Abstract
The successful translation of cytotoxic T lymphocyte antigen-4 (CTLA-4) blockade has revolutionized the concept of cancer immunotherapy. Although monoclonal antibody therapeutics remain the mainstream in clinical practice, aptamers are synthetic oligonucleotides that encompass antibody-mimicking functions. Here, we report a novel high-affinity CTLA-4-antagonizing DNA aptamer (dissociation constant, 11.84 nM), aptCTLA-4, which was identified by cell-based SELEX and high-throughput sequencing. aptCTLA-4 is relatively stable in serum, promotes lymphocyte proliferation, and inhibits tumor growth in cell and animal models. Our study demonstrates the developmental pipeline of a functional CTLA-4-targeting aptamer and suggests a translational potential for aptCTLA-4.
Keywords: CTLA-4, aptamer, cancer, immunotherapy
Graphical Abstract
Introduction
The concept of immune checkpoint blockade has revolutionized anti-cancer strategies in nearly all cancer fields.1 Monoclonal antibodies targeting the cytotoxic T lymphocyte antigen-4 (CTLA-4) and programed cell death protein 1 (PD)-1/programmed death-ligand 1 (PD-L1) axes are now part of routine practice.2, 3, 4, 5 Although widely adopted clinically, monoclonal antibody (mAb)-based therapy still consists of several inevitable drawbacks that need to be made aware of. First of all, the manufacturing process of mAb requires a cell-based production system. This may influence production capacity and cause variation between batches.6, 7 Second, mAbs are protein molecules and are prone to induce T cell-dependent neutralizing antibodies after repetitive administrations. Although the humanization process has significantly reduced such a possibility, the presence of a neutralizing antibody that reduces the therapeutic efficacy has been documented even in whole humanized mAbs.6, 7, 8 Third, although a relatively long half-life of mAbs (days to weeks) can be an advantage in terms of drug dosing, its large molecular size (∼150 kDa) may limit its tissue-penetration efficiency.9 Most important of all, because severe immune-related adverse effects are not infrequently observed in patients receiving mAb-based immune checkpoint blockades, a long serum half-life of mAbs can be a disadvantage in this regard.4
Aptamers are small DNA or RNA molecules that form complex three-dimensional (3D) structures.10 Since its first emergence in 1990, the aptamer has been considered as an antibody surrogate due to its specific binding affinity and low immunogenicity. Aptamers are chemically synthesized. This potentiates aptamers to be produced in large scale with controllable batch-to-batch variations and be customized and modified for specific purposes.11, 12, 13, 14, 15, 16, 17 Over the past years, aptamers have been shown to serve as successful guiding molecules for targeted delivery, with good tissue penetration efficiency.13, 18, 19, 20 Recent aptamer-related research has further extended the field into functional aptamer therapeutics, especially in the field of immuno-oncology, an area full of new hope and uncertainty.10, 21 Because the immune checkpoint landscape encompasses multiple positive and negative regulators, aptamers may have several advantages over mAbs in cancer immunotherapy per se. For example, it is possible to engineer aptamers with dual desirable functions, either being a dual antagonist or an agonist-antagonist integrative.10, 22 In addition, the small size of the aptamer (6–30 kDa) facilitates its rapid renal clearance, with a half-life ranging from hours to 2 days.23, 24 This allows for better managing in case of side effects, which are now recognized an an important issue in cancer immunotherapy.21
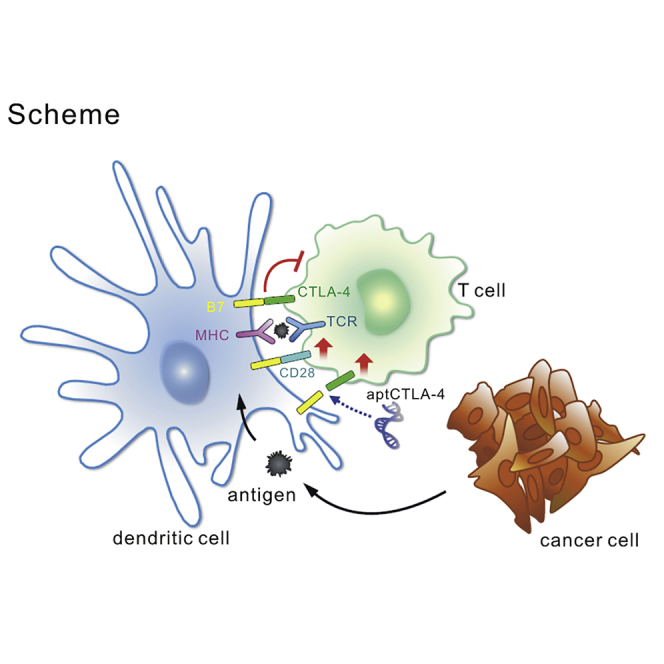
CTLA-4 is expressed on T cells. It is a homolog of CD28, which binds to the surface antigens B7-1 (CD80) and B7-2 (CD86).25 CTLA-4 expression is initiated upon T cell activation, which attenuates CD28 co-stimulation and inhibits signaling by competing for B7 binding.26 Blocking of CTLA-4 and B7 conjugation reshapes the host immune response and exerts a sustained anti-tumor effect in some cancer subpopulations.27, 28, 29 CTLA-4 and PD-1/PD-L1 axes blockade constitutes the backbone of the current cancer immunotherapy.5 As potential advantages of the aptamer are recognized, it is worth developing a CTLA-4-antagonizing aptamer. Although a tetrameric CTLA-4 RNA aptamer had been reported, it does not function as an effective antagonist in a monomeric form.30 It indeed, however, can serve as a carrier for targeting delivery, such as the case of the aptamer-siSTAT3 chimera against T-regulatory cells.31 Based on this unmet need, we developed a novel CTLA-4-antagonizing DNA aptamer, aptCTLA-4, by the integration of two high-throughput platforms: SELEX (systematic evolution of ligands by exponential enrichment) and next-generation sequencing (NGS). We showed that aptCTLA-4 is biologically functional and binds to CTLA-4 with high affinity and promotes T cell activity in the tumor microenvironment.
