Abstract
Heat stress proteins assist cellular proteins in the acquisition of native structure. The present research was conducted to study how thermo-tolerance is modulated by HSP90 and HSP70 gene polymorphism and its association with hemato-physio-biochemical parameters, supported by their expression profiles in Chokla, Magra, Marwari, and Madras Red sheep breeds. Least square analysis revealed significant effect (P < 0.05) of season and breed on all the physiological parameters, i.e., temperature, respiratory rate, and pulse rate (a.m. and p.m.), as well as hematological parameters like Hb, packed cell volume, total erythrocyte count (TEC), neutrophil/lymphocyte (N/L) ratio, and total leukocyte count (TLC). There was a significant influence (P < 0.05) of breed on biochemical parameters such as glucose, SGOT, phosphorous, triglyceride, and cholesterol. Eight fragments were amplified and sequenced in HSP90, and 70 genes and 13 single-nucleotide polymorphisms (SNPs) were identified. Tetra-primer amplification refractory mutation system PCR, PCR-RFLP, and allele-specific PCR genotyping protocols were developed for large-scale genotyping of five SNPs. A significant difference (P < 0.05) of rectal temperature (a.m.), respiratory rate (p.m.), triglyceride, and total protein was observed at SNP01; albumin at SNP2; pulse rate (p.m.) at SNP3; and rectal temperature (p.m.), pulse rate (p.m.), Hb (g/dL), and N/L ratio at SNP4 and TLC at SNP5. Gene expression analysis revealed lower expression in less adapted animals with Madras Red < Magra < Chokla < Marwari expression pattern. Predominant allele was found to be superior in most of the SNPs (SNP1–4) indicating the selection acting in directional manner (positive selection). Finally, it is concluded that TACCA haplotype combination of SNP1-SNP2-SNP3-SNP4-SNP5 might be of some selection advantage for the identification of animals more adaptable to heat stress.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-017-0770-4) contains supplementary material, which is available to authorized users.
Keywords: Heat stress proteins, Molecular chaperones, HSP90, HSP70, Polymorphism, Association study, HSP gene expression
Introduction
Heat stress proteins (HSPs) are molecular chaperones that prevent the formation of non-specific protein aggregates and assist cellular proteins in the acquisition of native structure. They play an important role in the cellular homeostasis. HSPs have been classified into several classes based on their molecular weight, such as HSP90 (85–90 kDa), HSP70 (68–73 kDa), HSP60, HSP47, and small HSPs (12–43 kDa) (Park et al. 2007). They are highly conserved across the species (Schlesinger 1990). These proteins are induced by heat and many other stresses like infection; inflammation; exercise; exposure of toxins like ethanol, arsenic, heavy metals, and pesticides; starvation; water stress; oxygen deprivation; cold; and salinity (Waters et al. 1996; Boston et al. 1996; Vierling 1991). Heat is one of the main sources of stress, which has an important impact on the production and reproduction of livestock species. Animals adapted to hot/cold climatic conditions should show least variation in traits when raised under such conditions. Animals often differ in their tolerance and susceptibility to the thermal stress. This variability can be explored at the DNA level to identify the genes/alleles underlying such traits of adaptability and further be utilized for the selection of animals tolerant to heat stress. Sheep and goat exposed to elevated temperatures results in a decreased body weight, average daily gain (ADG), growth rate, and body total solids, which is reflected by impaired reproduction (Maria et al. 2000; Shelton 2000; Abdel-Hafez 2002). This causes severe economic losses to sheep and goat keepers. Heat stress influences the physio-biochemical traits of sheep and goat (Ocak and Guney 2010; Alam et al. 2011; Phulia et al. 2010; Sivakumar et al. 2010; Helal et al. 2010; Sharma and Kataria 2011). In sheep, few reports are available about the HSP gene studies at molecular level. However, these are mainly confined to the genetic polymorphism, association with scrapie incubation period, adaptation to differential thermal condition, cloning, and expression (Salces-Ortiz et al. 2013, 2015; Marcos-Carcavilla et al. 2010a, b; Pawar et al. 2013). No association study has so far been conducted between HSP gene polymorphisms and physio-biochemical changes in Indian sheep breeds. Therefore, the present research was undertaken to explore single-nucleotide polymorphisms (SNPs) in HSP90 and 70 genes and to study the association of the identified SNPs with the physio-biochemical traits in four Indian sheep breeds. Real-time polymerase chain reaction study was also carried out to comprehend the effect of these polymorphisms on the expression profile of three HSP genes viz. HSP90AA1, HSPA1A, and HSPA8.
Materials and methods
Sample collection
A total of 211 animals of four sheep breeds [Chokla (CK) 47; Magra (MG) 52; Marwari (MW) 52, and Madras Red (MR) 60] were screened. Samples of CK, MG, and MW were collected from Arid Region Campus, CSWRI, Bikaner, and MR from Kattupakkam farm, TANUVAS, Chennai. Female animals of 1–1.5 years of age with almost similar body weight were only used in the investigation. Isolation of the plasma was carried out immediately after blood collection. The samples were brought to the laboratory while maintaining the cold chain. Blood samples were analyzed immediately for the hematological parameters whereas plasma samples were kept at −20 °C till further analysis.
DNA isolation, SNP identification, and polymorphism study
Genomic DNA of all samples was isolated by the phenol chloroform extraction method (Sambrook and Russell 2001). Five representative samples of each breed were initially sequenced, and SNPs were identified. PCR-RFLP, tetra-primer amplification refractory mutation system (TETRA-ARMS) PCR, and allele-specific PCR protocols were developed for the five identified SNPs for large-scale genotyping of all the samples.
Evaluation of physio-biochemical parameters
Same animals which were initially screened for the genomic DNA isolation were used for the estimation of hematological and physio-biochemical parameters under normal (lower temperature-humidity index of 65 and less in winter season) as well as heat stress condition (higher temperature-humidity index of 84 or more in summer season). THI during the study period was calculated by the following formula (LPHSI 1990):
where db°F is the dry bulb temperature in °F and RH is the relative humidity (RH %)/100.
