed slightly increased from 6 h under heat treatment and after 12 h showed down-regulation. In contrast, GmCBb transcript showed increased gradually over the 24 h period of treatment. Notably, the majority of genes exhibited a transient decrease during the first 1 h of heat treatment, which indicated that SUMOylation may be depressed temporarily at this time point in soybean roots.
After the ABA treatment, expression of most SUMO pathway members showed slightly up-regulation with the exception of GmSUMO2, GmSAE1a, GmOTSa, and GmOTSb1 genes which were maintained nearly basal levels during the 24 h period of treatment (Figure 6C). GmSUMO1 gene showed a transient positive response 6 h after ABA treatment and subsequently returned to initial levels. Conversely, GmSUMO5 transcript decreased significantly, thereby indicating a specific role for GmSUMO1 in ABA stress response. All of the E2 and E3 family genes transcript levels increased after 6 h of treatment, and GmMMS21 showed a transient up-regulation followed by restoration to basal levels. Among the ULP family, GmESD4a transcript showed up-regulation 1 h after the ABA treatment, indicating that GmESD4a may be responsible for a quick de-SUMOylation process after ABA treatment.
SUMOylation profiles in soybean roots under abiotic stress
In order to explore the regulation of SUMOylation in response to abiotic stress in soybean roots, western blot analysis was performed to monitor the expression changes on the translational level. To test whether anti-AtSUMO1 antibody could detect soybean SUMOs, GmSUMO1, GmSUMO2, and GmSUMO4 genes were amplified (Table S6) and cloned into pET32a expression vector for protein expression. The western blot analysis indicated that anti-AtSUMO1 antibody was able to specifically detect GmSUMO1 and GmSUMO2 proteins, but not for GmSUMO4 (Figure S6).
During salt treatment, roots accumulated large amount of low molecular weight SUMO conjugates as soon as 0.5 h after salt stimulus (Figure 7A), and the accumulation of high molecular weight SUMOylated products was detected from 0.5 to 1 h of salt treatment. At the same time, the level of free SUMO was greatly reduced from 1 h of treatment, and when treated for 24 h, most free SUMOs have been exhausted in soybean roots. Consistent with the results got from poplar (Reed et al., 2010), SUMOylation by salinity treatment in soybean roots was rapidly inducible and probably has some functional implications.
Figure 7.
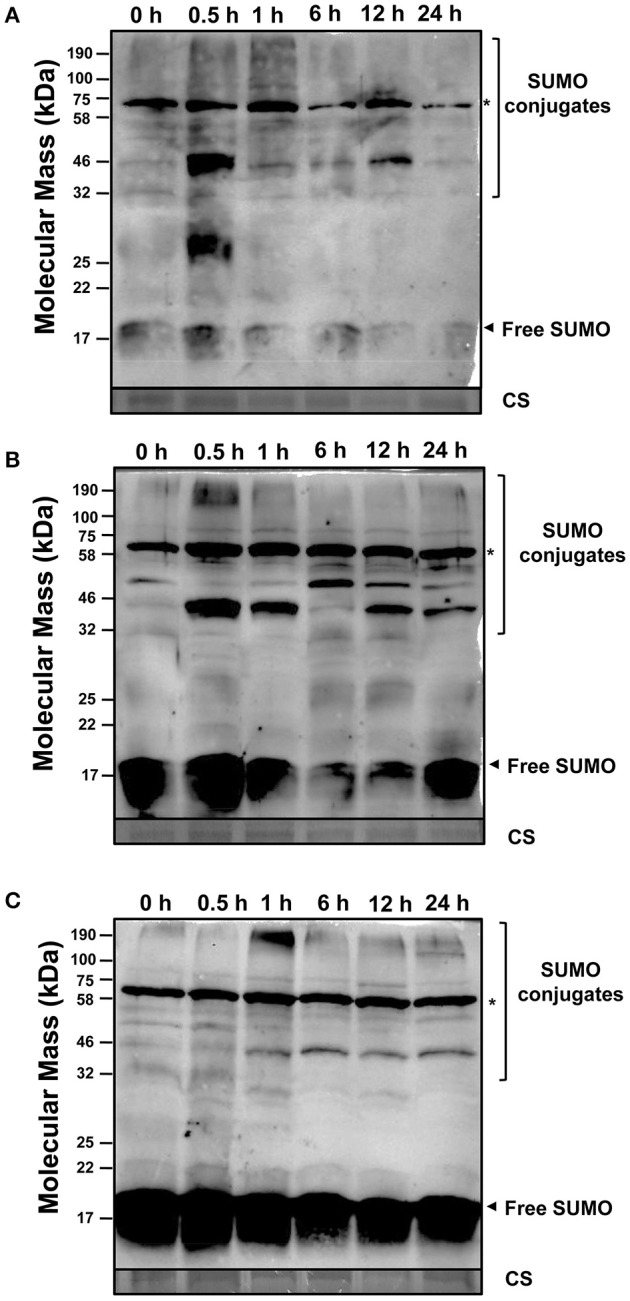
SUMOylation in soybean root is induced by stress. Fifteen microgram of total proteins extracts from the soybean roots at the second trifoliolate leaf stage were subjected to western blot analysis against anti-AtSUMO1. Coomassie stained of high abundant protein showed equal loading of protein samples. (A) NaCl (200 mM) (B) Heat stress (42°C) (C) ABA (100 μM). Free SUMO and conjugates are highlighted by the arrowheads and brackets respectively. The asterisk denotes a non-inducible immunoreactive product.