Results
Cell-Based SELEX and NGS for aptCTLA-4 Identification
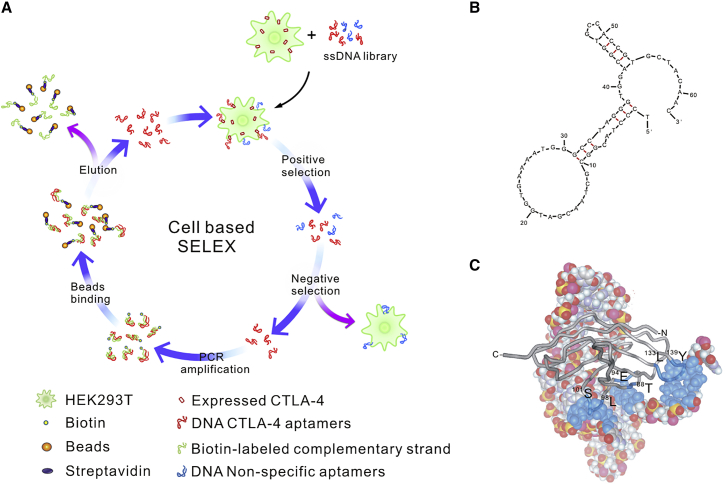
The CTLA-4-targeting aptamers were selected from a single-stranded DNA (ssDNA) library composed of 1015 molecules using 12 round cell-based SELEX (Figure 1A). The ssDNA pool enriched for CTLA-4-targeting sequences was subjected to NGS. The 1,623 NGS-derived sequence clusters were divided into five categories. There were 28 clusters in the ≥7,000-read category, with sequence-read numbers exceeding 50% of the total reads (Table S1A). Quadruplex-forming G-rich sequences (QGRS) prediction of these 28 clusters suggested a probable G-quadruplex structure within aptCTLA-4 (http://bioinformatics.ramapo.edu/QGRS/analyze.php) (Table S1B). M-fold structure-prediction software predicted that the aptCTLA-4 sequence formed a complex hairpin-bulge folding structure (Figure 1B). 3D-structure predictions and docking simulations further suggested that nucleotides 39-40, 54-57, and 60-61 of aptCTLA-4 bind to amino acids 98LDDS101, 88TYMMGNE94, and 133LMYPPPY139 of CTLA-4 protein (Figure 1C). Of note, the MYPPPYY sequence of CTLA-4 was shown to be the major binding region that interacts with B7 proteins.32 Peptide competition assays further suggested that aptCTLA-4 might have the same binding sites with B7-1 and B7-2, the two known ligands for CTLA-4 protein (Figure S1). On the other hand, the analyses with circular dichroism (CD) spectroscopy did not support the existence of a G-quadruplex within aptCTLA-4. The positive peak of aptCTLA-4 resided at 273 nm instead of 263 nm for a parallel or 295 nm for an anti-parallel G-quadruplex (Figure S2).33, 34, 35 Nevertheless, we continued our further analyses of aptCTLA-4 based on the results of structural prediction, docking simulation, and peptide competition assays.
Figure 1.
SELEX Workflow and aptCLTA4 Structural Prediction
(A) Schematic illustration of cell-based SELEX. (B) M-fold structure prediction of aptCTLA-4. (C) CTLA-4 docking. aptCTLA-4 was presented in a space-filling type; the blue color indicates aptCTLA-4 nucleotides that may interact with CTLA-4 protein, which is shown in a gray ribbon band.
aptCTLA-4 Binds to Human CTLA-4 Protein with Good Binding Affinity
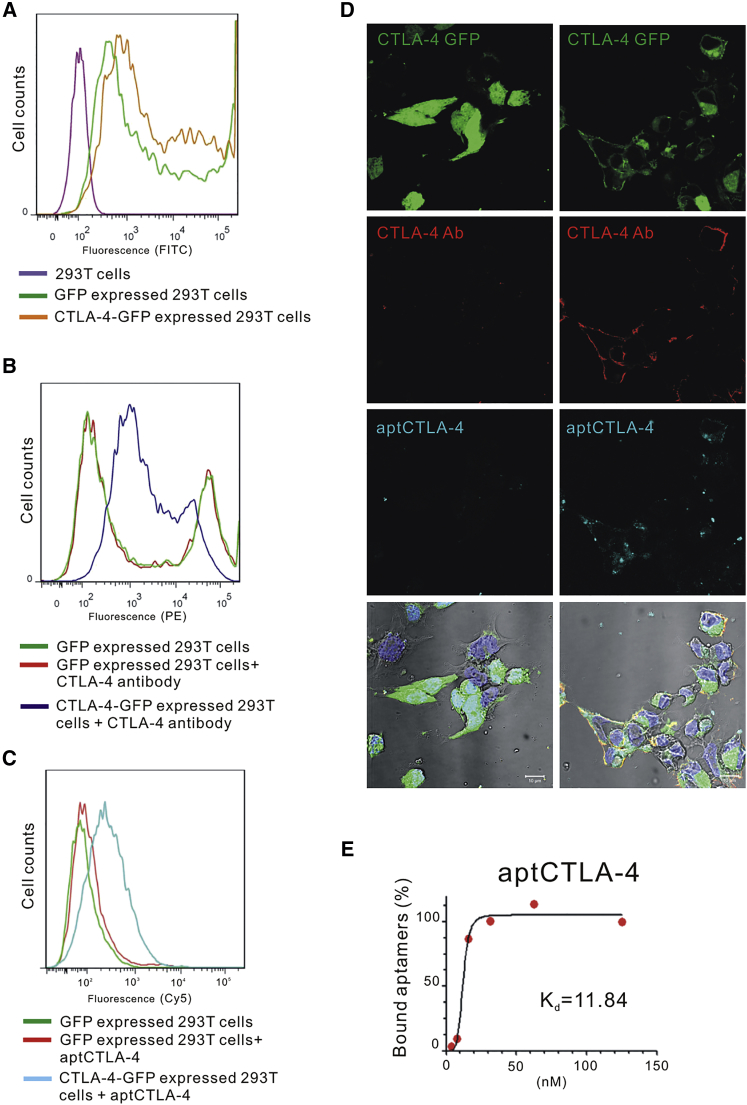
We first confirmed the binding between aptCTLA-4 and human CTLA-4. To this end, we incubated 20 nM Alexa Fluor 647-labeled aptCTLA-4 with human CTLA-4-GFP-overexpressing or GFP-overexpressing HEK293T cells, and analyzed fluorescence intensity by flow cytometry. The shift of fluorescence intensity with the treatment of aptCTLA-4 suggested that aptCTLA-4 was able to recognize CTLA-4-expressed HEK293T cells (Figures 2A–2C). This was further supported by the evidence that CTLA-4 knockdown could reverse the shift of fluorescence intensity induced by aptCTLA-4 or anti-CTLA-4 antibody treatment (Figure S3). Confocal microscopy demonstrated the accumulation of overexpressed CTLA-4-GFP protein on the HEK293T cell membranous region, where it was co-localized with aptCTLA-4 (indigo) and phycoerythrin (PE)-labeled anti-CTLA-4 antibody (red) (Figure 2D). The binding assay showed that aptCTLA-4 possessed good binding affinity toward CTLA-4 protein, with the dissociation constant (Kd) being 11.84 nM (Figure 2E). Taken together, our data implicated the potential functionality of aptCTLA-4.
Figure 2.