Thirty animals of each breed per season were analyzed as per the methods described by Singh et al. (2016). Briefly, 1.5 mL of blood sample collected with anticoagulant was analyzed for hematological parameters including hemoglobin (Hb-gm/dL), packed cell volume (PCV %), total erythrocyte count (TEC ×10 (Boston et al. 1996) per μL), total leukocyte count (TLC ×10 (Al-Murrani et al. 1997) per μL), and differential leukocyte count (DLC) as per the method described by Schalm et al. (1975). Biochemical analysis of the plasma and serum samples was carried out by semi-auto-analyzer (3000 Evolution, Biochemical Systems International, Italy) using commercially available kits of Erba (Transasia Biomedicals Ltd. Solan, India). The physiological parameters (temperature, respiratory rate, and pulse rate) were measured for 3 days in both morning (a.m.) and afternoon hours (p.m.) by standard procedures.
RNA isolation and cDNA synthesis
After large-scale screening of identified SNPs in four breeds, peripheral blood mononuclear cell (PBMC) fraction was separated from blood samples (n = 48) of representative animals of each breed (CK 5; MG 5; MW 5; MR 9) in two seasons by density gradient centrifugation using the Histopaque-1077 solution (Sigma-Aldrich). These animals were identified so that at least three or more animals are represented in each group (breed-wise, season-wise, and genotype-wise). RNA was subsequently isolated using the TRIzol reagent and purified by RNA isolation kit as per the standard procedure. Quantification of DNA and RNA was carried out by NanoDrop 1000 spectrophotometer. Good-quality samples with optical density for DNA within the range of 1.7–1.9 and RNA 1.9–2.1 were only used. The integrity of total RNA isolated was checked by visualization of 18S and 28S ribosomal RNA bands on 1% denaturing agarose gel. First -strand complementary DNA (cDNA) synthesis was prepared by using the RevertAid first-strand cDNA synthesis kit (Thermo Scientific). RNA was treated with RNAse-free DNAse I to remove DNA contamination, if any.
Quantitative real-time PCR
qPCR was performed using LightCycler 480 instrument (Roche, Germany) using SYBR Green I chemistry. The primers for qPCR were designed on the basis of prior sequence information available at NCBI and listed in the supplementary table (Table S1). Two endogenous reference genes were amplified (beta-actin and RPS-15), out of which beta-actin was found to be more stably expressed hence used in further expression analysis. Each sample had biological as well as technical triplicates.
Gene expression analysis
Gene expression analysis of HSP90AA1, HSPA1A, and HSPA8 genes was carried out breed-wise, season-wise, and genotype-wise taking at least three representative samples of each group and following the method as described by Pfaffl (2001). Median bootstrap analysis (Cleries et al. 2012; http://regstattools.net/br) was carried out to determine the significance of fold chain difference among groups.
Statistical analysis
The significant effect of genotypes on physio-biochemical traits was analyzed using the following model by SPSS software version 22.0.0.
where Y ijkl = physiological parameter of lth animal belonging to ith genotype in jth season of kth breed, μ = overall mean, G i = effect of ith genotype (i = 1 to n), S j = effect of jth season, B k = effect of kth breed (k = 1 to 4), and e ijkl = random error associated with Y ijkl observation and assumed to be NID (0, σ 2 e).
A genotype was considered to be superior over the other genotypes, for a particular locus, based on the minimum difference in mean values for physio-biochemical traits between a particular genotype and mean of all genotypes in normal condition (lower temperature-humidity index of 65 and less).
Results
PCR amplification and identification of SNPs
Eight different primer sets were designed for HSP-90 and HSP-70 gene, and PCR protocols were standardized. Four overlapping sets of HSP-70 gene primer pairs were successfully employed to amplify 544-, 609-, 559-, and 594-bp fragments covering the entire single exon of 1.93 kb. Four fragments of 490 bp (HSP90-exon 6–7), 258 bp (HSP90-exon 8), 354 bp (HSP90-exon 9), and 499 bp (HSP90AA1 promoter region) were successfully amplified for the characterization of HSP-90 gene. Five representative samples of each breed were sequenced, and the SNPs were identified. The details of the primer sets are given in the supplementary table (Table S2).
Development of genotyping protocols
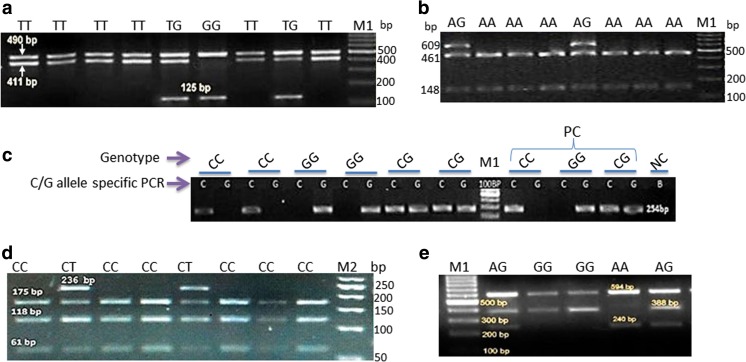
A total of 13 SNPs were initially identified. Genotyping protocols were developed for five SNPs viz. SNP01 (HSP90 ex6–7390T>G, 490 bp), SNP02 (HSP70 set2 459A>G, 609 bp), SNP03 (HSP90 ex9 180C>T, 354 bp), SNP04 (HSP90AA1 112G>C, 499 bp), and SNP5 (HSP70 set4 222G>A, 594 bp) employing tetra-primer ARMS-PCR, PCR-RFLP, and allele-specific PCR. The details of the primer sets used for genotyping SNP1 and 5 by tetra-primer ARMS-PCR, SNP2 and SNP3 by PCR-RFLP, and SNP4 by allele-specific PCR are given in Table S3. PCR-RFLP was standardized for SNP2 by HpyCH4 III restriction enzyme and for SNP3 by Taqα I. Figure 1a–e depicts the electrophoretic patterns (1.5% agarose gel electrophoresis) obtained by genotyping of SNP1, 2, 4, 3, and 5, respectively.
Fig. 1.
