Abstract
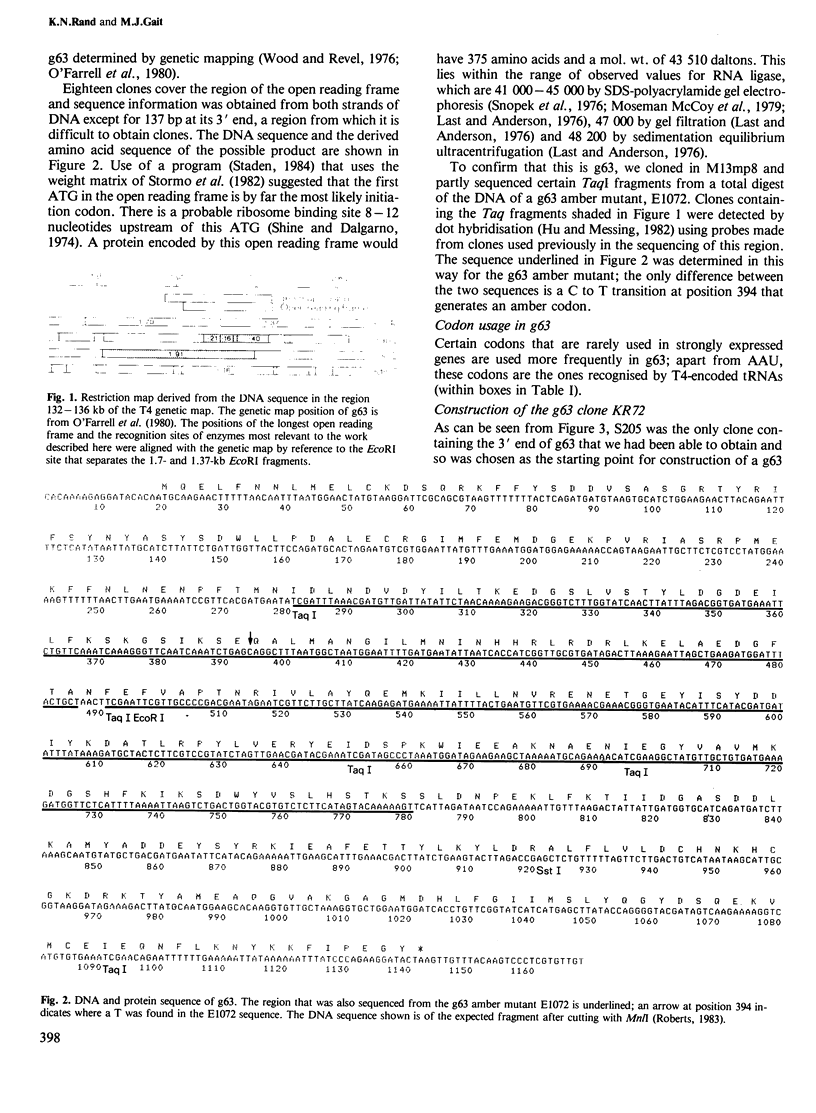
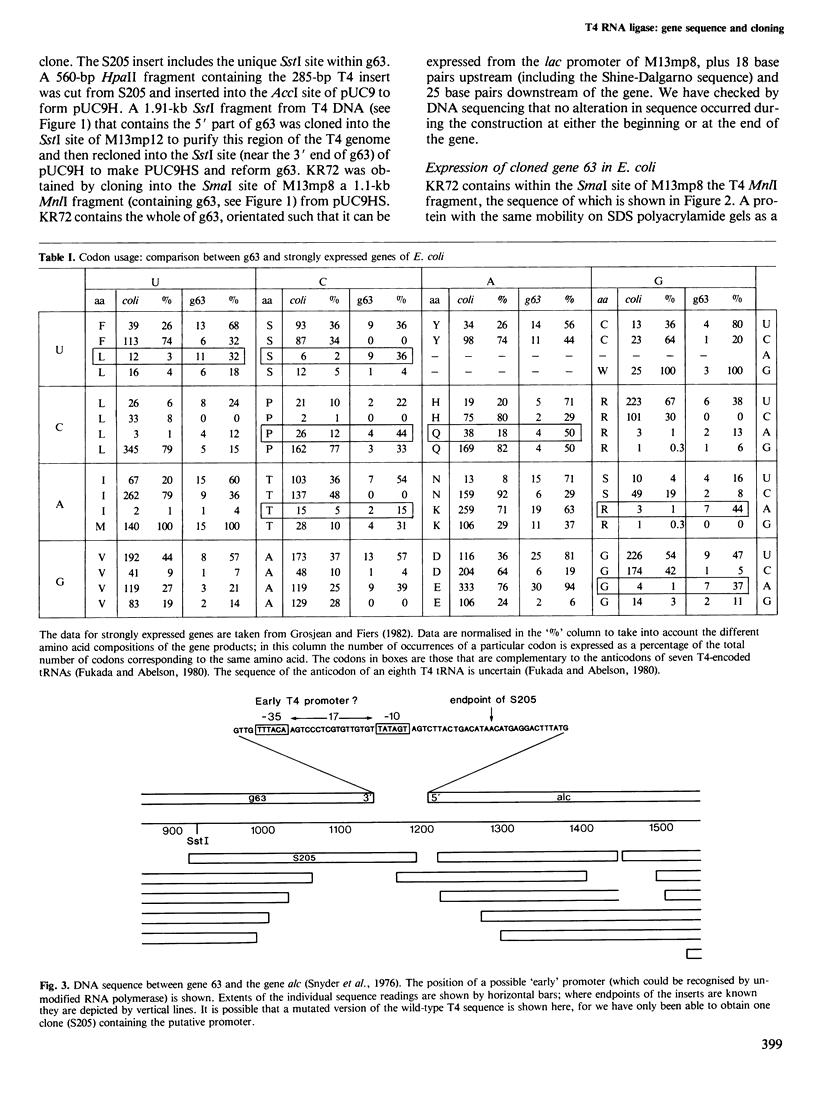
The sequence of gene 63 of bacteriophage T4 was determined by a shotgun approach. Small DNA fragments, derived by sonication of a restriction fragment that encompasses the region of gene 63, were cloned in M13 vectors and sequenced by the 'dideoxy' method. The position of the gene was established by comparison with the sequence of a gene 63 amber mutant. Knowledge of the DNA sequence of gene 63 and surrounding regions has allowed the construction of a clone of gene 63 in which RNA ligase production is under the control of the lac promoter of bacteriophage M13mp8. Infected E. coli cells can be induced to produce a protein indistinguishable from commercially available RNA ligase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. A., Manthey A. E., Gumport R. I. Using T4 RNA ligase with DNA substrates. Methods Enzymol. 1983;100:38–52. doi: 10.1016/0076-6879(83)00044-0. [DOI] [PubMed] [Google Scholar]
- Brown D. M., Frampton J., Goelet P., Karn J. Sensitive detection of RNA using strand-specific M13 probes. Gene. 1982 Dec;20(2):139–144. doi: 10.1016/0378-1119(82)90032-4. [DOI] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978 Oct;5(10):3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouikh Y., Volovitch M., Yot P. A simple and fast electrophoretic method for elution of nucleic acids from gels. Mol Biol Rep. 1979 Dec 31;5(4):237–239. doi: 10.1007/BF00782896. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- David M., Borasio G. D., Kaufmann G. Bacteriophage T4-induced anticodon-loop nuclease detected in a host strain restrictive to RNA ligase mutants. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7097–7101. doi: 10.1073/pnas.79.23.7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M., Vekstein R., Kaufmann G. RNA ligase reaction products in plasmolyzed Escherichia coli cells infected by T4 bacteriophage. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5430–5434. doi: 10.1073/pnas.76.11.5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Fukada K., Abelson J. DNA sequence of a T4 transfer RNA gene cluster. J Mol Biol. 1980 May 25;139(3):377–391. doi: 10.1016/0022-2836(80)90136-9. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Geballe A. P., Snopek T. J., Sugino A., Cozzarelli N. R. Bacteriophage T4 RNA ligase: preparation of a physically homogeneous, nuclease-free enzyme from hyperproducing infected cells. Nucleic Acids Res. 1977 Sep;4(9):3175–3186. doi: 10.1093/nar/4.9.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Jayaraman K., Müller-Hill B., Rickenberg H. V. Inhibition of the synthesis of beta-galactosidase in Escherichia coli by 2-nitrophenyl-beta-D-fucoside. J Mol Biol. 1966 Jul;18(2):339–343. doi: 10.1016/s0022-2836(66)80251-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Last J. A., Anderson W. F. Purification and properties of bacteriophage T4-induced RNA ligase. Arch Biochem Biophys. 1976 May;174(1):167–176. doi: 10.1016/0003-9861(76)90335-0. [DOI] [PubMed] [Google Scholar]