Rapid stimulation of SUMOylation in response to heat shock had been reported in several plant species (Chaikam and Karlson, 2010; Lopez-Torrejon et al., 2013; Augustine et al., 2016). In this study, SUMO conjugates were detected to be accumulated as soon as 0.5 h after heat stress, which was conformed with previous studies (Cai et al., 2017). Accordingly, the free SUMO pool showed decreased after 1 h of heat shock, and restored to nearly the original level after 24 h of treatment (Figure 7B).
After treating with ABA, the high molecular weight SUMO conjugates accumulated significantly at 1 h after treatment, but slightly decreased at later time points, and increased from 12 to 24 h after treatment (Figure 7C), which indicated that ABA treatment effected SUMOylated protein accumulation in soybean roots. These above results further supported the hypothesis that the rapid protein SUMOylation was an essential functional component in response to abiotic stress in soybean.
Discussion
Previous studies in Arabidopsis connected the protein SUMOylation to plant development and defense against environmental challenges (Castro et al., 2012). Thus, manipulation of this post-translational modification system might provide opportunities to modulate the plant stress resistance with lower inputs. However, only a few examinations have been made of SUMO pathway members in soybean (Cai et al., 2017). In this study, 40 SUMO pathway genes in soybean were identified. The soybean genome contained gene models for SUMO and its respective conjugation and de-conjugation machinery including E1, E2, E3, and SUMO protease components that were closely related to homologs in Arabidopsis. Transcriptional and translational expression profiling of soybean SUMOylation system allowed us to identify the cascade members that have crucial roles in the soybean responses to salt, heat and ABA stresses, and explain the role of SUMOylation in soybean abiotic stress responses.
Compared with the eight putative SUMO copies in Arabidopsis genome, soybean genome contained six members shared a similar ubiquitin-like structural conformation characterized by a β-grasp fold. In soybean, three canonical SUMO genes were present that expressed polypeptides homologous to canonical members in other eukaryote species, which have been well recorded to conjugate to a variety of target proteins in response to environmental stimuli (Kurepa et al., 2003; Saracco et al., 2007). Among these SUMO genes in soybean, GmSUMO1 was constitutively expressed in most organs and seed developmental stages according to the RNA seq data (Figure 5), which showed similar expression patterns with the counterpart genes in Arabidopsis (van den Burg et al., 2010). On the contrary, the expression of non-canonical SUMO GmSUMO4 in soybean appeared relatively weak based on transcriptional analysis. Moreover, GmSUMO1 transcript showed a sustained up-regulation 1 h after ABA treatment, while GmSUMO2 and -5 were not induced (Figure 6), suggesting a specific role of GmSUMO1 in ABA response. Based on the high sequence similarity and overlapping expression profiles, we speculated that GmSUMO1, -2, and -3 might be functional redundant, while whether the biological functions of GmSUMO1/2/3 were essential as their homologs in Arabidopsis still need to be identified. The roles of SUMOs under abiotic stress were inferred by the detection of increasing SUMOylation during stress (Catala et al., 2007; Conti et al., 2008; Golebiowski et al., 2009; Miura et al., 2011). In this study, rapid regulations of SUMO-conjugates in soybean roots were identified upon salinity, heat and ABA elicitation (Figure 7). Although most target proteins bear a single SUMO moiety, some are modified with a poly-SUMO chain onto a single Lys in the substrate. Previous study demonstrated that the residue Lys10 in Arabidopsis SUMO2 was required for self-SUMOylation to form the multimeric chains in vitro (Colby et al., 2006). It can be deduced that soybean SUMO proteins except for GmSUMO4 may be able to form poly-SUMO chains, given the existence of conserved Lys residue in these soybean SUMO proteins (Figure 1C). Notably, there were indeed SUMOylated products with the molecular mass of approximately 30 and 45 kDa in soybean SUMOylation profiles (Figure 7), which were speculated to be potential SUMO dimers and trimers in soybean. The poly-SUMO chain was reported to serve as degradation signal, channeling chain-modified target proteins into degradation by SUMO chain binding ubiquitin ligases (Plechanovová et al., 2011; Praefcke et al., 2012). Thus, the appearance of SUMO dimers and trimers in soybean was supposed to be an early response to stimuli that promoted reallocation of SUMO moieties to target proteins.
