Abstract
The eukaryotic ribosomal 5S RNA–protein complex (5S rRNP) is formed by a co-translational event that requires 5S rRNA binding to the nascent peptide chain of eukaryotic ribosomal protein L5. Binding between 5S rRNA and the nascent chain is specific: neither the 5S rRNA nor the nascent chain of L5 protein can be substituted by other RNAs or other ribosomal proteins. The region responsible for binding 5S rRNA is located at positions 35–50 with amino acid sequence RLVIQDIKNKYNTPKYRM. Eukaryotic 5S rRNA binds a nascent chain having this sequence, but such binding is not substantive enough to form a 5S-associated RNP complex, suggesting that 5S rRNA binding to the nascent chain is amino acid sequence dependent and that formation of the 5S rRNP complex is L5 protein specific. Microinjection of 5S rRNP complex into the cytoplasm of Xenopus oocytes results in both an increase in the initial rate and also in the extent of net nuclear import of L5. This suggests that the 5S rRNP complex enhances nuclear transport of L5. We propose that 5S rRNA plays a chaperone-like role in folding of the nascent chain of L5 and directs L5 into a 5S rRNP complex for nuclear entry.
INTRODUCTION
A 5S rRNA–protein complex (5S rRNP) is commonly found in both eukaryotic and prokaryotic ribosomes. The eukaryotic 5S rRNP comprises a single ribosomal protein L5 (L5) and 5S rRNA (1), whereas the prokaryotic 5S rRNP is made up of three ribosomal proteins with its own 5S rRNA (2). Unlike the prokaryotic 5S rRNP, in vitro constitution of the eukaryotic 5S rRNP from its constituents has been difficult. This problem of reconstitution of eukaryotic 5S rRNP has made progress in understanding the eukaryotic ribosome excruciatingly slow. One simple reason for such difficulty is the complicated processing of the eukaryotic ribosome during biogenesis (3,4). The requirement for nucleo-cytoplasmic shuttling of ribosomal proteins (5) or 5S rRNA (6,7) accounts for why making a reconstituted eukaryotic 5S rRNP is difficult.
In most eukaryotic cells 5S rRNA is synthesized in the nucleoplasm with excess copies over other ribosomal components. The synthesized 5S rRNA is then in the form of a small, non-ribosomal nuclear protein complex or ribosomal RNPs. Early pulse–chase experiments indicated that 5S rRNA is first formed as a non-ribosomal 7S RNP particle (8) which is then exported to the cytoplasm (9). Why 5S rRNA or 5S rRNA-associated complexes need to be exported to the cytoplasm is unclear because 5S rRNA must then re-enter the nucleus to ensure that the ribosome can be fully assembled (5,7). The most plausible interpretation for cytosolic export of 5S rRNA may lie in control of 5S rRNA synthesis, as suggested by the current view of a feedback regulation mechanism (10). The process of re-entry of 5S rRNA has been suggested as being selective (11) and energy dependent (6). The nuclear re-entry of 5S rRNA is exclusively mediated by ribosomal protein L5 (6,12,13), implying that formation of a 5S rRNP complex could be a necessary step for nuclear re-entry of 5S rRNA. Such co-entry of 5S rRNA and L5 would ensure stoichiometric amounts of the rRNA and ribosomal protein for production of the large ribosomal subunit (7). Thus, knowing how and when 5S rRNA and L5 react with each other would enhance our understanding of the process of ribosome biogenesis.
Here we have adapted an in vitro translation system to investigate how and when 5S rRNA triggers formation of the eukaryotic RNP complex. We also determine the region in L5 where specific binding to 5S rRNA occurs. The specificity of recognition between L5 and 5S rRNA is revealed. The advantage of forming a 5S rRNP complex for nuclear entry of L5 was demonstrated using Xenopus oocyte microinjection. This study tentatively suggests that 5S rRNA might play a chaperone-like role in the co-translational folding of L5 into a viable 5S-associated RNP complex.
MATERIALS AND METHODS
The L5 gene and truncated mutant L5 plasmid construction
The gene coding for human ribosomal protein L5 (L5) was obtained from a human liver cDNA library (a kind gift of Dr Peter Hsai, Institute of Genetics, National Yang-Ming University, Taipei, Republic of China). This was amplified by PCR using primers derived from the published sequence of rat liver ribosomal protein L5 (GenBank accession no. X17419) (14). The primers were 5′-catatggctcctgtgaaaaa-3′ (N-primer, NdeI site underlined) and 5′-gaattctaatcctcgtcttcc-3 (C-primer, EcoRI site underlined). The PCR fragment was subsequently cloned into vector pGEM(T) to make plasmid pGEM(T)/L5. Plasmid pGEM(T)/L5 was used as the template for construction of the N-terminal truncated mutants of L5 by the same PCR strategy. All constructions were verified by DNA sequencing.