- McCoy M. I., Gumport R. I. T4 ribonucleic acid ligase joins single-strand oligo(deoxyribonucleotides). Biochemistry. 1980 Feb 19;19(4):635–642. doi: 10.1021/bi00545a005. [DOI] [PubMed] [Google Scholar]
- McCoy M. I., Lubben T. H., Gumport R. I. The purification of nuclease-free T4-RNA ligase. Biochim Biophys Acta. 1979 Mar 28;562(1):149–161. doi: 10.1016/0005-2787(79)90134-5. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Mei-hao H., Ai W., Hui-fen H. Purification of T4 RNA ligase by dextran blue-Sepharose 4B affinity chromatography. Anal Biochem. 1982 Sep 1;125(1):1–5. doi: 10.1016/0003-2697(82)90375-x. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mileham A. J., Revel H. R., Murray N. E. Molecular cloning of the T4 genome; organization and expression of the frd--DNA ligase region. Mol Gen Genet. 1980;179(2):227–239. doi: 10.1007/BF00425449. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Bruce S. A., Murray K. Molecular cloning of the DNA ligase gene from bacteriophage T4. II. Amplification and preparation of the gene product. J Mol Biol. 1979 Aug 15;132(3):493–505. doi: 10.1016/0022-2836(79)90271-7. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H., Kutter E., Nakanishi M. A restriction map of the bacteriophage T4 genome. Mol Gen Genet. 1980;179(2):421–435. doi: 10.1007/BF00425473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1983 Jan 11;11(1):r135–r167. [PMC free article] [PubMed] [Google Scholar]
- Runnels J. M., Soltis D., Hey T., Snyder L. Genetic and physiological studies of the role of the RNA ligase of bacteriophage T4. J Mol Biol. 1982 Jan 15;154(2):273–286. doi: 10.1016/0022-2836(82)90064-x. [DOI] [PubMed] [Google Scholar]
- Runnels J., Snyder L. Isolation of a bacterial host selective for bacteriophage T4 containing cytosine in its DNA. J Virol. 1978 Sep;27(3):815–818. doi: 10.1128/jvi.27.3.815-818.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber R., Malathi V. G., Hurwitz J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc Natl Acad Sci U S A. 1972 Oct;69(10):3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin K., Wei J., Snyder L. T4 Bacteriophage-coded RNA polymerase subunit blocks host transcription and unfolds the host chromosome. Nature. 1977 Jan 6;265(5589):28–32. doi: 10.1038/265028a0. [DOI] [PubMed] [Google Scholar]