Electrophoretic patterns obtained by genotyping of sheep breeds at SNP1–5. a SNP1 genotyped by tetra-primer ARMS PCR. b SNP2 by PCR- RFLP with HpyCH4 III restriction enzyme. c SNP4 by allele-specific PCR. d SNP3 by PCR- RFLP with Taq α I restriction enzyme. e SNP5 by tetra-primer ARMS PCR; M1 100-bp DNA marker, M2 50-bp DNA marker
Gene and genotype frequencies
The gene and genotype frequencies of all the SNPs are given in Tables 1 and 2, respectively. The overall frequency of major allele was >80% for all the SNPs except SNP5 where it was 65%. The minor allele frequencies varied from 3% in SNP3 to 34% in SNP5. In MR sheep, alleles A and C were found to be fixed in SNP2 and SNP3, respectively. SNP5 had appreciably higher minor allele frequency with a value of 34%.
Table 1.
Allele frequencies of five different SNPs in four different sheep breeds
| SNP no. | SNP1 | SNP2 | SNP3 | SNP4 | SNP5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Breed/allele | T | G | A | G | C | T | G | C | G | A |
| Chokla | 0.84 | 0.16 | 0.83 | 0.17 | 0.94 | 0.05 | 0.85 | 0.14 | 0.60 | 0.39 |
| Marwari | 0.81 | 0.18 | 0.95 | 0.04 | 0.98 | 0.02 | 0.90 | 0.09 | 0.84 | 0.15 |
| Magra | 0.91 | 0.08 | 0.82 | 0.17 | 0.95 | 0.04 | 0.83 | 0.16 | 0.74 | 0.26 |
| Madras Red | 0.69 | 0.30 | 1.00 | 0.00 | 1.00 | 0.00 | 0.65 | 0.34 | 0.43 | 0.57 |
| Overall | 0.81 | 0.18 | 0.90 | .09 | 0.97 | 0.03 | 0.81 | 0.18 | 0.65 | 0.34 |
Table 2.
Genotype frequencies of five different SNPs in four different sheep breeds
| Breed | SNP1 | SNP2 | SNP3 | SNP4 | SNP5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | TG | GG | AA | AG | GG | CC | CT | TT | GG | GC | CC | GG | GA | AA | |
| Chokla (47) | 0.68 | 0.31 | 0 | 0.66 | 0.34 | 0 | 0.89 | 0.11 | 0 | 0.70 | 0.29 | 0.00 | 0.47 | 0.25 | 0.27 |
| Marwari (52) | 0.63 | 0.36 | 0 | 0.90 | 0.09 | 0 | 0.96 | 0.04 | 0 | 0.80 | 0.19 | 0.00 | 0.79 | 0.11 | 0.09 |
| Magra (52) | 0.82 | 0.17 | 0 | 0.65 | 0.34 | 0 | 0.92 | 0.08 | 0 | 0.67 | 0.32 | 0.00 | 0.60 | 0.28 | 0.11 |
| Madras Red (60) | 0.50 | 0.38 | 0.12 | 1.00 | 0.00 | 0 | 1.00 | 0.00 | 0 | 0.48 | 0.35 | 0.16 | 0.17 | 0.51 | 0.31 |
| Overall (211) | 0.657 | 0.30 | 0.03 | 0.80 | 0.19 | 0 | 0.94 | 0.05 | 0 | 0.66 | 0.29 | 0.04 | 0.50 | 0.29 | 0.20 |
Association study
Table S4a–f depicts the least square means with significant effect for all the five SNP genotypes. Only the traits with significant effect (P < 0.05) in the identified SNPs are discussed here.
Effect of SNP1
Genotypes or animals which show least deviation in their physio-biochemical traits between summer (stressed) and winter seasons (normal unstressed condition) are more adaptable to the heat stress (Habeeb et al. 1997; Marai and Habeeb 1998). For respiratory rate in afternoon (p.m.), animals of different genotypes and seasons had significantly different mean values. For the summer and winter seasons, the least square means were 56.13 ± 1.22 and 43.59 ± 1.15, respectively (Table S4a). The least square means for the TT, TG, and GG genotypes were 50.79 ± 1.01, 49.05 ± 0.99, and 51.19 ± 2.78, respectively (Table S4b). This indicates that mean values follow GG>TT>TG pattern, indicating the superiority of T allele over the G allele in terms of adaptability to heat stress as mean value was lower in winter season (normal condition) than in summer (heat-stressed condition). Likewise for total protein (g/dL), animals of different genotypes had significantly different mean values with GG>TG>TT again indicating the superiority of T allele over the G allele as mean value was lower in winter season (normal condition) than in summer season (heat-stressed condition). However, for triglyceride (mg/dL), animals of summer season had significantly reduced mean values (14.79 ± 5.50) than of winter season (27.23 ± 5.14). The least square means for the TT, TG, and GG genotypes were 29.60 ± 3.95, 21.98 ± 4.53, and 11.59 ± 1.02, respectively. This shows that different genotypes had significantly different mean values with TT>TG>GG pattern, again indicating the superiority of T allele over the G allele as mean value was higher in winter season (normal condition) than in summer season (heat-stressed condition). Overall, for SNP1, results indicate the superiority of T allele over G allele in terms of adaptability to heat stress.
Effect of SNP2
For albumin (g/dL), animals of different genotypes had significantly different mean values with AG (3.16 ± 0.19)>AA (2.88 ± 0.16) indicating the superiority of AA genotype or A allele over the AG genotype or G allele as the mean value was lower in winter season (2.51 ± 0.18) than in summer season (3.53 ± 0.16).
Effect of SNP3
For pulse rate, in afternoon, animals of different genotypes had significantly different mean values with CT (93.55 ± 1.89)>CC (89.82 ± 1.10) indicating the superiority of CC genotype or C allele over the T allele as mean value was higher in summer season (96.17 ± 1.40) than in winter season (87.19 ± 1.32).
Effect of SNP4
For rectal temperature in afternoon (p.m.), animals of different genotypes had significantly different mean values with GC (102.98 ± 0.07)>GG (102.91 ± 0.05)>CC (102.40 ± 0.24) indicating the superiority of CC genotype or C allele over the G allele as mean value was higher in summer season (103.16 ± 0.14) than in winter season (102.26 ± 0.13). Likewise for pulse rate, in afternoon (p.m.), animals of different genotypes had significantly different mean values with GG (90.04 ± 0.63)>GC (88.57 ± 0.89)>CC (74.33 ± 3.08) indicating the superiority of CC genotype or C allele over the G allele as mean value was higher in summer season (96.17 ± 1.40) than in winter season (87.19 ± 1.32). For Hb (g/dL), also animals of different genotypes had significantly different mean values with GC>GG>CC indicating the superiority of GC genotype over GG and CC as mean value was higher in winter season (normal condition) than in summer season (heat-stressed condition). For neutrophil/lymphocyte ratio (N/L ratio), animals of different genotypes had significantly different mean values with GG (1.01)>GC (0.94)>CC (0.50) trend indicating the superiority of CC genotype over GG and GC as N/L ratio increases in response to stress and circulatory glucocorticoids (Davis et al. 2008). For M (%), CC>GC>GG trend was obtained indicating the superiority of CC genotype as mean value was non-significantly higher in normal condition.