Besides SUMOs, soybean genome encoded all components participate in SUMOylation and de-SUMOylation cascade. The SUMO-activating enzyme E1 (SAE) enzymes facilitate conjugation of SUMO proteins through a series of reactions including adenylation, formation of E1-SUMO thioester and transfer of thioester from E1 to E2 conjugating proteins (Lois and Lima, 2005). Previous study has demonstrated that SAE2 formed a tight heterodimer with SAE1 and controlled the localization of the SUMO SAE heterodimer to the nucleus (Truong et al., 2012). In this study, nuclear locations of SAE subunits were also found in soybean genome (Table 1). In Arabidopsis, loss-of-function mutant sae2-1 exhibited developmental arrest at the early stages of embryogenesis, suggesting the essential role of SAE2 (Saracco et al., 2007). Given the comparable expression patterns between SAE1a and SAE1b, SAE2a and SAE2b (Figure 5), we suggested that there may be functionally redundancy between these paralogous SAE polypeptide. Strikingly, GmSAE1a gene was found to be enhanced by eight-fold after heat treatment for 12 h, which was presumed to contribute to the accumulation of SUMO-conjugates in responding to unfavorable environment.
GmSCEs belongs to conjugating enzyme family, characterized by a conserved UBC domain that spans most of the protein. In soybean, four sce genes showed high similarity at the amino acid levels (Figure 2), which indicated that these orthologous genes may be a result of duplication events. Putative cis-elements in 1.5 kb region upstream of GmSCE genes possessed higher abundance of stress-related elements like HSE, MBS, ARE, TC-rich repeats and CGTCA/TGACG-motif, which provided useful cues for determining the responsiveness of GmSCE genes to environmental stimuli. Indeed, our analyses showed that GmSCE transcripts were significantly up-regulated under heat, salt and ABA treatments, which was in conformity with the observations made earlier in rice (Nigam et al., 2008). Recent study provided evidence that overexpression of SaSce9 gene from a grass halophyte in plant can improve salinity and drought stress tolerance demonstrated the potential of using this gene in crop improvement (Ratna and Prasanta, 2012). Thus, biological functions of GmSCEa and GmSCEd under abiotic stress are deserved further study.