Selected DNA Aptamers Displayed Binding Specificity for Human CTLA-4 Protein
(A) Evaluation of GFP and CTLA4-GFP expression in HEK293T cells. (B and C) The PE-labeled CTLA-4 antibody (10 μg/mL) (B) and the Alexa-Fluor-647-labeled aptCTLA-4 (20 nM each) (C) were incubated with CTLA-4-GFP- or GFP-overexpressing HEK293T cells (negative controls). Fluorescence intensities were analyzed by flow cytometry. (D) The confocal image showed that the PE-labeled CTLA-4 antibody and Alexa-Fluor-647-labeled aptCTLA-4 (20 nM each) could identify CTLA-4-expressed HEK293T cells. (E) Dissociation constant of aptCTLA-4 was 11.84 nM. The starting point of aptCTLA-4 serial dilution was 125 nM.
aptCTLA-4 Binds to Mouse CTLA-4 Protein and Promotes In Vitro T Cell Activity
Because tumor microenvironment studies are often carried out in murine syngeneic tumor models, we examined the sequence homology between human and mouse CTLA-4 proteins. The analysis revealed that human and mouse CTLA-4 sequences exhibit 76% homology at the amino acid level (Figure S4A). We subsequently incubated fluorescein isothiocyanate (FITC)-labeled aptCTLA-4 with mouse T lymphocytes, which were co-stained with PE-labeled anti-mouse CTLA-4 antibody. The confocal microscopy results showed that aptCTLA-4 could recognize mouse CTLA-4 protein (Figure S4B).
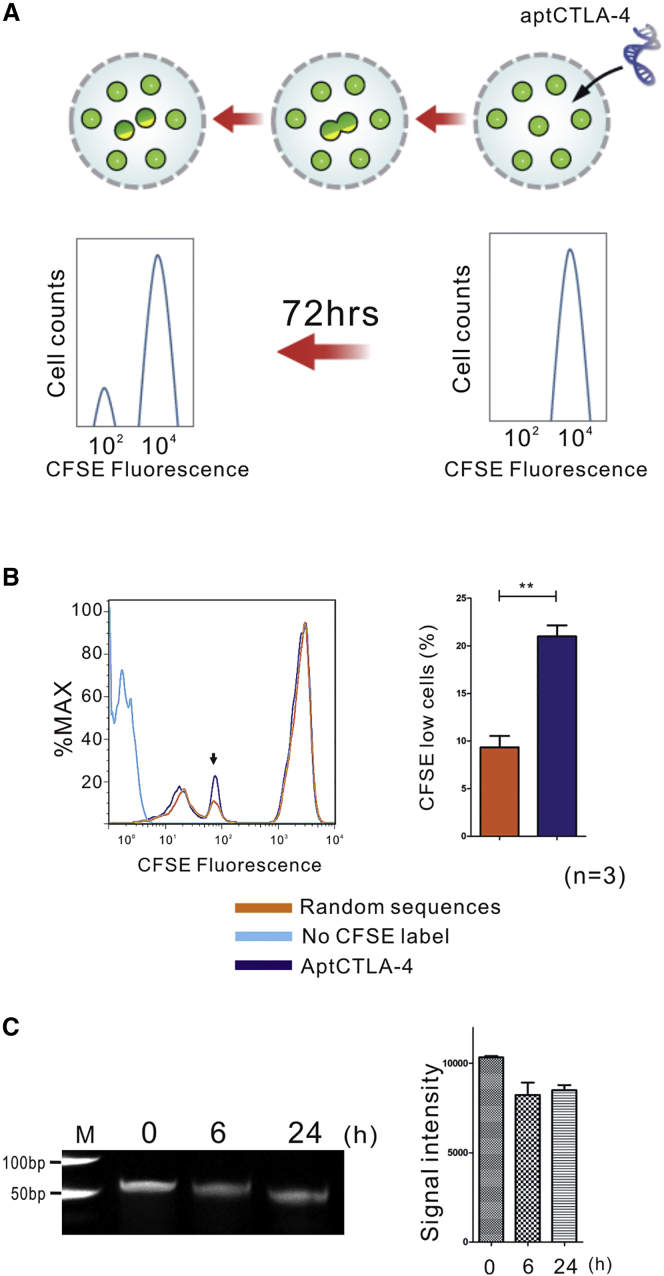
We then examined the effect of aptCTLA-4 on immune cells. The carboxyfluorescein succinimidyl ester (CFSE)-labeled lymphocytes derived from the BALB/c mouse strain were co-cultured with irradiated lymphocytes derived from the C57BL/6 mouse strain, which served as T cell activators.36 These cells were incubated with aptCTLA-4 or random sequences for 72 hr and then analyzed by flow cytometry (Figure 3A). The experimental protocol and dosage of aptCTLA-4 at 200 nM for cell treatment was based on the study from Santulli-Marotto and colleagues.30 The results showed that aptCTLA-4 treatment increased lymphocyte proliferation rates up to 21%, whereas the change of proliferation rate in the random sequences control group was about 9% (Figure 3B). The data suggested that aptCTLA-4 binds to CTLA-4 and promotes T cell activity.
Figure 3.
aptCTLA-4 Promoted Mouse Lymphocyte Function
(A) The diagram showed a decrease of fluorescence intensity of CFSE-labeled mouse lymphocytes upon cell divisions. (B) Histogram obtained by flow cytometry for CFSE-labeled allogeneic mouse lymphocytes after 72-hr treatment with random sequences or aptCTLA-4. aptCTLA-4 treatment increased lymphocyte proliferation rates up to 21%, whereas the change of proliferation rate in the control group was about 9%. (C) The gel electrophoresis revealed that aptCTLA-4 remained nearly intact after a 24-hr serum incubation period at 37°C. These data are presented as mean ± SEM and were analyzed by Student’s t test. Asterisks denote statistical significant differences (p < 0.01).
aptCTLA-4 Is Relatively Stable in Serum and Suppresses In Vivo Tumor Growth
Although short serum half-life has been considered as a shortcoming for aptamer in terms of therapeutics, this is a manageable issue.10 To evaluate the feasibility for a subsequent in vivo animal study, we first investigated the stability of aptCTLA-4 with in vitro serum incubation. aptCTLA-4 was incubated with serum for 0, 6, and 24 hr at 37°C, respectively. The gel electrophoresis results revealed that aptCTLA-4 remained nearly intact after a 24-hr serum incubation period (Figure 3C). The data suggested that even before structural or base modification, aptCTLA-4 is relatively stable.