- Snopek T. J., Sugino A., Agarwal K. L., Cozzarelli N. R. Catalysis of DNA joining by bacteriophage T4 RNA ligase. Biochem Biophys Res Commun. 1976 Jan 26;68(2):417–424. doi: 10.1016/0006-291x(76)91161-x. [DOI] [PubMed] [Google Scholar]
- Snopek T. J., Wood W. B., Conley M. P., Chen P., Cozzarelli N. R. Bacteriophage T4 RNA ligase is gene 63 product, the protein that promotes tail fiber attachment to the baseplate. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3355–3359. doi: 10.1073/pnas.74.8.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder L., Gold L., Kutter E. A gene of bacteriophage T4 whose product prevents true late transcription on cytosine-containing T4 DNA. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3098–3102. doi: 10.1073/pnas.73.9.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L., Ehrenfeucht A. Use of the 'Perceptron' algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stueber D., Bujard H. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1982;1(11):1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Snoper T. J., Cozzarelli N. R. Bacteriophage T4 RNA ligase. Reaction intermediates and interaction of substrates. J Biol Chem. 1977 Mar 10;252(5):1732–1738. [PubMed] [Google Scholar]
- Sugiura M., Suzuki M., Ohtsuka E., Nishikawa S., Uemura H., Ikehara M. Purification of T4 RNA ligase by 2', 5'-ADP sepharose chromatography. FEBS Lett. 1979 Jan 1;97(1):73–76. doi: 10.1016/0014-5793(79)80055-1. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Walker G. C., Uhlenbeck O. C., Bedows E., Gumport R. I. T4-induced RNA ligase joins single-stranded oligoribonucleotides. Proc Natl Acad Sci U S A. 1975 Jan;72(1):122–126. doi: 10.1073/pnas.72.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Molecular cloning of the DNA ligase gene from bacteriophage T4. I. Characterisation of the recombinants. J Mol Biol. 1979 Aug 15;132(3):471–491. doi: 10.1016/0022-2836(79)90270-5. [DOI] [PubMed] [Google Scholar]
- Wood W. B., Henninger M. Attachment of tail fibers in bacteriophage T4 assembly: some properties of the reaction in vitro and its genetic control. J Mol Biol. 1969 Feb 14;39(3):603–618. doi: 10.1016/0022-2836(69)90148-x. [DOI] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- Wood W. B., Revel H. R. The genome of bacteriophage T4. Bacteriol Rev. 1976 Dec;40(4):847–868. doi: 10.1128/br.40.4.847-868.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- Young J. F., Desselberger U., Palese P., Ferguson B., Shatzman A. R., Rosenberg M. Efficient expression of influenza virus NS1 nonstructural proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1983 Oct;80(19):6105–6109. doi: 10.1073/pnas.80.19.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]