Effect of SNP5
For total leukocyte count (TLC/cmm), animals of different genotypes had significantly different mean values with AA (12,784 ± 8.3212)>AG (11,804 ± 774.01)>GG (11,354 ± 722.93) indicating the superiority of AA genotype or A allele over the G allele as mean value was higher in winter season (12,420 ± 736.63) than in summer season (10,850 ± 726.89).
Gene expression analysis
Gene expression analysis was carried out breed-wise, season-wise, and genotype-wise by the method described by Cleries et al. (2012). The graphical representations of the median expression ratios are given in Figs. 2, 3, and 4.
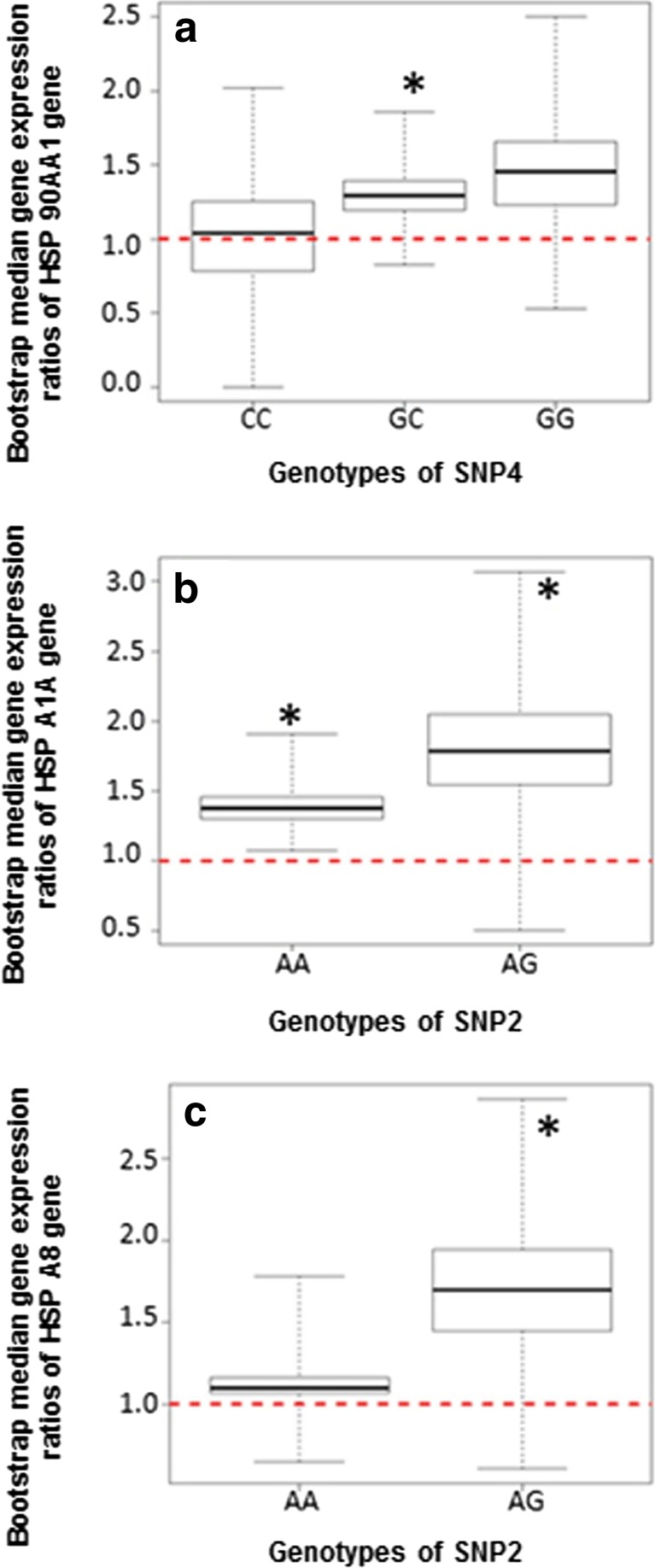
Fig. 2.
Genotype-wise bootstrap median gene expression ratios. a HSP90AA1-SNP4. b HSPA1A (HSP70 SETII genotypes)-SNP2. c HSPA8 gene (HSP70 SETII genotypes)-SNP2. Asterisks indicate P < 0.05
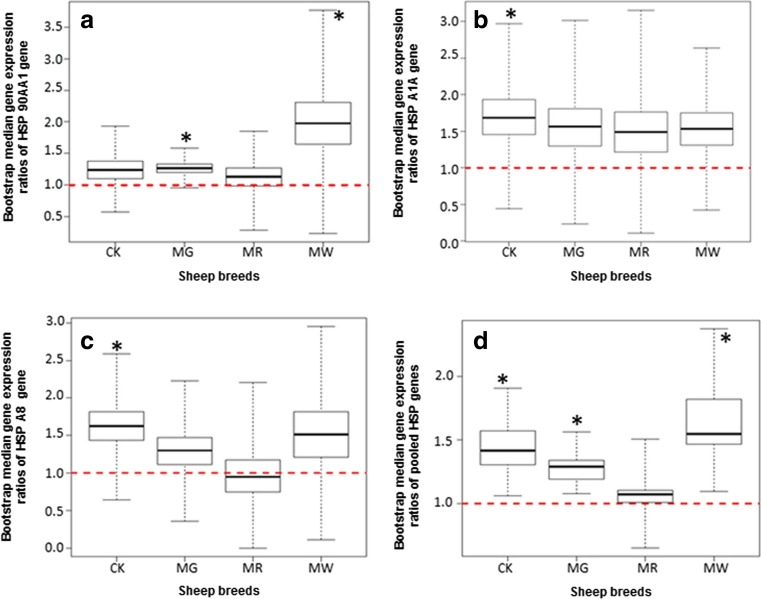
Fig. 3.