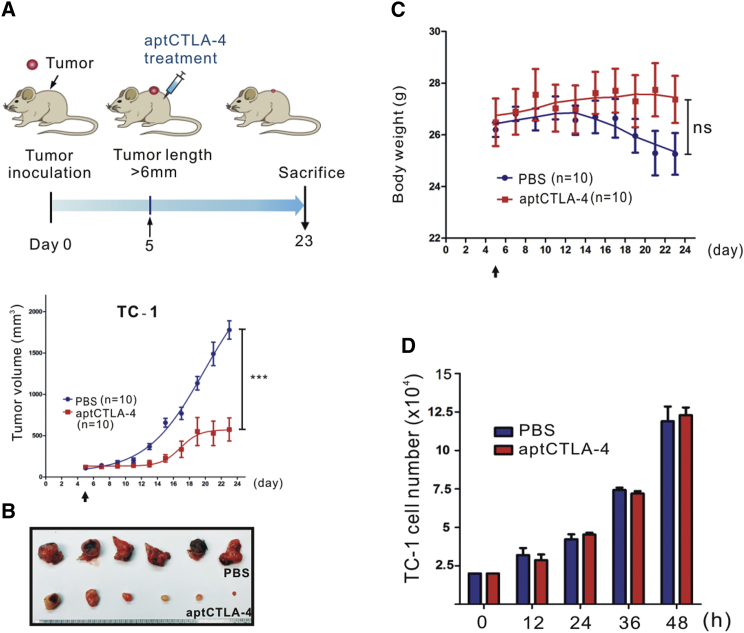
We then investigated whether aptCTLA-4 is biologically functional. We first performed in vivo studies using a mouse syngeneic tumor model created by subcutaneously inoculating C57BL/6 mice with TC-1 murine lung tumor cells. After inoculated tumors had reached a size of ∼6 mm (long axis), a single shot of aptCTLA-4 at the dosage of 0.2 mg/kg was administered intraperitoneally. The results showed that aptCTLA-4 effectively inhibited in vivo tumor growth and the body weight of mice remained stable throughout the experimental period (Figures 4A–4C). Direct toxicity of aptCTLA-4 against TC-1 cells was excluded by in vitro cell proliferation assays, which further suggested that the anti-tumor effect of aptCTLA-4 is through immune modulation mediated by its interaction with CTLA-4 (Figure 4D). Nevertheless, this low dose at one shot did not work with the aggressive cell type Lewis lung (Figure S5). We thus escalated the dosage of aptCTLA-4 to 2 mg/kg and adjusted the dosing schedule to one shot daily in 4 to 5 successive days. This protocol amendment was based on the study from Allison and colleagues when they developed anti-CTLA-4 antibody.27 With this modified dosing protocol, the aptCTLA-4 effectively suppressed in vivo tumor growth in both CT26 (BALB/c) and Lewis lung (C57BL/6) mouse syngeneic tumor models (Figure S6).
Figure 4.
aptCTLA-4 Inhibited In Vivo Tumor Growth in a Murine Syngeneic Tumor Model
C57BL/6 mice were subcutaneously inoculated with mouse TC-1 lung cancer cells (3 × 105). Tumor volumes were measured, and aptCTLA-4 (0.2 mg/kg) treatment was begun on day 5 after tumor inoculation. (A) aptCTLA-4 inhibited tumor growth (2 mg/kg, n = 10 mice/group). (B) Selected tumor pictures from sacrificed mice on day 23. (C) The body weights of mice in the aptCTLA-4-treated group remained relatively constant, whereas those of mice in the PBS-treated group decreased as tumor size increased. (D) aptCTLA-4 did not exert direct cytotoxicity against TC-1 cells (n = 3 in each group). These data are presented as mean ± SEM and were analyzed by Student’s t test. Asterisks denote statistical significant differences (p < 0.001).
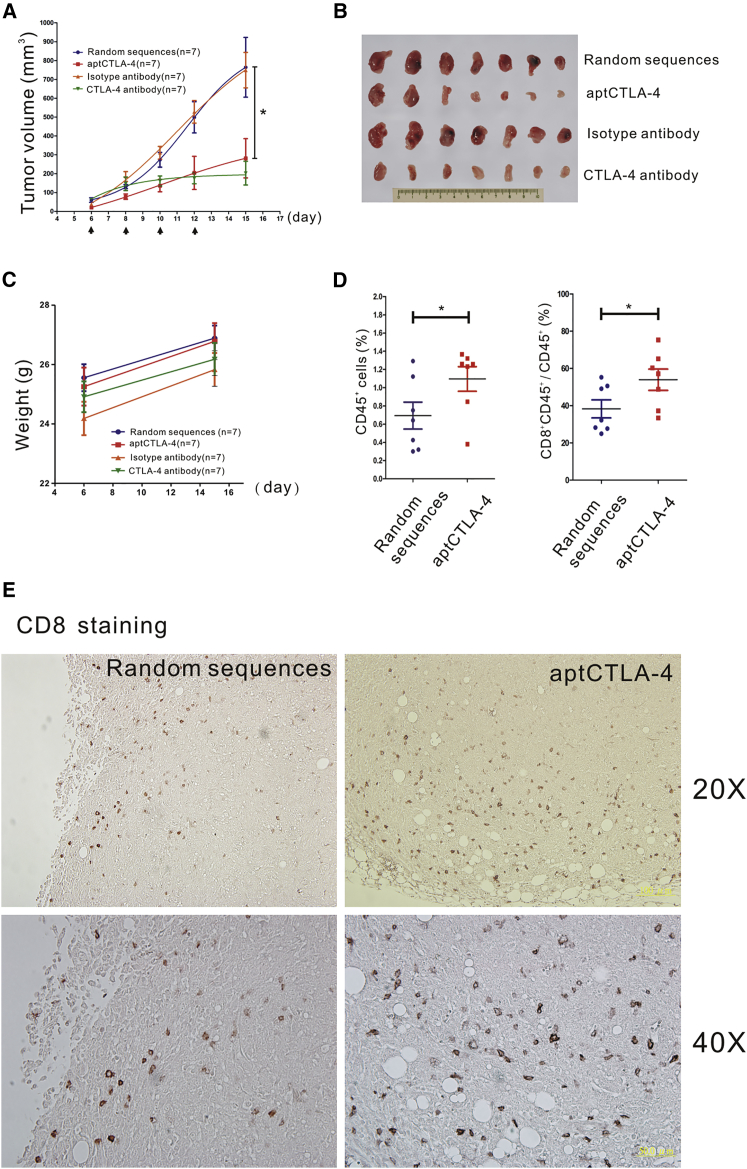
We next compared anti-tumor effects of aptCTLA-4 with an anti-mouse CTLA-4 mAb using the CT26 mouse syngeneic model. The data showed that aptCTLA-4 had identical tumor-suppressive effects to the anti-mouse CTLA-4 mAb but led to a lesser degree of body weight loss (Figures 5A–5C). Moreover, aptCTLA-4 did not induce measurable liver or renal toxicities, as represented by stable aspartate aminotransferase (GOT), alanine aminotransferase (GPT), and blood urea nitrogen (BUN) levels in groups with and without aptCTLA-4 treatment (Figure S7). On the other hand, the flow cytometry analyses showed that aptCTLA-4 treatment significantly increased the number of tumor-infiltrating lymphocytes (CD45+) and the percentage of cytotoxic T lymphocytes (CTLs) (CD45+ and CD8+) (Figure 5D). This was further verified by an immunohistochemistry study, which indicated the increase of CD8+ cells in tumors treated with aptCTLA-4 (Figure 5E). These pre-clinical results suggested a translational potential for aptCTLA-4.