Making the 5S rRNP complex by translating L5 in the presence of 5S rRNA
Translation was carried out in a rabbit reticulocyte translation system (Promega, Madison, WI) in 25 µl of lysate with 10 µg L5 mRNA and 10 µg 5S rRNA in the presence of [35S]methionine (Amersham). Amino acids, an energy generating system, placental RNase inhibitor and protease inhibitor were included in the reaction. Unless specifically indicated otherwise, all translation reactions were carried out for 45 min at 30°C. The translated proteins were analyzed by 15% SDS–PAGE and formation of RNP was assayed on an 8% native polyacrylamide gel or a composite gel. Radioactive proteins and radioactive RNP complexes were visualized using a phosphorimager. Radioactivities of the synthesized L5 and RNP were determined by cutting out the gel band. The percentage incorporation of synthesized protein into the RNP complex was calculated after normalization for the radioactivity of synthesized protein.
The L5 and truncated L5 mutant mRNAs used in this study were prepared with a T7 promoter-driven RiboMAX system (Promega) on a large scale. The corresponding plasmids were used as templates. Milligram amounts of mRNA were obtained. Preparations of radioactive human, Escherichia coli and Xenopus laevis 5S rRNA and radioactive yeast snU5 RNA used for the polysome binding assay were radioactively labeled using the same RiboMAX system in the presence of [α-32P]GTP. Plasmids pGEM(T)/5Sh (human 5S rRNA), pGEM(T)/5SXeno (Xenopus 5S rRNA) and pGEM(T)/5SE.coli (E.coli. 5S rRNA) were constructed previously and were used as templates. The yeast snU5 RNA plasmid was provided by Dr Soo-Chen Cheng (Institute of Molecular Biology, Academic Sinica, Taiwan, Republic of China).
Polysome binding assay
A typical co-translational reaction was carried out with 10 µg L5 mRNA in the presence of 10 µg radioactive [32P]5S rRNA (sp. act. 1.5 × 103 c.p.m./µg). The reaction mixture was incubated for 10 min at 30°C, then analyzed on a 12 ml 0.5–1.5 M sucrose gradient containing T20K50M3 buffer (20 mM Tris–HCl, pH 7.6, 50 mM KCl and 3 mM MgCl2). Gradients were centrifuged at 35 000 r.p.m. for 165 min in a Beckman SW 41 rotor at 4°C. Fractions (0.6 ml) were collected and monitored for radioactivity. To determine the distribution of radioactive 5S rRNA, an aliquot was taken from each fraction and co-precipitated with 10 µg tRNA and analyzed by autoradiography on a 2% agarose gel. Puromycin-induced release of the nascent chain from the polysome was carried out by treatment of the 10 min reaction mixture with puromycin (final concentration 1 mM) for 15 min at 30°C.
The standard polysome profile was obtained by analyzing a total cell lysate from HeLa (1 × 107) cells under the same gradient conditions and monitored by absorbency at 254 mM.
Photo-crosslinking of iodouracil-containing 5S rRNA to the nascent chain of L5
The preparation of iodouracil-containing [32P]5S rRNA used the RiboMax system (Promega) in the presence of iodouracil 5′-triphosphate and [α-32P]GTP. In vitro translation of L5 and the ΔN35 and ΔN50 mutants was essentially as described above except that iodouracil-[32P]5S rRNA replaced the [32P]5S rRNA. The reaction was carried out for 10 min, then treated with puromycin (final concentration 1 mM) for 15 min at 30°C. After puromycin treatment the reaction mixture was placed on parafilm under long wavelength UV irradiation (15) for 2 h at 4°C. After irradiation the reaction mixture was layered on top of a 0.8 ml cushion of 0.5 M sucrose in T20K50M3 buffer in a 3 ml centrifuge tube, then centrifuged at 60 000 r.p.m. in a Beckman Ti101 rotor for 50 min at 4°C. This process removed ribosomes. The top fraction of 100 µl was collected and analyzed on a urea-containing polyacrylamide gel. Detection of iodouracil-[32P]5S rRNA or its crosslinking product was by autoradiography of the composite gel using a phosphorimager.
Microinjection and nuclear localization
Stage V or VI oocytes (according to the Dumont classification) of X.laevis in general received a 30 nl injection of [35S]L5–5S rRNP or translated [35S]L5 using an automated injector (model 1000XL; EFD, USA). The specific activities of [35S]L5–5S rRNP and [35S]L5 were adjusted to fall in the range 1.2–1.3 × 102 c.p.m./ng for each injection. The microinjected oocytes were healed on ice for 30 min before being incubated in OR-2 medium at 20°C.
To measure nuclear transport each individual oocyte was dissected manually into nuclear and cytoplasmic fractions by microscopic surgery. Total proteins in the nuclear and cytoplasmic fractions of a pool of 18 oocytes were precipitated with 15% TCA and analyzed on a 15% polyacrylamide gel containing SDS. The presence of radioactive L5 was determined using a phosphorimager. The presence of nucleolin in the nuclear or cytoplasmic fraction was detected using a monoclonal anti-nucleolin antibody (kindly provided by Dr Ning-Hsing Yeh, Institute of Microbiology and Immunology, National Yang-Ming University, Taipei, Republic of China). The amount of nucleolin detected by western blotting was used to normalize the amount of L5 being transported.