Breed-wise bootstrap median gene expression ratios of HSP genes in different sheep breeds. a HSP90AA1. b HSPA1A. c HSPA8. d Pooled of all the three HSP genes together (HSP90AA1, HSPA1A, and HSPA8). CK Chokla breed, MG Magra breed, MR Madras Red breed, MW Marwari breed. Asterisks indicate P < 0.05
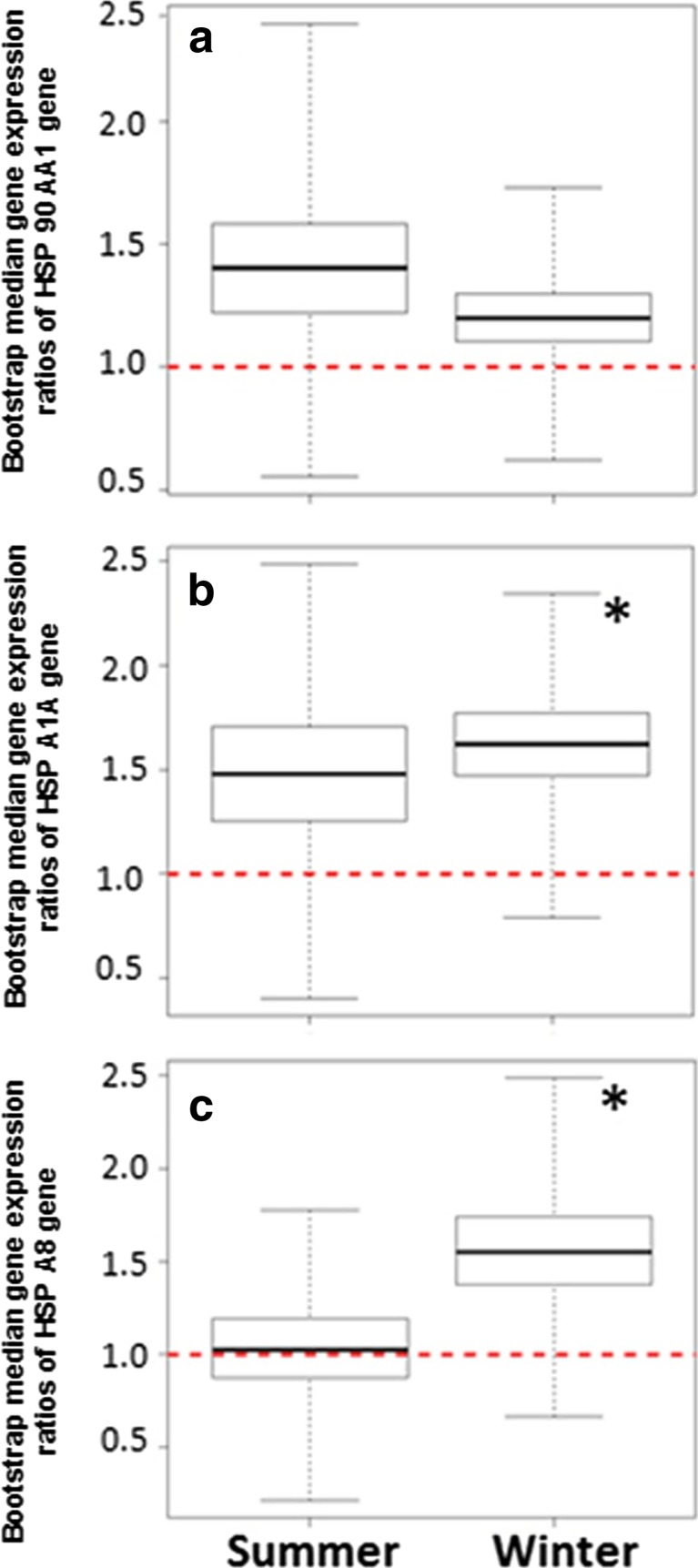
Fig. 4.
Season-wise bootstrap median gene expression ratios of different HSP genes. a HSP90AA1. b HSPA1A. c. HSPA8 Asterisks indicate P < 0.05
Genotype-wise expression profile
Genotype-wise expression profiles of HSP90AA1, HSPA1A, and HSPA8 are given in Fig. 2. For HSP90AA1 expression profile, the median boot strap ratio was higher in summer than in winter and animals of CC genotypes had lower expression than GC and GG genotypes (GG>GC>CC) (Fig. 2a; Table S4). This indicates that the animals of superior genotypes (CC or C allele) had lower expression for this gene. For expression profile of HSPA1A, the median boot strap ratio was higher in winter than in summer and animals of AG genotypes had higher expression than AA genotypes (Fig. 2b; Table S4). So, the animals of superior genotype with better adaptability (AA) had lower expression of HSPA1A than the animals of AG genotypes. Similarly, for HSPA8 expression profile observed, median boot strap ratio was higher in winter compared to summer season and animals of AA genotypes had lower expression than of AG genotypes (Fig. 2c; Table S4). This again shows that the animals of superior genotype with better adaptability (AA) had lower expression of HSPA1A than the animals of AG genotypes.
Breed-wise expression profile
The expression profile of HSP genes showed breed-specific expression pattern for three HSP genes viz. HSP90AA1, HSPA1A, and HSPA8 (Fig. 3). For HSP90AA1, the order of the expression profile pattern was MR < CK < MG < MW (Fig. 3a). For HSPA1A, MR < MW < MG < CK expression profile pattern was obtained (Fig. 3b) whereas for HSPA8, MR < MG < MW < CK pattern was obtained (Fig. 3c). Overall, the pooled data analysis revealed the MR < MG < CK < MW expression pattern (Fig. 3d). Results shows highest expression of HSP genes in MW and least in MR. Among the sheep breeds of arid and semi-arid regions of Rajasthan, MG breed had lowest expression ratio, hence highest adaptability, which is in consonance with our previous finding (Singh et al. 2016) in which the adaptability % was found to be highest in MG sheep (83.29%).
Season-wise expression profile
Comparatively higher expression (P > 0.05) of HSP90AA1 gene was observed in summer than in winter season (Fig. 4a). On the contrary, HSP90A1A and HSPA8 genes were expressed significantly (P < 0.05) more in winter than in summer season (Fig. 4b, c).