Besides, soybean genome encoded three types of SUMO E3 ligases containing the signature SP-RING domain (Figure 2). The functions of E3 ligases in the regulation of environmental stress responses and developmental processes have been extensively characterized in Arabidopsis (Lois, 2010; Miura and Ohta, 2010) and rice (Li et al., 2013; Mishra et al., 2017). Recently, two identical SIZ1 homologs in soybean, GmSIZ1a and GmSIZ1b, were identified, which were reported to mediate SUMO modification of nuclear proteins and regulate vegetative growth in soybean (Cai et al., 2017). In this study, SIZ1 gene in soybean was significantly up-regulated in response to heat and salt stress treatments (Figure 6). However, whether GmSIZ1a/b could regulate environmental stress responses in soybean is still needed to be determined. The second type of E3 ligases MMS21 was reported to function as an important regulator of cell proliferation and cytokinin in signaling during root development in Arabidopsis (Huang et al., 2009). Previous work has reported that Arabidopsis plants exposed to ABA accumulate increased levels of SUMOylated proteins dependent on MMS21 activity, because the accumulation of SUMO conjugates is significantly lower in mms21 mutant, and higher in MMS21-OE compared with WT plants (Zhang et al., 2013). In this study, we found that the expression levels of GmMMS21 gene were induced by ABA and salt treatment (Figure 6), it was presumed that GmMMS21 gene could act a pivotal part in salt stress response in soybean by regulating gene expression in the ABA-dependent manner. Moreover, there is another type of E3 ligases containing SP-RING domain and SUMO interaction motifs (SIM) which designated as PIAL1 and 2 that function as SUMO ligases specialized in SUMO chain formation. Mutant studies in Arabidopsis indicated that PIAL1 and 2 functioned in the regulation of salt and osmotic stress responses by removal of SUMOylation substrates, and may contribute to connection of SUMOylation pathway to ubiquitin proteolytic pathway (Tomanov et al., 2014). However, the biological functions and molecular mechanisms of PIAL1 and 2 in SUMOylation cascade are still largely unknown in soybean. The up-regulation of most of E2 and E3 genes in soybean under NaCl, heat and ABA stimuli (Figure 6) may account for stress induced accumulation of SUMO conjugates which detected by western blot analysis (Figure 7).
Compared with other SUMO pathway components, there are a larger number of plant SUMO proteases (Figure 3) which cleave precisely between the terminal Gly of SUMO and the substrate Lys, and play a vital regulatory role during de-SUMOylation (Hickey et al., 2012). So far only a few bona fide SUMO proteases like ESD4 and OTS1/OTS2 have been characterized using genetic, physiological, and biochemical approaches. The Arabidopsis esd4 mutant showed extreme early flowering and alterations in shoot development, suggesting that SUMOylation played roles in plant development which regulated by ESD4 (Murtas et al., 2003). Previous study suggested that the OTS class of ULP-like SUMO proteases may provide routes for boosting crop growth under stress because over-expression of OTS gene led to salt tolerance (Conti et al., 2008). However, SUMO proteases remain largely unstudied, especially in soybean and other crop plants. Similar with AtOTS1, OTS gene expression was enhanced in soybean under heat and salt stress would suggest that soybean stimulated the de-SUMOylation process in response to abiotic stresses, which had been shown in the soybean SUMOylation profiles (Figure 7).
Conclusions
In summary, a total of 40 genes involved in SUMOylation system from the soybean genome were identified. We investigated the classification, characterization and tissue-specific expression of these SUMO pathway genes, revealing SUMO pathway members broadly participate in the regulation of plant tissue development. Meanwhile, qRT-PCR analysis to 13 SUMO pathway genes suggested that these genes functioned at the transcriptional level during adaption to adverse environments. Moreover, the investigation of SUMOylation profiles in response to salt, heat and ABA treatment demonstrated that soybean responded to abiotic stress treatments by regulating the SUMO conjugated proteins. These results may contribute to the existing knowledge on the complexity of soybean post translational modifications that occur in response to environmental stresses. Further proteome wide identification of various targets will be paramount to defining how SUMOylation ultimately participates in stress protection.
Author contributions
YL performed the analysis and laboratory assays and wrote the manuscript. GW and ZX performed the phylogentic analysis and the RNA-seq analysis. JL, MS, and JG provide help in analysis of qRT-PCR and western blot. WJ conceived and designed the experiments, facilitated the project, and assisted in manuscript preparation. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Natural Science Foundation of Heilongjiang Province of China (C201419), and “Young Talents” Project of Northeast Agricultural University in China (16QC28). We appreciate Dr. Jiechao Yin for the assistance in Western blotting.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01458/full#supplementary-material
Sequence alignment of SAE2s from Arabidopsis and soybean. Adenylation, cysteine, and ubiquitin-fold domains are displayed below the sequence. Red asterisk indicates the catalytic cysteine residue. Blue asterisk indicates the predicted SUMOylation site. Gray and black boxes identify similar and identical amino acids, respectively. Dashes denote gaps.