Figure 5.
aptCTLA-4 Inhibits Tumor Growth and Augments Tumor-Infiltrating T Cells in a CT26 Murine Syngeneic Tumor Model
(A) Anti-mouse CTLA-4 mAb (10 mg/kg, n = 7 mice/group) or aptCTLA-4 (2 mg/kg, n = 7 mice/group) were intraperitoneally administered on day 6, 8, 10, and 12 after tumor inoculation (as indicated by arrows). (B) Selected tumor pictures from sacrificed mice on day 15. (C) The body weights of mice in the aptCTLA-4 or random sequence-treated groups remained relatively constant, whereas those of mice in the anti-mouse CTLA-4 mAb or isotype antibody-treated groups decreased as tumor size increased (n = 7 mice/group). (D) The flow cytometry studies showed that aptCTLA-4 treatment increased the number of tumor-infiltrating CD8+ cells (n = 7 mice/group). (E) The immunohistochemistry staining revealed the increase of tumor-infiltrating CD8+ cells after aptCTLA-4 treatment (n = 7 mice/group).
Discussion
aptCTLA-4 is a novel DNA aptamer that promotes T cell proliferation and inhibits tumor growth. The developmental pipeline adopted an integrated high-throughput technology, and aptCTLA-4 was functionally validated in both cell and animal models.
In the current study, we adopted a DNA aptamer selection system using cell-based SELEX. Although RNA aptamers have more diverse 3D conformations and stronger intra-strand RNA-RNA interactions that may increase binding affinity and specificity, RNAs are vulnerable to nuclease-mediated degradation, an unavoidable problem in a natural biological environment.10, 37 Sequence or structural modifications are almost always needed to improve RNA aptamer stability. Commonly used methods include substitutions of the 2′-OH functional group by 2′-fluoro, 2′-amino, or 2′-O-methyl motifs and/or changes of the phosphodiester backbone to the boranophosphate or phosphorothioate backbones or use of locked nucleic acids in the SELEX.38, 39, 40, 41, 42, 43, 44 Nevertheless, these modifications may lead to loss of function due to structural variation, toxicity due to extensive base modifications, and a substantial increase of manufacturing costs. DNA aptamers, on the other hand, are inherently more stable and the related manufacturing costs are lower.10 These make the DNA aptamer system more feasible in terms of translational research and clinical therapeutics development.
Cell-based SELEX allows identification of specific aptamers that are capable of selectively binding to the target protein in its native conformation. The selection is theoretically performed with living cells to ensure that target proteins on the cell surface remain in their active conformation throughout the selection process. Nevertheless, non-specific binding of ssDNAs to dead cells is almost an unavoidable shortcoming with this process, which may impede effective enrichment of the target-specific sequences.10, 45 Therefore, although SELEX round required for targeted sequence identification can be theoretically reduced to less than 10 when NGS is incorporated into the pipeline, the characteristic of cell-based SELEX prohibits this possibility.46
Functional DNA or RNA aptamers usually contain particular structures, such as duplexes, hairpins, triplexes, pseudoknots, or G-quadruplexes, which assemble the functional structural conformation for their biological roles.47 Therefore, we started with structural predictions with M-fold and QGRS Mapper web programs in the initial candidate identification. The G-score of the QGRS Mapper is the assigned score calculated based on the potential of G-quadruplex formation.48 Interestingly, the G-score of aptCTLA-4 was 19, which was closed to that of two aptamers with confirmed G-quadruplex structures, the TBA (thrombin-binding aptamer) with a G-score of 20 and the IBA (insulin-binding aptamer) with a G-score of 21. Nevertheless, the CD spectrum analyses did not support the existence of a G-quadruplex structure within aptCTLA-4. Although such inconsistency has also been observed in several previous studies, computer structural prediction still does help to some extent in initial candidate screening.49, 50, 51, 52, 53
The aptCTLA-4 identified in the current study possesses good binding affinity and fair serum half-life without further modification. Although the half-life of aptCTLA-4 can be theoretically shorter than an antibody, our study showed that aptCTLA-4 had identical anti-tumor effects to an anti-mouse CTLA-4 mAb. Similar tumor-suppressive effects between a functional PD-L1-targeting aptamer, the aptPD-L1, and an anti-mouse PD-L1 mAb were also reported in our previous study.54 The observed differential treatment responses to aptCTLA-4 in TC-1, Lewis Lung, and CT26 models may be partly attributable to differences in their neoantigen landscapes and antigen presentation. TC-1 cells were transformed from C57BL/6 mouse lung epithelial cells with HPV-16 E6 and E7.55 The alloantigen presented may thus augment the aptCTLA-4 anti-tumor effect observed. Clinically, about 20% of solid tumors respond to ipilimumab, and it is hypothesized that this subset of patients presents distinct neoantigens that are recognized as non-self and elicit a sustained anti-tumor immune response.5 Our results in part support these clinical observations and genomic studies.
In summary, our data support the translational development of DNA aptamers with therapeutic applications. Further investigations following the current proof-of-concept study may determine the role of aptCTLA-4 in immuno-oncology.
Materials and Methods
Ethics Statement
All animal experimental procedures were approved by the Institutional Animal Care and Use Committee of the Institute of Biomedical Science, Academia Sinica (series number ibms-CRC100-P02).
Oligonucleotides
Aptamer libraries and modified aptamers were synthesized by IDT (Integrated DNA Technologies, Coralville, IA, USA) or Purigo Biotech (Taipei, Taiwan). The primer pair R9 (forward primer: 5′-TCCCTACGGCGCTAAC-3′; reverse primer: 5′-GTTGTAGCACGGTGGC-3′), for aptamer amplification, as well as non-modified aptamers were purchased from Purigo Biotech. Human CTLA-4 siRNA (ID: 146607; Thermo Fisher). The CTLA-4 forward primer: 5’-TGGCTTGCCTTGGATTTCAGC-3’ and CTLA-4 reverse primer: 5’-ACACACAAAGCTGGCGATGC-3’ were purchased from Purigo Biotech.
Cell Culture and Construction of Stable Cell Lines
HEK293T cells stably expressing SV40 large T-antigen was cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA) and 50 μg/mL Primocin (Thermos Fisher, Waltham, MA, USA). Stable cell lines expressing CTLA-4 were created from HEK293T cells by selection in G418 antibiotic (800 μg/mL). CT26 and Lewis lung cells, established from murine colon and lung tumors, respectively, were cultured in RPMI supplemented with 10% FBS. Mouse TC-1 cells, generated by transduction of C57BL/6 (B6) primary lung epithelial cells with a retroviral vector expressing HPV16 E6/E7, were cultured in RPMI supplemented with 10% FBS. All cell lines were cultured at 37°C in a humidified 5% CO2 environment.