Discussion
Among the 22 different physio-biochemical parameters studied in the present study, ten parameters viz. respiratory rate (a.m.), pulse rate (a.m.), rectal temperature (a.m.), triglyceride, total protein, albumin, hemoglobin %, N/L ratio, monocytes (%), and total leukocytes cells (TLC) were significantly affected (P < 0.05) by the SNPs identified (Table S4). Therefore, 45.45% traits were significantly affected by the five SNPs. Respiratory rate (a.m.), pulse rate (a.m.), and rectal temperature (a.m.) are the three most important physiological parameters which increased significantly due to the SNP effect of SNP1, SNP3, and SNP4, respectively, during heat stress. Results indicated the superiority of T, C, and C alleles of SNP1, SNP3, and SNPP4, respectively, in terms of thermo-tolerance. Lower values in these parameters indicate an improved thermo-tolerance. Charoensook et al. (2012) observed the significant effect on RR (a.m.) due to the SNPs present in the intron9 (g.4338T>C) and exon 11 (g.5007T>C) of HSP90AB1 gene in Thai indigenous cattle. Furthermore, exon 11 (g.5007T>C) had significant effect on RT (a.m.). This is in consonance with our present findings in which both respiratory rate (a.m.) and rectal temperature (a.m.) had significant effect on the SNP4 and SNP1, respectively.
Neutrophils and lymphocytes make up the majority (about 80%) of TLC in mammals, birds, amphibians, and reptiles. Neutrophils are primary phagocytic leukocytes which proliferate in circulation in response to infection, inflammation, and stress (Jain 1993) whereas lymphocytes are involved in the immunoglobulin production and modulation (Campbell 1996). Higher N/L ratio is an indication of heat stress. This ratio increases in response to heat stress and circulatory glucocorticoids (Davis et al. 2008). Al-Murrani et al. (1997) used heterophil/lymphocyte ratio in poultry as a selection criterion for the grouping of the resistant and susceptible birds and observed a significantly higher effect on egg size, production, and mortality among birds of resistant group. Similarly, Alsamarrae (2015) also observed significantly higher N/L ratio (winter) in local Awasi sheep breed indicating winter stress. In our study, we observed lower N/L ratios in MR sheep breed (0.49) and CC genotype (0.50) indicating the superiority of MR breed and C allele for heat stress resistance.
The present study revealed a superiority of T allele (SNP1), T allele (SNP1), and A allele (SNP2) for triglyceride, total protein, and albumin, respectively. In our earlier study, we also observed the significant effect of heat stress on triglyceride, total protein, and albumin in different sheep breeds of arid zone (Singh et al. 2016). Likewise, “A” allele of SNP5 was found to be superior over G allele when TLC of different genotypes was considered.
When animals are exposed to thermal stress, HSP proteins are usually rapidly synthesized. However, in our study, we observed an increased expression of HSP90AA1 and lower expression of HSPA1A and HSPA8 during summer season (Fig. 4). This might be due to the fact that a long-term chronic exposure results in a decline in the expression level of protein due to cell lesion development (Lei et al. 2009). Moreover, HSPA8 is a constitutively expressed chaperone protein. It typically makes up 1 to 3 % of total cellular protein whereas HSP90AA1 is an inducible form of the protein. Similar results were also reported in poultry strains (Rimoldi et al. 2015; Felver-Gant et al. 2012) where reduced expression of HSP 90 and 70 genes during heat stress was observed. They inferred adaptation of such strains at elevated temperature to be the probable cause. In the present study, the well-adapted breeds of the sheep might have shown similar stress response due to prolong thermal stress in their native environment.
Expression of HSP90, HSP60, and UBQ was significantly higher (P < 0.05) in all age groups during peak summer season as compared to peak winter season in both tropical and temperate region goats (Dangi et al. 2012). However, in tropical goats, only HSP70 expression was significantly higher (P < 0.05) during summer season as compared to winter season. Similarly, seasonal variation has been witnessed in expression profile of HSP70 gene family in different breeds of goats adapted to diverse agro climatic conditions (Banerjee et al. 2014). During summer, they observed high-level expression of HSPA8, HSPA6, and HSPA1A genes whereas the expression levels of HSPA1L and HSPA2 were found to be lower. The expression of HSPA1A and HSPA8 was higher during winter in both heat- and cold-adapted goats but downregulated in case of other HSPs. Therefore, both heat and cold stresses induced the overexpression of HSP70 genes. An interesting finding that emerged from the study is the higher expression of HSP70 genes in cold-adapted goats during summer and in heat-adapted goats during winter. The results indicate that the expression pattern of HSP70 genes is species and breed specific, most likely due to variations in thermal tolerance and adaptation to different climatic conditions (Banerjee et al. 2014). This is in agreement with our finding in which breed-specific expression pattern of HSP90 and 70 genes was obtained (Fig. 3).
Two SNPs in HSP90AA1 G/C-660 and A/G-444 were associated with gene overexpression resulting from heat stress in Manchego Spanish sheep (Salces-Ortiz et al. 2013). CC genotype at −660 position was found to be superior to GG/GC in terms of gene expression rate under heat stress. However, in the present study, the superior genotypes were having lower expression than the less adapted genotypes. This might be due to the difference in the experimental design. We have considered a genotype to be superior over the other genotypes, for a particular locus, on the basis of minimum difference in mean values for physio-biochemical traits between a particular genotype and mean of all genotypes in normal condition. However, no such comparison was made in the study of Salces-Ortiz and coworkers to designate the genotypes as superior or inferior. Our results are in consonance with the expression profiles of different HSP genes generated in different cattle and buffalo breeds in which lower HSP expression was observed in more adaptable native animals during periods of stress (Kumar et al. 2015; Kishore et al. 2016). Overall, the pooled data analysis revealed the MR < MG < CK < MW expression pattern (Fig. 3d). Among breeds of arid region, MG had the lowest expression, hence highest adaptability, which is in agreement with our earlier findings (Singh et al. 2016) in which the adaptability % was found to be highest in MG sheep. Comparative smaller size (MR), white and lustrous coat (MG), dark skin color (MR), and hairy coat (MR) make the animals of MG and MR more adaptable to heat stress as compared to other breeds under investigation. Moreover, being the hairy sheep, MR had higher adaptability than the wooly sheep (CK, MG, and MW). This is in accordance with the findings of Titto et al. (2016) in which higher adaptability of hairy sheep in response to heat stress as compared to wooly sheep was obtained. In their study, hairy breeds (Santa Ines and Morada Nova) had lower respiratory rate (P < 0.05) than wooly breeds (Texel, Suffolk, and Ile de France). Further, MR sheep had significantly lower mean values (P < 0.05) for respiratory rate (a.m.) and pulse rate (both a.m. and p.m.) than the wooly sheep, which further supports this hypothesis. Predominant allele was found to be superior in SNP1–4 indicating the selection acting in directional manner (positive selection). Higher frequency of lesser adapted allele obtained in SNP5 indicates frequency-dependent selection (balancing selection). Overall, TACCA haplotype combination of SNP1-SNP2-SNP3-SNP4-SNP5 might be of some selection advantage for the identification of animals more adaptable to heat stress.