RESULTS
Formation of 5S rRNP is a translational event with participation by 5S rRNA during translation of L5
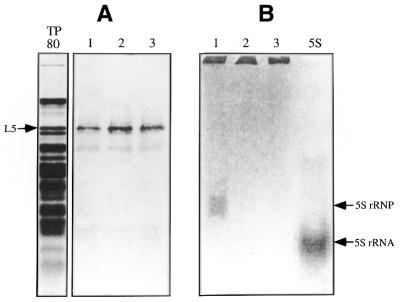
Standard in vitro translation of L5 was carried out in a rabbit reticulocyte lysate system containing [35S]methionine, except that a molar excess of 5S rRNA was added at the beginning of translation or at 45 min post-translation. The amount of synthesized radioactive [35S]L5 protein was determined by SDS–PAGE and found to be independent of the timing of addition of 5S rRNA and occurred even without 5S rRNA (Fig. 1A). However, the [35S]L5–5S rRNP complex was found only if 5S rRNA was present at the beginning of the translation reaction (Fig. 1B) when the same reaction mixture was analyzed by native composite gel electrophoresis. The [35S]L5–5S rRNP complex was not observed if 5S rRNA was added after completion of synthesis of L5 (Fig. 1B), suggesting that early participation of 5S rRNA was essential for formation of an RNP complex.
Figure 1.
Timing requirements for 5S rRNA in the formation of a 5S rRNP complex during translation of L5. (A) The in vitro synthesis of radioactive L5. Radioactive L5 was synthesized in a rabbit reticulocyte translation system with [35S]methionine in the presence of 5S rRNA added at the beginning of the reaction (lane 1) or 45 min after completion of translation (lane 2) or without 5S rRNA (lane 3). Each lane contains 25% of the total reaction mixture. (B) Formation of the 5S rRNP complex. This analysis was carried out with the remaining reaction mixture by 8% native PAGE. Formation of the 5S rRNP complex was observed as an up-shifted band (as indicated) by autoradiography. Lane TP80 is a Coomassie staining of total ribosomal protein extracted from rat liver 80S ribosomes. Lane 5S is [32P]5S rRNA, used as a marker. Both the L5 and the 5S rRNP complexes were detected using a phosphorimager.
From the measurement of [35S]L5 incorporated into [35S]L5–5S rRNP complexes it was determined that ∼12% of the total newly synthesized wild-type L5 was bound in the 5S rRNP complex (Table 1). His-tagged L5 was also found to participate in formation of the 5S rRNP complex, however with slightly lower efficiency (Fig. 2 and Table 1).
Table 1. Incorporation of L5 protein in RNP complexes and of 5S rRNA bound to the ribosome-associated nascent chain.
| Protein co-translated
with 5S rRNAa |
[35S]protein
incorporated
into RNP complexb (%) |
Radioactivity (c.p.m. × 103)
of [32P]5S rRNA associated with polysomesc |
| L5* | 12.38 | 16.52 |
| L5 | 9.39 | 12.07 |
| ΔN10 | 6.45 | 7.75 |
| ΔN17 | 7.08 | 9.88 |
| ΔN35 | 4.58 | 7.50 |
| ΔN50 | – | – |
| N1–93 | – | 9.62 |
| [31–51]ΔN114 | – | 5.25 |
Values for [35S]protein incorporated into RNP complex and radioactivity of [32P]5S rRNA associated with polysomes are the averages of two experiments.
–, negative finding.
aThe different mRNA-encoded proteins participating with 5S rRNA in the co-translational event. All mRNAs, except wild-type L5 (L5*), were derived from plasmid pET28a, which carries a His-tagged peptide at the N-terminus of the coded sequence.
bTotal radioactivity (c.p.m.) in RNP against the total radioactivity of synthesized 35S-labeled proteins after normalizing for the total amount of radioactive protein synthesized.
cThe determined radioactivity (c.p.m.) represents the sum of radioactivity from three sucrose gradient fractions located in the polysome region.
Figure 2.
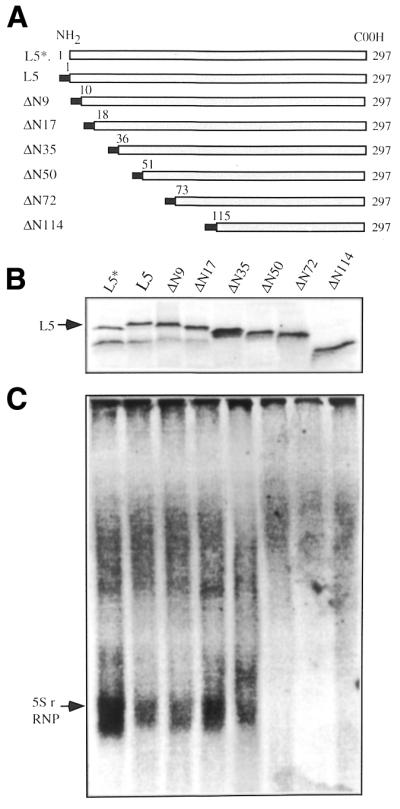
Formation of the 5S rRNP complex by N-terminal truncated mutants. (A) A schematic illustration of the coding region of the truncated L5 mutant proteins constructed for this study. The filled bar represents the His peptide sequence tag at the N-terminus of the coded protein. (B) Synthesis of 35S-labeled mutant proteins in the presence of 5S rRNA. (C) Formation of the RNP complex by the truncated mutant proteins. The translation reactions were carried out in the presence of human 5S rRNA added at the beginning of the translation reaction. Detection of radioactive proteins and RNP complex was as described in Figure 1. The radioactivity in L5 and 5S rRNP was determined by excising the bands from the gel. The obtained radioactivity was used to calculate the percentage of [35S]L5 participating in 5S rRNP, as presented in Table 1.