Cell-Based SELEX and NGS
The cell-based SELEX was carried out based on the protocol reported by Sefah et al., with modification.45 In the first round (R1) of positive selection, CTLA4-expressing HEK293T cells at a cell density of 80%–90% confluence were incubated with a ssDNA library composed of 1015 molecules in SELEX buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 5 mM MgCl2) at 4°C for 30 min with rotation. The cells were washed twice with SELEX buffer to remove unbound ssDNAs. Subsequently, the cells were suspended in 100 μL of SELEX buffer, lysed by heating at 95°C, and then cooled at 4°C. Total lysates were centrifuged at 500 × g for 5 min. The supernatant containing the eluted ssDNAs was incubated with HEK293T cells in the eppendorf tube at 4°C for 30 min with rotation for negative selection. After centrifugation, the supernatant was collected for PCR amplification using R9 forward primers and biotin-labeled R9 reverse primers (200 nM each) in PCR buffer (10 mM Tris-HCl, pH 8.9, 50 mM NaCl, 1 mM MgCl2, 10 mM betaine, and 1% DMSO) containing 200 nM each of dNTP and 2 U of Taq DNA polymerase. The PCR amplicons were captured with streptavidin-coated magnetic beads (1X SSC buffer incubation at 37°C for 3 hr with rotation) and eluted with heating at 95°C for 2 min. After the 12th round of the cell-based SELEX, the eluted ssDNAs enriched for CTLA4-targeting sequences were subjected to NGS on an Illumina MiSeq system (Illumina, San Diego, CA). The data were processed using Galaxy and FASTA aptamer software.56
Flow Cytometric Analyses
The specificity of isolated aptamers was first evaluated by flow cytometry. A total of 1 × 106 GFP and CTLA-4-GFP-overexpressing HEK293T cells were resuspended in 50 μL of PBS. The cells were then incubated with 20 nM Alexa Fluor 647-labeled selected aptamers for 30 min at 4°C. Next, cells were washed and resuspended in 0.5 mL of 4% paraformaldehyde (PFA), and analyzed by flow cytometry (4-color FACSCaliber; BD Biosciences, San Jose, CA, USA). A total of 20,000 cells were collected for each analysis.
Determination of Dissociation Constants
The 2-fold serial diluted aptCTLA-4 (starting at 125 nM) was incubated with CTLA-4-GFP-overexpressing HEK293T cells for 30 min at 4°C. After PBS washing, the cell-bounded aptCTLA-4 was eluted and quantified by RT-qPCR. The Kd values were calculated by nonlinear regression of the relationship, Y = Bmax × X/(Kd + X), using the equation: one site-specific binding, saturating binding from GraphPad Prism software (GraphPad Software, La Jolla, CA, USA). Bmax is the maximal binding at equilibrium, Kd is the ligand concentration that binds to half the receptor sites at equilibrium, and Bmax is the maximum number of binding sites.
Mouse In Vitro Lymphocyte Proliferation Assay
For the preparation of mouse spleen cells, the spleen was cut into pieces and passed through a steel mesh using a plunger. Cells were subsequently flushed into a Petri dish containing PBS, and then were treated with collagenase in red blood cell lysis buffer (10 mM Tris-HCl, 10 mM KCl, 10 mM MgCl2, 2 mM EDTA, and 2% NP-40). After centrifugation at 1,800 rpm, the supernatant was removed and the pellet was suspended and cultured in RPMI containing 10% FBS. This procedure yielded approximately 5 × 107 to 1 × 108 cells per spleen. A total of 1 × 105 lymphocytes was labeled with 2 μM CFSE (Thermos Fisher, Waltham, MA, USA) at 37°C for 20 min, and then seeded into 96-well plates. CFSE-labeled lymphocytes were mixed with unlabeled, allogeneic lymphocytes and then treated with CTLA4 aptamers (200 nM) or random sequences control (200 nM). After a 72-hr incubation, cell proliferation was examined by flow cytometry.
Tumor Immunotherapy Study
C57BL/6 mice were subcutaneously inoculated with mouse TC-1 (3 × 105) or Lewis lung (1 × 105) cancer cells. BALB/c mice were subcutaneously inoculated with CT26 (2 × 105) cancer cells. Tumor volumes were measured beginning on day 4 or 5 after tumor inoculation according to the relationship (L × D2)/2, where L is the long dimension and D is the short dimension. The aptCTLA-4, anti-mouse CTLA-4 antibody (9H10; Bio-cell), isotype antibody (Syrian Hamster immunoglobulin G [IgG]; Bio-cell), random sequences, or PBS was administered intraperitoneally once the long axis of tumors had reached ∼6 mm, as measured by digital caliper.
Confocal Image
aptCTLA-4 was incubated with CTLA-4-GFP-expressed HEK293T cells or mouse lymphocytes in culture medium (DMEM+10% FBS) at 37°C for 30 min. After washing by PBS and fixing the cell by 4% PFA for 10 min, the samples were blocked in PBS containing 1% normal goat serum and 2% BSA. PE-conjugated anti-human CTLA-4 (1:400) and PE-conjugated anti-mouse CTLA-4 (1:400) antibodies (eBioscience, San Diego, CA, USA) were prepared in 1% normal goat serum and 2% BSA in PBS. After PBS washing for three times, samples were stained by DAPI (Invitrogen) for cell nucleus identification. Images of cross-sections were acquired using a Zeiss LSM710 confocal microscope (Carl Zeiss Microimage, Thornwood, NY, USA).
CD Spectroscopy
CD spectra experiments were performed with Jasco J-815 CD Spectropolarimeter (JASCP, USA). Aptamers (10 μM) were prepared in SELEX buffer (pH 7.5), denatured at 95°C, and then kept at 25°C overnight prior to CD spectra experiments. CD spectra data were obtained from 200 to 350 nm at a step size of 1 nm, a 0.2-s time per point, and a bandwidth of 1 nm. Each spectrum was an average of three scans at room temperature and was corrected with buffer baseline.
Blood Biochemistry Analyses
The amount of GOT, GPT, and BUN was detected by Fuji Dri-Chem 4000i (Fujifilm, Tokyo, Japan) under technical support of the Taiwan Mouse Clinic.
Flow Cytometry Analyses and Immunohistochemical Staining of the Tumors
Tumors were digested with collagenase, and cells in the tumors were isolated following the procedures built by Zabel et al.57 Antibodies against mouse CD45 and CD8 and relative isotype control antibodies (BioLegend) were used to stain these isolated cells. Formalin-fixed, paraffin-embedded tissues were rehydrated and subjected to an antigen-retrieval process in citrate buffer (pH 6.0). A primary antibody against mouse CD8 (Thermo Scientific) was used at the concentration of 1:100 dilution.