Conclusion
In the present investigation, it was observed that the better adapted genotypes had lower expression of HSP90AA1, HSPA1A, and HSPA8. Overall, the pooled data analysis revealed the MR < MG < CK < MW expression pattern. Among breeds of arid region, MG had lowest expression, hence highest adaptability. Comparative smaller size (MG and MR), white and lustrous coat (MG), dark skin color (MR), and hairy coat (MR) make the animals of MG and MR more adaptable to heat stress. Predominant allele was found to be superior in most of the SNPs (SNP1–4), indicating the selection acting in directional manner (positive selection). TACCA haplotype combination of SNP1-SNP2-SNP3-SNP4-SNP5 might be of some selection advantage for the identification of animals more adaptable to heat stress.
Electronic supplementary material
(DOCX 14 kb)
(DOCX 16 kb)
(DOCX 16 kb)
(DOCX 42 kb)
Acknowledgements
The authors are thankful to the director, ICAR-NBAGR, and Incharge ARC, CSWRI, Bikaner, for providing the necessary facilities for the research work.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-017-0770-4) contains supplementary material, which is available to authorized users.
An erratum to this article is available at https://doi.org/10.1007/s12192-017-0803-z.
References
- Abdel-Hafez MAM (2002) Studies on the reproductive performance in sheep. Ph.D. thesis, Faculty of Agriculture, Zagazig University, Zagazig
- Alam MM, Hashem MA, Rahman MM, Hossain MM, Haque MR, Sobhan Z, Islam MS. Effect of heat stress on behaviour, physiological and blood parameters of goat. Progress Agric. 2011;22(1&2):37–45. [Google Scholar]
- Al-Murrani WK, Kassab AK, Alsam HZ, Al-Athuri AK. The use of H/L ratio as a selection criterion for heat resistant in fowls. Br Poult Sci. 1997;38(2):159–163. doi: 10.1080/00071669708417962. [DOI] [PubMed] [Google Scholar]
- Alsamarrae SH. Genetic variation in stress resistance according to some blood values in Iraqi sheep. Inter Journal of Agri Innovations and research. 2015;3(5):2319–1473. [Google Scholar]
- Banerjee D, Upadhyay RC, Chaudhary UB, Kumar R, Singh SV, Ashutosh RGJ, Polley S, Mukherjee A, Das TK, De S. Seasonal variation in expression pattern of genes under HSP70 family in heat and cold-adapted goats (Capra hircus) Cell Stress and Chaperones. 2014;19:401–408. doi: 10.1007/s12192-013-0469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boston RS, Viitanen PV, Vierling E. Molecular chaperones and protein folding in plants. Plant Mol Biol. 1996;32(1–2):191–222. doi: 10.1007/BF00039383. [DOI] [PubMed] [Google Scholar]
- Campbell TW (1996) Clinical pathology Mader DR (ed): Reptile medicine and surgery. WB Saunders, Philadelphia, 1996, pp 248-257.
- Charoensook R, Gatphayak K, Sharifi AR, Chaisongkram C, Brenig B, Knorr C. Polymorphisms in the bovine HSP90AB1 gene are associated with heat tolerance in Thai indigenous cattle. Trop Animal Health Prod. 2012;44:921–928. doi: 10.1007/s11250-011-9989-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleries R, Galvez J, Espino M, Ribes J, Nunes V, Herediac MLD. BootstRatio: a web-based statistical analysis of fold-change in qPCR and RT-qPCR data using resampling methods. Comput Biol Med. 2012;42:438–445. doi: 10.1016/j.compbiomed.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Dangi SS, Gupta M, Maurya D, et al. Expression profile of HSP genes during different seasons in goats (Capra hircus) Trop Anim Health Prod. 2012;44(8):1905–1912. doi: 10.1007/s11250-012-0155-8. [DOI] [PubMed] [Google Scholar]
- Davis AK, Maney DL, Maerz JC. The use of leukocyte profile to measure stress in vertebrates: a review for ecologists. Funct Ecol. 2008;22:760–772. doi: 10.1111/j.1365-2435.2008.01467.x. [DOI] [Google Scholar]
- Felver-Gant JN, Mack LA, Dennis RL, Eicher SD, Cheng HW. Genetic variations alter physiological responses following heat stress in 2 strains of laying hens. Poult Sci. 2012;91:1542–1551. doi: 10.3382/ps.2011-01988. [DOI] [PubMed] [Google Scholar]
- Habeeb AA, Marai IFM, Kamal TH, Qwen JB. Proceedings of the international conference on animal, poultry and rabbit production and health. Cairo: Dokki; 1997. Genetic improvement of livestock for heat adaptation in hot climate; pp. 11–16. [Google Scholar]
- Helal A, Hashem ALS, Abdel-Fattah MS, El-Shaer Effects of heat stress on coat characteristics and physiological responses of Balady and Damascus goat in Sinai Egypt. American Eurasian Journal of Agriculture & Environmental Science. 2010;7(1):60–69. [Google Scholar]
- Jain NC. Essentials of veterinary hematology. Philadelphia: Blackwell Publishing; 1993. [Google Scholar]
- Kishore A, Sodhi M, Sharma A, Shandilya UK, Mohanty A, Verma P, Mann S, Mukesh M. Transcriptional stability of heat shock protein genes and cell proliferation rate provides an evidence of superior cellular tolerance of Sahiwal (Bos indicus) cow PBMCs to summer stress. Research & Reviews: Journal of Veterinary Sciences. 2016;2(1):34–40. [Google Scholar]
- Kumar A, Ashraf S, Goud TS, Grewal A, Singh SV, Yadav BR, Upadhyay RC. Expression profiling of major heat shock protein genes during different seasons in cattle (Bos indicus) and buffalo (Bubalus bubalis) under tropical climatic condition. J Therm Biol. 2015;51:55–64. doi: 10.1016/j.jtherbio.2015.03.006. [DOI] [PubMed] [Google Scholar]