Formation of the 5S rRNP complex requires the N-terminal region of L5
To determine the region of L5 responsible for formation of RNP, degenerate deletions of the N-terminus of the protein (Fig. 2A) were constructed and their participation in formation of the 5S rRNP complex was examined. When 5S rRNA was initially present in the in vitro translation reaction these mutant proteins were qualitatively and quantitatively produced (Fig. 2B). Formation of the RNP complex was only observed with mutant proteins that were missing the first 9, 17 or 35 amino acids (Fig. 2C) and was not detectable for mutant proteins missing the first 50 amino acid residues or more (Fig. 2C). Incorporation of mutant protein into RNP complex was found to be much lower than that of wild-type L5, in the range 4–6.5% generally being observed (Table 1). Despite such a low percentage incorporation, these results indicate that the first 50 amino acid residues of L5 are essential for formation of 5S rRNP and might be the binding site for 5S rRNA. Together with the finding of a requirement for early 5S rRNA participation, the process of forming the 5S rRNP complex might be initiated by 5S rRNA binding to the nascent chain of L5.
5S rRNA associates with the nascent chain of L5
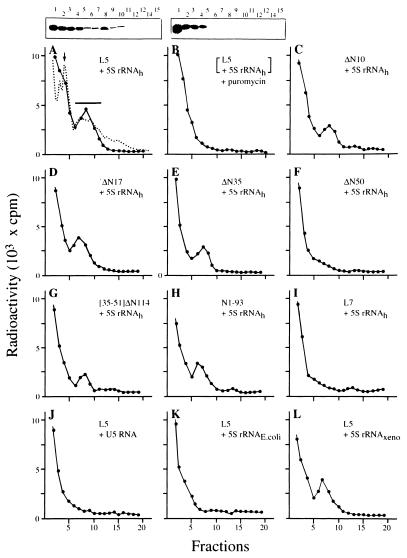
To determine whether 5S rRNA binds to the nascent chain or not we examined this possibility by a polysome-associated nascent chain assay (16,17). This was done by analyzing a 10 min translation mixture containing L5 mRNA with an excess of radioactive [32P]5S rRNA. Possible association of radioactive 5S rRNA with the nascent chain was determined with a polysome sucrose density gradient. The radioactive profile of the polysome gradient showed two distinguishable regions. One was at the top of the gradient, which contained the majority of the radioactivity, and the other was a distinct radioactive peak in the polysome region (Fig. 3A). Autoradiographs of a gel analysis of the gradient (Fig. 3A and top insert) showed that intact radioactive 5S rRNA was predominantly found in the top fractions, but also in the polysome peak fraction, suggesting that 5S rRNA was bound to polysome-associated nascent chains. Furthermore, when the same 10 min translation reaction was treated with puromycin we found that the radioactive peak in the polysome region was no longer observed (Fig. 3B and top insert), implying that the radioactive 5S rRNA was co-released with the nascent chain. Thus, an association between 5S rRNA and the nascent chain of L5 was established.
Figure 3.
Binding of 5S rRNA to the polysome-associated nascent chain. (A–I) The distribution of radioactive human [32P]5S rRNA (5S rRNAh) in the polysome gradient used to analyze the reaction mixture from translation of L5 or mutant protein mRNA with radioactive [32P]5S rRNA (each mRNA is listed on the right side). All mRNAs carry a sequence encoding the His peptide. The translation reaction was carried out for 10 min at 30°C in the presence of 10 µg human [32P]5S rRNA (1.5 × 103 c.p.m./µg). The dashed line in (A) represents the standard polysome profile derived from analysis of total HeLa cell lysate under the same conditions and monitored for absorbency at 254 nm wavelength. (B) The result of a 10 min reaction from (A) that was treated with puromycin prior to polysome assay. The boxes above (A) and (B) show autoradiograms of agarose gels analyzing each sucrose fraction (indicated by number above). The ribosome monomers and the region containing polysomes are marked by an arrow and a bar, respectively. (I) Translation of L7 mRNA with early participation of radioactive human [32P]5S rRNA (5S rRNAh). (J–L) Translation of L5 mRNA with either radioactive yeast snU5 RNA (U5 rRNA), E.coli 5S rRNA (5S rRNAE.coli) or Xenopus 5S rRNA (5S rRNAXeno), respectively. All RNAs except mRNA were uniformly labeled with radioactive [α-32P]GTP. The radioactive specificities of U5 rRNA, 5S rRNAE.coli and 5S rRNAXeno were adjusted to 1.5 × 103 c.p.m./µg by adding the corresponding cold RNA. Each assay was carried out with the same amount of RNA radioactivity for the purpose of comparison.