Soybean genome encodes four E2 genes. (A) Structure-based sequence alignment of GmSCEa/b/c/d along with SCEs from Arabidopsis and rice reveals conserved domain and motif. UBCC (Ub-Conjugating Enzyme Catalytic) domain is indicated by dotted line. Asterisk highlights the activate-site Cys. Green rectangles and blue arrows indicate helices and strands. Gray and black boxes identify similar and conserved amino acids, respectively. Dashes denote gaps. (B) E2 protein sequences from Arabidopsis thaliana, Zea mays, Oryza sativa, Solanum lycopersium, Populus trichocarpa and Glycine max (Table S2) were used to construct the phylogenetic tree by the neighbor-joining method in MEGA.
Phylogenetic tree of full-length ULP proteins from soybean and other plants. ULP family protein sequences from Arabidopsis thaliana, Zea mays, Oryzasativa, Solanum lycopersium, Populus trichocarpa and Glycine max (Table S2) were used to construct the phylogenetic tree. These ULPs can be classified into four groups.
Sequence alignment of the GmESD4s. Red bars above the sequences highlight predicted SIM motif. Asterisks indicate SUMOylation sites. The extent of the Peptidase_C48 domain is indicated below the sequence. Green arrowheads indicate the His-Asp-Cys catalytic triad. Gray and black boxes identify similar and conserved amino acids, respectively. Dashes denote gaps.
Alignment of SUMO chain binding proteins in soybean. RING finger domain is indicated by the dotted box. Red lines indicate the SIM motifs. Gray and black boxes identify similar and conserved amino acids, respectively. Dashes denote gaps.
Detection of soybean SUMO proteins using anti-AtSUMO1 antibody. (A) SDS-PAGE analysis of His:GmSUMO1/2/4 fusion protein expressed in E. Coli strain BL21. M, protein marker; 1, pET32a vector control; 2, pET32a-GmSUMO1; 3, pET32a-GmSUMO2; 4, pET32a-GmSUMO4. (B) SDS PAGE analysis of purified His:GmSUMO1/2/4 fusion protein using Ni-NTA. M, protein marker; 1, pET32a vector control; 2, pET32a-GmSUMO1; 3, pET32a-GmSUMO2; 4, pET32a-GmSUMO4. (C) Western-blot analysis of purified GmSUMO1/2/4 protein against anti-AtSUMO1 antibody. M, protein marker; 1, pET32a vector control; 2, GmSUMO1 protein; 3, GmSUMO2 protein; 4, GmSUMO4 protein.
Primers used for quantitative RT-PCR of soybean SUMO pathways genes.
SUMO pathway genes in six species.
Sequence of 1,500 bp upstream of SUMO pathway genes in soybean.
Detailed information of cis-acting element.
Normalized data (Reads/kilobase/million normalization of the raw data) of SUMO pathway genes in soybean.
CDS sequence and amplified primers for GmSUMO1/2/4 genes.
References
- Aguilar-Martinez E., Chen X., Aaron W., Mould A. P., Anne S., Hay R. T., et al. (2015). Screen for multi-SUMO–binding proteins reveals a multi-SIM–binding mechanism for recruitment of the transcriptional regulator ZMYM2 to chromatin. Proc. Natl. Acad. Sci. U.S.A. 112, 4854–4863. 10.1073/pnas.1509716112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine R. C., York S. L., Rytz T. C., Vierstra R. D. (2016). Defining the SUMO system in maize: SUMOylation is up-regulated during endosperm development and rapidly induced by stress. Plant Physiol. 171, 2191–2210. 10.1104/pp.16.00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitz E. (2000). TeXshade: shading and labeling of multiple sequence alignments using LaTeX2e. Bioinformatics 16, 135–139. 10.1093/bioinformatics/16.2.135 [DOI] [PubMed] [Google Scholar]
- Belamkar V., Weeks N., Bharti A., Farmer A., Graham M., Cannon S. (2014). Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. Biomed. Central 15:950. 10.1186/1471-2164-15-950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossis G., Melchior F. (2006). SUMO: regulating the regulator. Cell Div. 1:13. 10.1186/1747-1028-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Cai B., Kong X., Zhong C., Sun S., Zhou X. F., Jin Y. H., et al. (2017). SUMO E3 Ligases GmSIZ1a and GmSIZ1b regulate vegetative growth in soybean. J. Integr. Plant Biol. 59, 2–14. 10.1111/jipb.12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro P. H., Bejarano E. R., Tavares R. M., Azevedo H. (2012). SUMO, a heavyweight player in plant abiotic stress responses. Cell. Mol. Life Sci. 69, 3269–3283. 10.1007/s00018-012-1094-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R., Ouyang J., Abreu I. A., Hu Y., Seo H., Zhang X., et al. (2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19, 2952–2966. 10.1105/tpc.106.049981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaikam V., Karlson D. T. (2010). Response and transcriptional regulation of rice SUMOylation system during development and stress conditions. BMB Rep. 43, 103–109. 10.5483/BMBRep.2010.43.2.103 [DOI] [PubMed] [Google Scholar]