Author Contributions
B.T.H. conceived the work, designed and performed the experiments, and analyzed the data. W.Y.L. designed and performed the experiments and analyzed the data. Y.C.C. conceived the work and designed the experiments. J.W.W. performed the experiments. S.D.Y. advised the work. E.P.Y.L. conceived the work, analyzed the data, and wrote the manuscript. P.C.Y. conceived the work, and critically reviewed and supervised the study.
Conflicts of Interest
The authors report no conflicts of interest.
Acknowledgments
We dedicate this work to Dr. Konan Peck, a respected researcher in the fields of high-throughput biotechnology, biophysics, and cancer genomics. Shortly after initiating this project, Dr. Peck unfortunately succumbed to a rapidly progressing cancer. It is our fervent hope that the research platform established by Dr. Peck will result in the development of new diagnostic and therapeutic options that benefit cancer patients. We also thank the National Center for Genome Medicine for the technical and bioinformatics support. This project was supported by grants from the National Science Council (NSC99-2320-B-001-012-MY3 and MOST104-2314-B-002-228-MY3).
Footnotes
Supplemental Information includes seven figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.08.006.
Contributor Information
Emily Pei-Ying Lin, Email: pylin@ibms.sinica.edu.tw.
Pan-Chyr Yang, Email: pcyang@ntu.edu.tw.
Supplemental Information
References
- 1.Sharma P., Allison J.P. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Errico A. Immunotherapy: PD-1-PD-L1 axis: efficient checkpoint blockade against cancer. Nat. Rev. Clin. Oncol. 2015;12:63. doi: 10.1038/nrclinonc.2014.221. [DOI] [PubMed] [Google Scholar]
- 3.Boussiotis V.A. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N. Engl. J. Med. 2016;375:1767–1778. doi: 10.1056/NEJMra1514296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutros C., Tarhini A., Routier E., Lambotte O., Ladurie F.L., Carbonnel F., Izzeddine H., Marabelle A., Champiat S., Berdelou A. Safety profiles of anti-CTLA-4 and anti-PD-1 antibodies alone and in combination. Nat. Rev. Clin. Oncol. 2016;13:473–486. doi: 10.1038/nrclinonc.2016.58. [DOI] [PubMed] [Google Scholar]
- 5.Nishino M., Ramaiya N.H., Hatabu H., Hodi F.S. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat. Rev. Clin. Oncol. 2017 doi: 10.1038/nrclinonc.2017.88. Published online June 27, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samaranayake H., Wirth T., Schenkwein D., Räty J.K., Ylä-Herttuala S. Challenges in monoclonal antibody-based therapies. Ann. Med. 2009;41:322–331. doi: 10.1080/07853890802698842. [DOI] [PubMed] [Google Scholar]
- 7.Chames P., Van Regenmortel M., Weiss E., Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. Br. J. Pharmacol. 2009;157:220–233. doi: 10.1111/j.1476-5381.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harding F.A., Stickler M.M., Razo J., DuBridge R.B. The immunogenicity of humanized and fully human antibodies: residual immunogenicity resides in the CDR regions. MAbs. 2010;2:256–265. doi: 10.4161/mabs.2.3.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabrizi M., Bornstein G.G., Suria H. Biodistribution mechanisms of therapeutic monoclonal antibodies in health and disease. AAPS J. 2010;12:33–43. doi: 10.1208/s12248-009-9157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:440. doi: 10.1038/nrd.2017.86. [DOI] [PubMed] [Google Scholar]
- 11.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 12.Ellington A.D., Szostak J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 13.Jayasena S.D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 14.Pendergrast P.S., Marsh H.N., Grate D., Healy J.M., Stanton M. Nucleic acid aptamers for target validation and therapeutic applications. J. Biomol. Tech. 2005;16:224–234. [PMC free article] [PubMed] [Google Scholar]
- 15.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagalkot V., Farokhzad O.C., Langer R., Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew. Chem. Int. Ed. Engl. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 17.Niu W., Chen X., Tan W., Veige A.S. N-heterocyclic carbene-gold(I) complexes conjugated to a leukemia-specific DNA aptamer for targeted drug delivery. Angew. Chem. Int. Ed. Engl. 2016;55:8889–8893. doi: 10.1002/anie.201602702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monaco I., Camorani S., Colecchia D., Locatelli E., Calandro P., Oudin A., Niclou S., Arra C., Chiariello M., Cerchia L. Aptamer functionalization of nanosystems for glioblastoma targeting through the blood-brain barrier. J. Med. Chem. 2017;60:4510–4516. doi: 10.1021/acs.jmedchem.7b00527. [DOI] [PubMed] [Google Scholar]
- 19.Cheng C., Chen Y.H., Lennox K.A., Behlke M.A., Davidson B.L. In vivo SELEX for identification of brain-penetrating aptamers. Mol. Ther. Nucleic Acids. 2013;2:e67. doi: 10.1038/mtna.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lao Y.H., Phua K.K., Leong K.W. Aptamer nanomedicine for cancer therapeutics: barriers and potential for translation. ACS Nano. 2015;9:2235–2254. doi: 10.1021/nn507494p. [DOI] [PubMed] [Google Scholar]
- 21.Pastor F. Aptamers: a new technological platform in cancer immunotherapy. Pharmaceuticals (Basel) 2016;9:E64. doi: 10.3390/ph9040064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pastor F., Soldevilla M.M., Villanueva H., Kolonias D., Inoges S., de Cerio A.L., Kandzia R., Klimyuk V., Gleba Y., Gilboa E. CD28 aptamers as powerful immune response modulators. Mol. Ther. Nucleic Acids. 2013;2:e98. doi: 10.1038/mtna.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tucker C.E., Chen L.S., Judkins M.B., Farmer J.A., Gill S.C., Drolet D.W. Detection and plasma pharmacokinetics of an anti-vascular endothelial growth factor oligonucleotide-aptamer (NX1838) in rhesus monkeys. J. Chromatogr. B Biomed. Sci. Appl. 1999;732:203–212. doi: 10.1016/s0378-4347(99)00285-6. [DOI] [PubMed] [Google Scholar]
- 25.Walker L.S., Sansom D.M. The emerging role of CTLA4 as a cell-extrinsic regulator of T cell responses. Nat. Rev. Immunol. 2011;11:852–863. doi: 10.1038/nri3108. [DOI] [PubMed] [Google Scholar]