Using the same polysome assay, binding of radioactive 5S rRNA to the nascent chain of mutant proteins was measured. The results show that radioactive 5S rRNA bound to the nascent chains of mutant proteins ΔN9, ΔN17 and ΔN35 (Fig. 3C–E, respectively), but did not bind the nascent chain of mutant proteins that had been deleted by 50 residues (Fig. 3F) or more (shown below). The binding of mutant proteins to 5S rRNA agreed with the results from formation of RNP when the same truncated mutant proteins were used, as does the percentage of 5S rRNA binding to the nascent chain of truncated mutant proteins (Table 1).
According to the data from the polysome assays the region of the nascent chain required for binding 5S rRNA can be identified as between positions 35 and 50, which encompasses the sequence RLVIQDIKNKYNTPKYRM. To further validate the hypothesis that the elucidated sequence has the capability of binding 5S rRNA we constructed a mutant protein, [35–51]ΔN114, which carried this sequence at the N-terminal end of the ΔN114 mutant. The presence of the additional sequence on mutant [35–51]ΔN114 allowed binding of 5S rRNA, however at lower efficiency (Fig. 3G), despite the fact that the nascent chain of ΔN114 was unable to bind 5S rRNA (data not shown). However, no RNP complex could be detected for [35–51]ΔN114 mutant protein (data not shown). This suggests that formation of the rRNP complex requires a cooperative effort by other parts of L5. This suggestion was further supported by a similar result from an assay of 5S rRNA binding to the nascent chain of N1–93 mutant protein. The nascent chain of N1–93 (containing the first 93 amino acids of L5) is positively bound to 5S rRNA (Fig. 3H and Table 1), but does not form an RNP complex (data not shown).
Next, whether specific recognition of 5S rRNA by the nascent chain of L5 is specific was investigated. Human 5S rRNA binding to the nascent chain of another ribosomal protein, L7, was tested, with negative results (Fig. 3I), implying that the proposed co-translational 5S rRNA binding is specific to L5 and is amino acid sequence dependent. Whether the nascent chain of L5 is capable of binding other RNA molecules was also tested. The results showed that the nascent chain of human ribosomal protein L5 failed to bind a similar size (150 nt) RNA molecule, U5 snRNA (Fig. 3J), or 5S rRNA from E.coli (Fig. 3K). It does bind 5S rRNA from X.laevis (Fig. 3L). These results show that the interaction between the nascent chain and 5S rRNA seems to be eukaryote specific.
Crosslinking of 5S rRNA to the nascent chain of L5
Specific recognition of 5S rRNA by the nascent peptide chain of L5 has been further demonstrated by a photo-crosslinking experiment. Photoactive iodouracil-[32P]5S rRNA was introduced into the translation reaction with L5 and deletion mutant protein mRNAs. The 10 min reaction mixture treated with puromycin was UV irradiated. The results of UV crosslinking showed that a radioactive smear was up-shifted from the position of iodouracil-[32P]5S rRNA (Fig. 4) when reaction mixtures containing L5 or mutant protein ΔN35 (lacking the first 35 residues) were analyzed. These products represent different sizes of nascent peptides crosslinked to radioactive iodouracil-[32P]5S rRNA. Under the same experimental conditions no radioactive zone was detected if the reaction mixture contained ΔN50 (Fig. 4). The lack of binding by the ΔN50 mutant provided a good negative control. In particular, the results agreed with the negative findings of the polysome assay for ΔN50, as described above (Fig. 3F). Moreover, it confirmed the observation of crosslinking of 5S rRNA to L5 and to ΔN35, because our experiment was conducted with an excess of photoactive iodouracil-[32P]5S rRNA, which may cause non-specific crosslinkage. The data confirm that binding of 5S rRNA and the nascent chain of L5 requires specific recognition.
Figure 4.
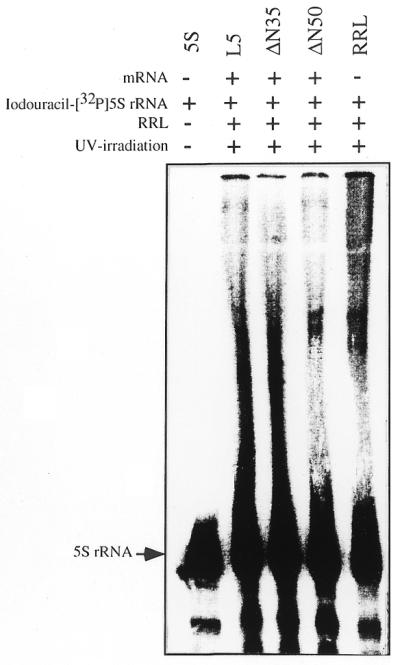
Crosslinking of photoactivated radioactive 5S rRNA to the nascent chain of L5. Translation was carried out separately with L5, ΔN35 or ΔN50 mutant mRNA in the presence of iodouracil-[32P]5S rRNA. After 10 min reaction the reaction mixture was treated with puromycin and photoactivated with UV irradiation as described in the text. The crosslinked product was then analyzed by urea-containing sequencing gel electrophoresis and autoradiographed using a phosphorimager. The position of iodouracil-[32P]5S rRNA is indicated. RRL represents the photo-crosslinking product from rabbit reticulocyte lysate, which had gone through the same procedure but without any mRNA.