- Lei L, Yu J, Bao E. Expression of heat shock protein 90 (Hsp90) and transcription of its corresponding mRNA in broilers exposed to high temperature. Br Poult Sci. 2009;50:504–511. doi: 10.1080/00071660903110851. [DOI] [PubMed] [Google Scholar]
- LPHSI . Livestock and poultry heat stress indices agriculture engineering technology guide. Clemson: Clemson Uni; 1990. [Google Scholar]
- Marai IFM, Habeeb AA. Adaptation of Bos Taurus cattle under hot climate conditions. Ann Arid Zone. 1998;37:253–281. [Google Scholar]
- Marai IFM, Bahgat LB, Shalaby TH, Abdel-Hafez MA. Fattening performance, some behavioural traits and physiological reactions of male lambs fed concentrates mixture alone with or without natural clay, under hot summer of Egypt. Ann Arid Zone. 2000;39(4):449–460. [Google Scholar]
- Marcos-Carcavilla A, Moreno C, Serrano M, Laurent P, Cribiu EP, Andreoletti O, Ruesche J, Jean-Louis W, Calvo JH, Moazami-Goudarzi K. Polymorphisms in the HSP90AA1 5′ flanking region are associated with scrapie incubation period in sheep. Cell Stress and Chaperones. 2010;15:343–349. doi: 10.1007/s12192-009-0149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos-Carcavilla A, Mutikainen M, Gonzalez C, Calvo JH, Kantanen J, Sanz A, Nurbiy S, Marzanov MD, Perez-Guzman SM. A SNP in the HSP90AA1 gene 5′ flanking region is associated with the adaptation to different thermal conditions in the ovine species. Cell Stress and Chaperones. 2010;15:67–81. doi: 10.1007/s12192-009-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocak S, Guney O. Physiological responses and some blood parameters of bucks under Mediterranean climate conditions. Anadolu J Agric Sci. 2010;25(2):113–119. [Google Scholar]
- Park H, Ahn I-Y, Lee HE. Expression of heat shock protein 70 in the thermally stressed Antarctic clam Laternula elliptica. Cell Stress and Chaperones. 2007;12(3):275–282. doi: 10.1379/CSC-271.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar HN, Agrawal RK, Ramneek BGS. Expression, purification and characterization of recombinant heat shock protein 70 (HSP70) from sheep and goat species. Int J Curr Microbiol App Sci. 2013;2(11):440–452. [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phulia SK, Upadhyay RC, Jindal SK, Misra RP. Alteration in surface body temperature and physiological responses in Sirohi goats during day time in summer season. Indian Journal of Animal Science. 2010;80(4):340–342. [Google Scholar]
- Rimoldi S, Lasagna E, Sarti FM, Marelli SP, Cozzi MC, Bernardini G, Terova G. Expression profile of six stress-related genes and productive performances of fast and slow growing broiler strains reared under heat stress conditions. Meta Gene. 2015;6:17–25. doi: 10.1016/j.mgene.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salces-Oritz J, Gonzalez C, Martinez M, Mayoral T, Calvo JH, Serrano MM. Looking for adaptive footprints in the HSP90AA1 ovine gene. BMC Evol Biol. 2015;15:7. doi: 10.1186/s12862-015-0280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salces-Ortiz J, González C, Moreno-Sánchez N, Calvo JH, Pérez-Guzmán MD, Serrano MM. Ovine HSP90AA1 expression rate is affected by several SNPs at the promoter under both basal and heat stress conditions. PLoS One. 2013;8(6):e66641. doi: 10.1371/journal.pone.0066641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schalm OW, Jain NC, Carrol E. Veterinary haematology. 3. Philadelphia: Lea and Febiger; 1975. pp. 160–121. [Google Scholar]
- Schlesinger MJ. Heat shock proteins. J Biol Chem. 1990;265(21):12111–12114. [PubMed] [Google Scholar]
- Sharma AK, Kataria N. Effects of extreme hot climate on liver and serum enzymes in Marwari goat. Indian Journal of Animal Science. 2011;81(3):293–295. [Google Scholar]
- Shelton M. Reproductive performance of sheep exposed to hot environments. In: Malik RC, Razzaque MA, Al-Nasser AY, editors. Sheep production in hot and arid zones. Kuwait: Published by the Kuwait Institute for Scientific Research; 2000. pp. 155–162. [Google Scholar]
- Singh KM, Singh S, Ganguly I, Ganguly A, Raja KN, Chopra A, Narula HK. Evaluation of Indian sheep breeds of arid zone under heat stress condition. Small Rumin Res. 2016;141:113–117. doi: 10.1016/j.smallrumres.2016.07.008. [DOI] [Google Scholar]
- Sivakumar AVN, Singh G, Varshney VP. Antioxidants supplementation on acid base balance during heat stress in goats. Asian-Australian Journal of Animal Science. 2010;23(11):1462–1468. doi: 10.5713/ajas.2010.90471. [DOI] [Google Scholar]
- Titto CG, Veríssimo CJ, Pereira AMF, Geraldo ADM, Katiki LM, Evaldo Antonio Lencioni Titto (2016) Thermoregulatory response in hair sheep and shorn wool sheep. Small Ruminant Research 144:341–345
- Vierling E (1991) The roles of heat shock proteins in plants. Annual Review of Plant Physiology and Plant Molecular Biology 42:579–620
- Waters ER, Lee GJ, Vierling (1996) Evolution, structure and function of the small heat shock proteinsin plants. Journal of Experimental Botany 47:325–338
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 14 kb)
(DOCX 16 kb)
(DOCX 16 kb)
(DOCX 42 kb)