- 26.Krummel M.F., Allison J.P. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 1995;182:459–465. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leach D.R., Krummel M.F., Allison J.P. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 28.Yang J.C., Hughes M., Kammula U., Royal R., Sherry R.M., Topalian S.L., Suri K.B., Levy C., Allen T., Mavroukakis S. Ipilimumab (anti-CTLA4 antibody) causes regression of metastatic renal cell cancer associated with enteritis and hypophysitis. J. Immunother. 2007;30:825–830. doi: 10.1097/CJI.0b013e318156e47e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolchok J.D., Neyns B., Linette G., Negrier S., Lutzky J., Thomas L., Waterfield W., Schadendorf D., Smylie M., Guthrie T., Jr. Ipilimumab monotherapy in patients with pretreated advanced melanoma: a randomised, double-blind, multicentre, phase 2, dose-ranging study. Lancet Oncol. 2010;11:155–164. doi: 10.1016/S1470-2045(09)70334-1. [DOI] [PubMed] [Google Scholar]
- 30.Santulli-Marotto S., Nair S.K., Rusconi C., Sullenger B., Gilboa E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003;63:7483–7489. [PubMed] [Google Scholar]
- 31.Herrmann A., Priceman S.J., Swiderski P., Kujawski M., Xin H., Cherryholmes G.A., Zhang W., Zhang C., Lahtz C., Kowolik C. CTLA4 aptamer delivers STAT3 siRNA to tumor-associated and malignant T cells. J. Clin. Invest. 2015;125:2547. doi: 10.1172/JCI82555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamper C.C., Zhang Y., Tobin J.F., Erbe D.V., Ikemizu S., Davis S.J., Stahl M.L., Seehra J., Somers W.S., Mosyak L. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature. 2001;410:608–611. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- 33.Víglaský V., Tlučková K., Bauer L. The first derivative of a function of circular dichroism spectra: biophysical study of human telomeric G-quadruplex. Eur. Biophys. J. 2011;40:29–37. doi: 10.1007/s00249-010-0625-8. [DOI] [PubMed] [Google Scholar]
- 34.Verdian-Doghaei A., Housaindokht M.R., Bozorgmehr M.R., Abnous K. Conformational switch of insulin-binding aptamer into G-quadruplex induced by K+ and Na+: an experimental and theoretical approach. J. Biomol. Struct. Dyn. 2015;33:1153–1163. doi: 10.1080/07391102.2014.935482. [DOI] [PubMed] [Google Scholar]
- 35.Fialová M., Kypr J., Vorlícková M. The thrombin binding aptamer GGTTGGTGTGGTTGG forms a bimolecular guanine tetraplex. Biochem. Biophys. Res. Commun. 2006;344:50–54. doi: 10.1016/j.bbrc.2006.03.144. [DOI] [PubMed] [Google Scholar]
- 36.Quah B.J., Parish C.R. New and improved methods for measuring lymphocyte proliferation in vitro and in vivo using CFSE-like fluorescent dyes. J. Immunol. Methods. 2012;379:1–14. doi: 10.1016/j.jim.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 38.Keefe A.D., Cload S.T. SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 2008;12:448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 39.Burmeister P.E., Lewis S.D., Silva R.F., Preiss J.R., Horwitz L.R., Pendergrast P.S., McCauley T.G., Kurz J.C., Epstein D.M., Wilson C. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 2005;12:25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 40.Kuwahara M., Obika S. In vitro selection of BNA (LNA) aptamers. Artif. DNA PNA XNA. 2013;4:39–48. doi: 10.4161/adna.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y., Qiu Q., Gill S.C., Jayasena S.D. Modified RNA sequence pools for in vitro selection. Nucleic Acids Res. 1994;22:5229–5234. doi: 10.1093/nar/22.24.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruckman J., Green L.S., Beeson J., Waugh S., Gillette W.L., Henninger D.D., Claesson-Welsh L., Janjić N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 43.Veedu R.N., Wengel J. Locked nucleic acid nucleoside triphosphates and polymerases: on the way towards evolution of LNA aptamers. Mol. Biosyst. 2009;5:787–792. doi: 10.1039/b905513b. [DOI] [PubMed] [Google Scholar]
- 44.Meek K.N., Rangel A.E., Heemstra J.M. Enhancing aptamer function and stability via in vitro selection using modified nucleic acids. Methods. 2016;106:29–36. doi: 10.1016/j.ymeth.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Sefah K., Shangguan D., Xiong X., O’Donoghue M.B., Tan W. Development of DNA aptamers using cell-SELEX. Nat. Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 46.Sun H., Zhu X., Lu P.Y., Rosato R.R., Tan W., Zu Y. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol. Ther. Nucleic Acids. 2014;3:e182. doi: 10.1038/mtna.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayer G. The chemical biology of aptamers. Angew. Chem. Int. Ed. Engl. 2009;48:2672–2689. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- 48.Kikin O., D’Antonio L., Bagga P.S. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong H.M., Stegle O., Rodgers S., Huppert J.L. A toolbox for predicting g-quadruplex formation and stability. J. Nucleic Acids. 2010;2010 doi: 10.4061/2010/564946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Furoukh N., Goffart S., Szibor M., Wanrooij S., Braun T. Binding to G-quadruplex RNA activates the mitochondrial GTPase NOA1. Biochim. Biophys. Acta. 2013;1833:2933–2942. doi: 10.1016/j.bbamcr.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 51.Mittelberger F., Meyer C., Waetzig G.H., Zacharias M., Valentini E., Svergun D.I., Berg K., Lorenzen I., Grötzinger J., Rose-John S. RAID3--an interleukin-6 receptor-binding aptamer with post-selective modification-resistant affinity. RNA Biol. 2015;12:1043–1053. doi: 10.1080/15476286.2015.1079681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishiguro A., Kimura N., Watanabe Y., Watanabe S., Ishihama A. TDP-43 binds and transports G-quadruplex-containing mRNAs into neurites for local translation. Genes Cells. 2016;21:466–481. doi: 10.1111/gtc.12352. [DOI] [PubMed] [Google Scholar]
- 53.Murakami K., Zhao J., Yamasaki K., Miyagishi M. Biochemical and structural features of extracellular vesicle-binding RNA aptamers. Biomed. Rep. 2017;6:615–626. doi: 10.3892/br.2017.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai W.Y., Huang B.T., Wang J.W., Lin P.Y., Yang P.C. A novel PD-L1-targeting antagonistic DNA aptamer with antitumor effects. Mol. Ther. Nucleic Acids. 2016;5:e397. doi: 10.1038/mtna.2016.102. [DOI] [PubMed] [Google Scholar]
- 55.Lin K.Y., Guarnieri F.G., Staveley-O’Carroll K.F., Levitsky H.I., August J.T., Pardoll D.M., Wu T.C. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 1996;56:21–26. [PubMed] [Google Scholar]
- 56.Alam K.K., Chang J.L., Burke D.H. FASTAptamer: a bioinformatic toolkit for high-throughput sequence analysis of combinatorial selections. Mol. Ther. Nucleic Acids. 2015;4:e230. doi: 10.1038/mtna.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pachynski R.K., Scholz A., Monnier J., Butcher E.C., Zabel B.A. Evaluation of tumor-infiltrating leukocyte subsets in a subcutaneous tumor model. J. Vis. Exp. 2015 doi: 10.3791/52657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.