Nuclear entry of L5 in the form of a 5S rRNP complex
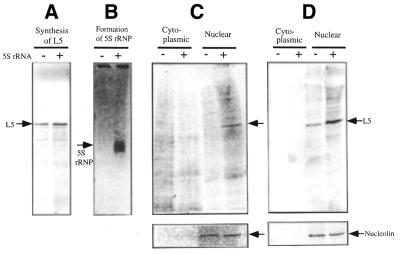
The importance of the 5S rRNP complex in nuclear import of L5 was measured by microinjection of prepared [35S]L5–5S rRNP or [35S]L5 (Fig. 5A and B) into the cytoplasm of Xenopus oocytes. After cellular dissection the presence of radioactive L5 in the nucleus was detected by autoradiography of an SDS–polyacrylamide gel of subcellular fractions. The results show that L5 in [35S]L5–5S rRNP reached the nucleus 12 h post-microinjection (Fig. 5C), whereas L5 from microinjected [35S]L5 did not appear until 16 h post-microinjection (Fig. 5C and D). The amount of radioactive L5 appearing in the nucleus was calculated after normalization with respect to the presence of nucleolin in the nuclear fraction (Fig. 5C and D). The calculation showed an ∼2.7-fold increase in L5 entering the nucleus when the L5–5S rRNA complex was injected as compared with injection of L5 alone. The initial rate and the extent of net nuclear import of L5 suggested that the 5S rRNP complex provides a significant advantage in nuclear import of L5. The results agree with findings previously reported with respect to the measurement of nuclear transport of 5S RNA in the form of a 5S rRNP complex (12,13,18,19).
Figure 5.
Nuclear entry of L5 from a co-translational form of 5S rRNP after microinjection into the cytoplasm of Xenopus oocytes. (A and B) Preparations of [35S]L5 and [35S]L5–5S rRNA complex analyzed by SDS-containing PAGE and 8% native gel electrophoresis, respectively. (C and D) The results of nuclear entry of radioactive L5 after injection of samples of [35S]L5–5S rRNA complex and [35S]L5 into the cytoplasm of Xenopus oocytes. The cytoplasmic and nuclear fractions were prepared from 18 pooled oocytes by manual dissection. Proteins were extracted from pooled fractions. The cellular distribution of radioactive [35S]L5 in cytoplasm and nucleus at 12 (C) and 16 h (D) was analyzed by autoradiography of SDS-containing 15% polyacrylamide gels. Detection of nucleolin in nuclei (shown in the lower panels C and D) was by western blot using a monoclonal anti-nucleolin antibody. The amount of nucleolin in the nuclear fraction was used for normalization of the amount of L5 in the nucleus. In general we have found that [35S]L5 underwent protein degradation post-microinjection if the protein did not enter the nucleus.
DISCUSSION
To reconstitute a eukaryotic 5S rRNP complex has been a long-standing problem in the study of eukaryotic ribosomes. The problem has been solved by the finding that assembly of the 5S rRNP complex takes place with newly synthesized L5 (20,21) or freshly prepared L5 protein (22). Such observations imply that the configuration of L5 and/or the timing of 5S rRNA participation during synthesis of L5 may be the key to making a eukaryotic 5S rRNP complex. In this study we have examined both possibilities by use of cell-free translation. We have shown that formation of 5S rRNP requires early participation of 5S rRNA. By polysome assay and crosslinking experiments we have shown that 5S rRNA binds the nascent chain during translation of L5. Therefore, we conclude that nascent chain binding of 5S rRNA and formation of the RNP complex are related events. Participation of 5S rRNA in in vitro formation of the RNP complex is therefore a co-translational event (23).
In this study the region of L5 that associates with 5S rRNA has been refined and narrowed down to 18 amino acid residues compared with that previously reported (19,24,25). The identified region encompasses the sequence RLVIQDIKNKYNTPKYRM and is located at positions 35–50. We have found that delineation of the 5S rRNA-binding sequence in this study has revealed a different RNA-binding motif to that previously identified (26,27). It is not present in any other RNA-binding protein. We therefore suspect that this sequence may be a new class of RNA-binding sequence motif that is specific for binding eukaryotic 5S rRNA.
It is noteworthy to mention the work of Pieler and colleagues (19), who described an L5 mutant (47–296) that lacked most of the assigned residues and was found to bind to 5S rRNA less efficiently. The discrepancy in the assigned region for 5S rRNA binding could be due to differences in the methods of detection. The C-terminal region of their mutant (47–296) may also contribute to positive binding of 5S rRNA (28,29) when a co-immunoprecipitation assay is applied. That the C-terminal region is essential for binding to 5S rRNA has been suggested by the fact that deletion of the C-terminus of yeast L5 is lethal in vivo (28,30). In addition, we have also shown that binding of 5S rRNA to a nascent chain carrying the binding sequence does not form a 5S rRNA-associated RNP complex if the rest of the L5 protein is not present. This supports the hypothesis that participation of the overall structure of L5 protein is required to form the RNP complex.