- Chen C. C., Chen Y. Y., Tang I. C., Liang H. M., Lai C. C., Chiou J. M., et al. (2011). ArabidopsisSUMO E3 ligase SIZ1 is involved in excess copper tolerance. Plant Physiol. 156, 2225–2234. 10.1104/pp.111.178996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong M. S., Park H. C., Hong M. J., Lee J., Choi W., Jin J. B., et al. (2009). Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes. Plant Physiol. 151, 1930–1942. 10.1104/pp.109.143719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby T., Matthai A., Boeckelmann A., Stuible H. P. (2006). SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 142, 318–332. 10.1104/pp.106.085415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L., Price G., O'Donnell E., Schwessinger B., Dominy P., Sadanandom A. (2008). Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20, 2894–2908. 10.1105/tpc.108.058669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau J. R., Lima C. D. (2010). The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871. 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiowski F., Matic I., Tatham M. H., Cole C., Yin Y., Nakamura A., et al. (2009). System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2:ra24. 10.1126/scisignal.2000282 [DOI] [PubMed] [Google Scholar]
- Hickey C. M., Wilson N. R., Hochstrasser M. (2012). Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13, 755–766. 10.1038/nrm3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Park K. J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35:W585. 10.1093/nar/gkm259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Yang S., Zhang S., Liu M., Lai J., Qi Y., et al. (2009). The ArabidopsisSUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 60, 666–678. 10.1111/j.1365-313X.2009.03992.x [DOI] [PubMed] [Google Scholar]
- Kraft E., Stone S. L., Ma L., Su N., Gao Y., Lau O. S., et al. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139, 1597–1611. 10.1104/pp.105.067983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J., Walker J. M., Smalle J., Gosink M. M., Davis S. J., Durham T. L., et al. (2003). The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis accumulation of sumo1 and-2 conjugates is increased by stress. J. Biol. Chem. 278, 6862–6872. 10.1074/jbc.M209694200 [DOI] [PubMed] [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Hu Q., Zhou M., Vandenbrink J., Li D., Menchyk N., et al. (2013). Heterologous expression of OsSIZ1, a rice SUMO E3 ligase, enhances broad abiotic stress tolerance in transgenic creeping bentgrass. Plant Biotechnol. J. 11, 432–445. 10.1111/pbi.12030 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lois L. M. (2010). Diversity of the SUMOylation machinery in plants. Biochem. Soc. Trans. 38(Pt 1), 60–64. 10.1042/BST0380060 [DOI] [PubMed] [Google Scholar]
- Lois L. M., Lima C. D. (2005). Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439–451. 10.1038/sj.emboj.7600552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Torrejon G., Guerra D., Catala R., Salinas J., del Pozo J. C. (2013). Identification of SUMO targets by a novel proteomic approach in plants(F). J. Integr. Plant Biol. 55, 96–107. 10.1111/jipb.12012 [DOI] [PubMed] [Google Scholar]
- Matic I., Schimmel J., Hendriks I. A., van Santen M. A., van de Rijke F., van Dam H., et al. (2010). Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 39, 641–652. 10.1016/j.molcel.2010.07.026 [DOI] [PubMed] [Google Scholar]
- Miller M. J., Barrett-Wilt G. A., Hua Z., Vierstra R. D. (2010). Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 16512–16517. 10.1073/pnas.1004181107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. J., Scalf M., Rytz T. C., Hubler S. L., Smith L. M., Vierstra R. D. (2013). Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol. Cell. Proteomics 12, 449–463. 10.1074/mcp.M112.025056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N., Sun L., Zhu X., Smith J., Prakash Srivastava A., Yang X., et al. (2017). Overexpression of the rice SUMO E3 Ligase Gene OsSIZ1 in cotton enhances drought and heat tolerance, and substantially improves fiber yields in the field under reduced irrigation and rainfed conditions. Plant Cell Physiol. 58, 735–746. 