This study shows that participation of L5 in formation of the RNP or binding of 5S rRNA to the nascent chain was relatively low (Table 1), particularly when the experiments were conducted with an excess of 5S rRNA. However, this may be due to availability of the correct form of the newly synthesized nascent chain acting as a limiting factor. Spatial arrangement of the nascent peptide chain generally takes place in much less then 1 s (16,31). It is expected that formation of a non-electrostatic stacking interaction (22) between the folding nascent chain of L5 and 5S rRNA has a very limited time window. Thus, the determined sequence motif on the nascent chain could represent a folding-competent state for binding 5S rRNA once it emerges from the ribosome. This type of nascent chain binding has been observed in the small chaperones of the heat shock protein 70/DnaJ/GrpE families, which first bind to the elongating nascent chain and prevent it from folding prematurely (17). It also mimics the behavior of NAC (nascent chain-associated complex) during elongation of a nascent chain to ensure proper folding and translocation of a secretory protein (32). In essence, correct binding of 5S rRNA at the sequence motif of the nascent chain provides a ‘safe’ condition for proper folding of L5 into the RNP complex. As our experimental data indicate, any mishap, such as lack of the binding site or late arrival of the 5S rRNA, would prevent 5S rRNA forming the RNP complex. Consequently, nuclear import of L5 protein or 5S rRNA would be retarded (6,12). 5S rRNA-assisted nuclear entry of L5 is similar to RNA-assisted nuclear transport of the meiotic regulator protein Mei2p in yeast (33).
Taking these concepts together with our findings, 5S rRNA might be considered to have chaperone-like behavior within the broad meaning of ‘molecular chaperone’, which defines the role of a chaperone as an entity modulating protein conformation (34). This implies that 5S rRNA participates in folding of the protein. Such an implication is extraordinary, but not unique, since 23S rRNA and ribosomal proteins have recently been suggested as helping misfolded proteins escape from kinetic traps, as well as isolating them from the cellular environment (23,35). However, this suggestion does fall within the concept that RNA might execute many of the functions that were once thought to be the sole role of proteins (36), evident in the discovery of the self-processing action of RNA molecules. This, too, is related to the long-standing paradox as to which was the original chaperone. The question as to how the original chaperone protein was folded could find an answer in RNA.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Professor Ralph Kirby of Rhodes University, South Africa, for critical reading of the manuscript. This work was supported in part by grants from the National Science Council (NSC89-2311-B010-004). A.L. is the recipient of a Medical Research and Advancement Foundation awarded in memory of Dr Chi-Shuen Tsou.
References
- 1.Blobel G. (1971) Isolation of a 5S RNA–protein complex from mammalian ribosomes. Proc. Natl Acad. Sci. USA, 68, 1881–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erdmann V.A., Pieler,T., Wolters,J., Digweed,M., Vogel,D. and Hartmann,R. (1985) In Hardesty,B. and Kramer,G. (eds), Structure, Function and Genetics of Ribosomes. Springer-Verlag, New York, NY, pp.164–183.
- 3.Rout M.P., Blobel,G. and Aitchison,J.D. (1997) A distinct nuclear import pathway used by ribosomal proteins. Cell, 89, 715–725. [DOI] [PubMed] [Google Scholar]
- 4.Kakel S. and Gorlich,D. (1998) Importin β, transportin, RanRP5 and RanRP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J., 17, 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steitz J.A., Berg,C., Hendrick,J.P., La Branche-Chabot,H., Metspalu,A., Rinke,J. and Yario,T. (1988) A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J. Cell Biol., 106, 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allison L.A., North,M.T., Murdoch,K.J., Romaniuk,P.J., Deschamps,S. and LeMaire,M. (1993) Structure requirements of 5S rRNA for nuclear transport, 7S ribonucleoprotein particle assembly and 60S ribosomal subunit assembly in Xenopus oocytes. Mol. Cell. Biol., 13, 6819–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dechampesme A.-M., Koroleva,O., Leger-Silvestre,I., Gas,N. and Camier,S. (1999) Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway. J. Cell Biol., 145, 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinke J. and Steitz,J.A. (1982) Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell, 29, 149–159. [DOI] [PubMed] [Google Scholar]
- 9.Guddat U., Bakken,A.H. and Pieler,T. (1990) Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in Xenopus oocytes. Cell, 60, 619–628. [DOI] [PubMed] [Google Scholar]
- 10.Pittman R.H., Andrews,M.T. and Setzer,D.R. (1999) A feedback loop coupling 5S rRNA synthesis to accumulation of a ribosomal protein. J. Biol. Chem., 274, 33198–33201. [DOI] [PubMed] [Google Scholar]