10.1093/pcp/pcx032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Ohta M. (2010). SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation. J. Plant Physiol. 167, 555–560. 10.1016/j.jplph.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Miura K., Jin J. B., Hasegawa P. M. (2007a). Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. 10, 495–502. 10.1016/j.pbi.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Miura K., Jin J. B., Lee J., Yoo C. Y., Stirm V., Miura T., et al. (2007b). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403–1414. 10.1105/tpc.106.048397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Sato A., Ohta M., Furukawa J. (2011). Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2. Planta 234, 1191–1199. 10.1007/s00425-011-1476-y [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D., Dasso M. (2007). Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32, 286–295. 10.1016/j.tibs.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Murtas G., Reeves P. H., Fu Y. F., Bancroft I., Dean C., Coupland G. (2003). A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell 15, 2308–2319. 10.1105/tpc.015487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Ito Y., Yamaguchi-Shinozaki K. (2009). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. 10.1104/pp.108.129791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam N., Singh A., Sahi C., Chandramouli A., Grover A. (2008). SUMO-conjugating enzyme (Sce) and FK506-binding protein (FKBP) encoding rice (Oryza sativa L.) genes: genome-wide analysis, expression studies and evidence for their involvement in abiotic stress response. Mol. Genet. Genomics 279, 371–383. 10.1007/s00438-008-0318-5 [DOI] [PubMed] [Google Scholar]
- Novatchkova M., Tomanov K., Hofmann K., Stuible H.-P., Bachmair A. (2012). Update on sumoylation: defining core components of the plant SUMO conjugation system by phylogenetic comparison. New Phytol. 195, 23–31. 10.1111/j.1469-8137.2012.04135.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. C., Choi W., Park H. J., Cheong M. S., Koo Y. D., Shin G., et al. (2011). Identification and molecular properties of SUMO-binding proteins in Arabidopsis. Mol. Cells 32, 143–151. 10.1007/s10059-011-2297-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. J., Kim W. Y., Park H. C., Lee S. Y., Bohnert H. J., Yun D. J. (2011). SUMO and SUMOylation in plants. Mol. Cells 32, 305–316. 10.1007/s10059-011-0122-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plechanovová A., Jaffray E., McMahon S., Johnson K., Navrátilová I., Naismith J., et al. (2011). Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat. Struct. Mol. Biol. 18, 1052–1059. 10.1038/nsmb.2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke G. J., Hofmann K., Dohmen R. J. (2012). SUMO playing tag with ubiquitin. Trends Biochem. Sci. 37, 23–31. 10.1016/j.tibs.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Ratna K., Prasanta K. S. (2012). A stress inducible SUMO conjugating enzyme gene (SaSce9) from a grass halophyte Spartina alterniflora enhances salinity and drought stress tolerance in Arabidopsis. BMC Plant Biol. 12:187. 10.1186/1471-2229-12-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J. M., Christopher D., Alison M. M., John M. D. (2010). The SUMO conjugation pathway in Populus_ genomic analysis. Planta 232, 51–59. 10.1007/s00425-010-1151-8 [DOI] [PubMed] [Google Scholar]
- Roden J., Eardley L., Hotson A., Cao Y., Mudgett M. B. (2004). Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant Microbe Interact. 17, 633–643. 10.1094/MPMI.2004.17.6.633 [DOI] [PubMed] [Google Scholar]
- Rodriguez M. S., Dargemont C., Hay R. T. (2001). SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654–12659. 10.1074/jbc.M009476200 [DOI] [PubMed] [Google Scholar]
- Saracco S. A., Miller M. J., Kurepa J., Vierstra R. D. (2007). Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 145, 119–134. 10.1104/pp.107.102285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Severin A. J., Woody J. L., Bolon Y. T., Joseph B., Diers B. W., Farmer A. D., et al. (2010). RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10:160. 10.1186/1471-2229-10-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. K., Zhang C., Yates G., Bailey M., Brown A., Sadanandom A. (2016). SUMO Is a critical regulator of salt stress responses in rice. Plant Physiol. 170, 2378–2391. 10.1104/pp.15.01530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., Naismith J. H., et al. (2001). Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374. 10.1074/jbc.M104214200 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Higgins D. G. (2002). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. Chapter 2:Unit 2.3. 10.1002/0471250953.bi0203s00 [DOI] [PubMed] [Google Scholar]