- 11.DeRobertis E.M., Lienhard,S. and Parisot,R.F. (1982) Intracellular transport of microinjected 5S and small nuclear RNAs. Nature, 295, 572–577. [DOI] [PubMed] [Google Scholar]
- 12.Rudt F. and Pieler,T. (1996) Cytoplasmic retention and nuclear import of 5S ribosomal RNA containing RNPs. EMBO J., 15, 1383–1391. [PMC free article] [PubMed] [Google Scholar]
- 13.Murdoch K.J. and Allison,L.A. (1996) A role for ribosomal protein L5 in the nuclear import of 5S rRNA in Xenopus oocytes. Exp. Cell Res., 227, 332–343. [DOI] [PubMed] [Google Scholar]
- 14.Chan Y.L., Lin,A., McNally,J. and Wool,I.G. (1987) The primary structure of rat ribosomal protein L5. J. Biol. Chem., 262, 12879–12886. [PubMed] [Google Scholar]
- 15.Willis M.C., Hicke,B.J., Uhlenbeck,O.C., Cech,T.R. and Koch,T.H. (1993) Photocrosslinking of 5-iodouracil-substituted RNA and DNA to proteins. Science, 262, 1255–1257. [DOI] [PubMed] [Google Scholar]
- 16.Fedorov A.N. and Baldwin,T.O. (1997) Cotranslational protein folding J. Biol. Chem., 272, 32715–32718. [DOI] [PubMed] [Google Scholar]
- 17.Langer T., Lu,C., Echols,H., Flanagan,J., Hayer,M.K. and Hartl,F.U. (1992) Successive action of Dnak, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature, 356, 683–689. [DOI] [PubMed] [Google Scholar]
- 18.Rosorius O., Fries,B., Stauber,R.H., Hirschmann,N., Bevec,D. and Hauber,J. (2000) Human ribosomal protein L5 contains defined nuclear localization and export signals. J. Biol. Chem., 275, 12061–12068. [DOI] [PubMed] [Google Scholar]
- 19.Claussen M., Rudt,F. and Pieler,T. (1999) Functional modules in ribosomal protein L5 for ribonucleoprotein complex formation and nucleocytoplasmic transport. J. Biol. Chem., 274, 33951–33958. [DOI] [PubMed] [Google Scholar]
- 20.Wormington W.M. (1989) Developmental expression and 5S rRNA-binding activity of Xenopus laevis ribosomal protein L5. Mol. Cell. Biol., 9, 5281–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeh L.-C.C. and Lee,J.C. (1995) An in vitro system for studying RNA–protein interaction: application to a study of yeast ribosomal protein L1 binding into 5S rRNA. Biochimie, 77, 167–173. [DOI] [PubMed] [Google Scholar]
- 22.Scripture J.B. and Huber,P.W. (1996) Analysis of the binding of Xenopus ribosomal protein L5 to oocyte 5S rRNA. J. Biol. Chem., 270, 27358–27365. [DOI] [PubMed] [Google Scholar]
- 23.Hardesty B., Tsalkova,T. and Kramer,G. (1999) Co-translational folding. Curr. Opin. Struct. Biol., 9, 111–114. [DOI] [PubMed] [Google Scholar]
- 24.Michael W.M. and Dreyfuss,G. (1996) Distinct domains in ribosomal protein L5 mediate 5S rRNA binding and nucleolar localization. J. Biol. Chem., 271, 11571–11574. [DOI] [PubMed] [Google Scholar]
- 25.Lin E., Liu,S.R. and Lin,A. (1999) Biochemical analysis of a 5S rRNA-associated sub-particle from trypsinized eukaryotic ribosomes. J. Biochem., 125, 1029–1033. [DOI] [PubMed] [Google Scholar]
- 26.Kenan D.J., Query,C.C. and Keene,J.D. (1991) RNA recognition: towards identify determinants of specificity. Trends Biochem. Sci., 16, 214–220. [DOI] [PubMed] [Google Scholar]
- 27.Fierro-Monti I. and Mathews,M.B. (2000) Protein binding to duplexed RNA: one motif, multiple functions. Trends Biochem. Sci., 25, 241–246. [DOI] [PubMed] [Google Scholar]
- 28.Deshmukh M., Stark,J., Yeh,L.-C.C., Lee,J.C. and Woolford,J.L. (1995) Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5S rRNA and assembly into ribosomes. J. Biol. Chem., 270, 30148–30156. [DOI] [PubMed] [Google Scholar]
- 29.Yeh L.-C.C., Deshmukh,M., Woolford,J.L. and Lee,J.C. (1996) Involvement of lysine 270 and lysine 271 of yeast 5S rRNA binding protein in RNA binding and ribosome assembly. Biochim. Biophys. Acta, 1308, 133–141. [DOI] [PubMed] [Google Scholar]
- 30.Deshmukh M., Tsay,Y.-F., Paulovich,A.G. and Woolford,J.L. (1993) Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol. Cell. Biol., 13, 2835–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roder H. and Colon,W. (1997) Kinetic role of early intermediates in protein folding. Curr. Opin. Struct. Biol., 7, 15–28. [DOI] [PubMed] [Google Scholar]
- 32.Lauring B., Sakai,H., Kreibich,G. and Wiedmann,M. (1995) Nascent polypeptide-associated complex protein prevents mistargeting of nascent chains to the endoplasmic reticulum. Proc. Natl Acad. Sci. USA, 92, 5411–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamashita A., Watanabe,Y., Nukina,N. and Yamamoto,M. (1998) RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell, 95, 115–123. [DOI] [PubMed] [Google Scholar]
- 34.Ellis R.J. (1997) Molecular chaperones: avoiding the crowd. Curr. Biol., 7, 531–533. [DOI] [PubMed] [Google Scholar]
- 35.Kudlicki W., Coffman,A., Kramer,G. and Hardesty,B. (1997) Ribosomes and ribosomal RNA as chaperones. Fold Des., 2, 101–108. [DOI] [PubMed] [Google Scholar]
- 36.Ellis R.J. (1997) Do molecular charperones have to be protein? Biochem. Biophys. Res. Commun., 238, 687–692. [DOI] [PubMed] [Google Scholar]