- Tomanov K., Zeschmann A., Hermkes R., Eifler K., Ziba I., Grieco M., et al. (2014). Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell 26, 4547–4560. 10.1105/tpc.114.131300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong K., Lee T. D., Li B., Chen Y. (2012). Sumoylation of SAE2 C terminus regulates SAE nuclear localization. J. Biol. Chem. 287, 42611–42619. 10.1074/jbc.M112.420877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg H. A., Kini R. K., Schuurink R. C., Takken F. L. (2010). Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 22, 1998–2016. 10.1105/tpc.109.070961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus A. A., Lima C. D. (2009). Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35, 669–682. 10.1016/j.molcel.2009.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Qi Y., Liu M., Yang C. (2013). SUMO E3 ligase AtMMS21 regulates drought tolerance in Arabidopsis thaliana(F). J. Integr. Plant Biol. 55, 83–95. 10.1111/jipb.12024 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., et al. (2014). GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42, W325–W30. 10.1093/nar/gku383 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of SAE2s from Arabidopsis and soybean. Adenylation, cysteine, and ubiquitin-fold domains are displayed below the sequence. Red asterisk indicates the catalytic cysteine residue. Blue asterisk indicates the predicted SUMOylation site. Gray and black boxes identify similar and identical amino acids, respectively. Dashes denote gaps.
Soybean genome encodes four E2 genes. (A) Structure-based sequence alignment of GmSCEa/b/c/d along with SCEs from Arabidopsis and rice reveals conserved domain and motif. UBCC (Ub-Conjugating Enzyme Catalytic) domain is indicated by dotted line. Asterisk highlights the activate-site Cys. Green rectangles and blue arrows indicate helices and strands. Gray and black boxes identify similar and conserved amino acids, respectively. Dashes denote gaps. (B) E2 protein sequences from Arabidopsis thaliana, Zea mays, Oryza sativa, Solanum lycopersium, Populus trichocarpa and Glycine max (Table S2) were used to construct the phylogenetic tree by the neighbor-joining method in MEGA.
Phylogenetic tree of full-length ULP proteins from soybean and other plants. ULP family protein sequences from Arabidopsis thaliana, Zea mays, Oryzasativa, Solanum lycopersium, Populus trichocarpa and Glycine max (Table S2) were used to construct the phylogenetic tree. These ULPs can be classified into four groups.
Sequence alignment of the GmESD4s. Red bars above the sequences highlight predicted SIM motif. Asterisks indicate SUMOylation sites. The extent of the Peptidase_C48 domain is indicated below the sequence. Green arrowheads indicate the His-Asp-Cys catalytic triad. Gray and black boxes identify similar and conserved amino acids, respectively. Dashes denote gaps.
Alignment of SUMO chain binding proteins in soybean. RING finger domain is indicated by the dotted box. Red lines indicate the SIM motifs. Gray and black boxes identify similar and conserved amino acids, respectively. Dashes denote gaps.
Detection of soybean SUMO proteins using anti-AtSUMO1 antibody. (A) SDS-PAGE analysis of His:GmSUMO1/2/4 fusion protein expressed in E. Coli strain BL21. M, protein marker; 1, pET32a vector control; 2, pET32a-GmSUMO1; 3, pET32a-GmSUMO2; 4, pET32a-GmSUMO4. (B) SDS PAGE analysis of purified His:GmSUMO1/2/4 fusion protein using Ni-NTA. M, protein marker; 1, pET32a vector control; 2, pET32a-GmSUMO1; 3, pET32a-GmSUMO2; 4, pET32a-GmSUMO4. (C) Western-blot analysis of purified GmSUMO1/2/4 protein against anti-AtSUMO1 antibody. M, protein marker; 1, pET32a vector control; 2, GmSUMO1 protein; 3, GmSUMO2 protein; 4, GmSUMO4 protein.
Primers used for quantitative RT-PCR of soybean SUMO pathways genes.
SUMO pathway genes in six species.
Sequence of 1,500 bp upstream of SUMO pathway genes in soybean.
Detailed information of cis-acting element.
Normalized data (Reads/kilobase/million normalization of the raw data) of SUMO pathway genes in soybean.
CDS sequence and amplified primers for GmSUMO1/2/4 genes.