Abstract
Usher syndrome (USH) is a rare autosomal recessive disease and the most common inherited form of combined visual and hearing impairment. Up to 13 genes are associated with this disorder, with USH2A being the most prevalent, due partially to the recurrence rate of the c.2299delG mutation. Excluding hearing aids or cochlear implants for hearing impairment, there are no medical solutions available to treat USH patients. The repair of specific mutations by gene editing is, therefore, an interesting strategy that can be explored using the CRISPR/Cas9 system. In this study, this method of gene editing is used to target the c.2299delG mutation on fibroblasts from an USH patient carrying the mutation in homozygosis. Successful in vitro mutation repair was demonstrated using locus-specific RNA-Cas9 ribonucleoproteins with subsequent homologous recombination repair induced by an engineered template supply. Effects on predicted off-target sites in the CRISPR-treated cells were discarded after a targeted deep-sequencing screen. The proven effectiveness and specificity of these correction tools, applied to the c.2299delG pathogenic variant of USH2A, indicates that the CRISPR system should be considered to further explore a potential treatment of USH.
Keywords: Usher syndrome, USH2A, c.2299delG, CRISPR, gene editing, RNPs
Introduction
Usher syndrome (USH) is an autosomal recessive disease involving sensorineural hearing loss, retinitis pigmentosa (RP), and, in some cases, vestibular dysfunction. It is a rare disorder with a prevalence ranging from 3 to 6.2 per 100,000 and it is the most common genetic cause combining hearing and vision loss,1 for which no clinical treatment is presently available. Three clinical forms of the disease can be distinguished according to severity and progression: USH type I (USH1), type II (USH2), and type III (USH3), with USH type II (USH2) being the most frequent clinical form. This type is defined by congenital moderate-severe hearing loss and RP of post-pubertal onset.1
USH is clinically and genetically heterogeneous, since it is associated to date with 13 genes.2 However, more than 50% of USH cases,3, 4 as well as 8% of non-syndromic autosomal recessive RP (ARRP) patients,5 harbor mutations in the USH2A gene, which is therefore the principle gene responsible for both diseases. This is due, in part, to the high prevalence of two mutations located 22 bp from each other in exon 13: c.2299delG/p.E767Sfs*21, which accounts for up to 31% of USH2 cases, and c.2276G>T/p.C759F, which is found in approximately 4.5% of ARRP cases.6, 7
The large size, up to 15 kb, of the USH2A coding sequence (GenBank: NM_206933) makes it difficult to develop a gene substitution therapy for patients with mutations in this gene. In addition, more than one essential isoform of the USH2A gene is expressed in the retinal tissue, and augmentation therapy by delivery of one single cDNA would be insufficient.8 Nevertheless, a suitable alternative now exists: genetic correction using gene editing strategies such as the CRISPR/Cas9 system, which holds enormous promise for in vivo and ex vivo genome editing-based therapies.9, 10, 11, 12, 13 Furthermore, the close proximity of the two highly prevalent mutations in exon 13 of USH2A is convenient for using this particular technology.
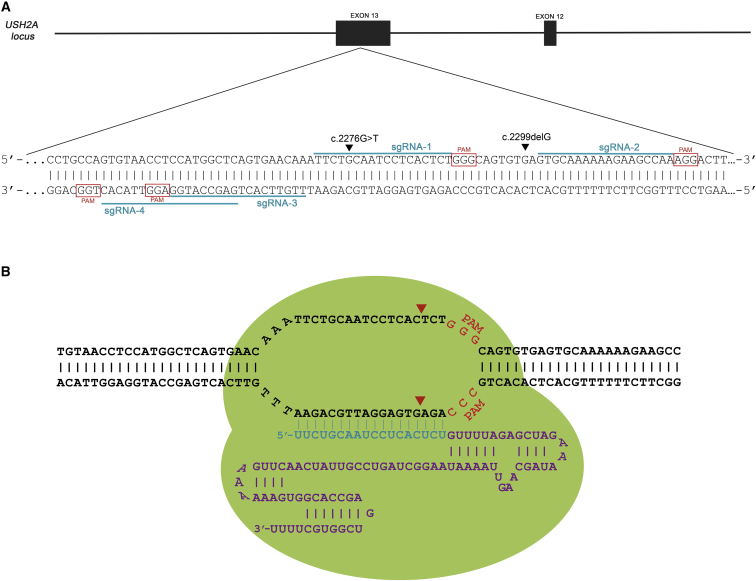
CRISPR-associated endonuclease Cas9 protein is an up-to-date technology that has recently been implemented for use in a broad spectrum of cell types and model organisms.14, 15, 16, 17 The system comprises two primary elements that form the RGEN (RNA-guided engineered nucleases) complex:18, 19 namely, the Cas9 nuclease and a single guide RNA molecule (sgRNA or guide RNA) (Figure 1A). The latter is specifically designed to be complementary to the target locus (Figure 1B).
Figure 1.
Diagram Showing the CRISPR Construct Designs for the USH2A Locus Editing
(A) Exon 13 of USH2A highlighting the location of mutations c.2299delG and c.2276G>T, as well as the four different sgRNAs (blue) selected to target the locus. Red boxes indicate PAM sequences 3′-adjacent to the sgRNAs. (B) The CRISPR/Cas9 system relies on the Cas9 nuclease and the sgRNA, and this last molecule consists of two RNA domains: crispr RNA (crRNA), which is specifically designed to be complementary to the target locus (blue), and trans-activating crRNA (tracrRNA), which is more a structural part for the binding with the nuclease (purple). The scheme represents the Cas9 endonuclease-sgRNA-1 complex (RGEN-1): the RNA couples with Cas9 endonuclease (green outline), forming the complex known as RGEN (RNA-guided engineered nucleases), and guides it to the sgRNA complementary sequence of the target DNA through Watson-Crick base pairing, enabling the Cas9 to produce a double-stranded break (DSB) in the DNA (red arrowheads). In addition, in order for the Cas9 to recognize and site-specifically cleave the DNA, a specific short pattern needs to be present 3′-adjacent to the target sequence: the protospacer adjacent motif (PAM), consisting of the 3 bases 5′-NGG (shown in red).
Upon cleavage by Cas9, the target locus typically undergoes one of two major pathways for DNA damage repair: the error-prone non-homologous end joining (NHEJ) or the homology-directed repair (HDR) pathway,20, 21 both of which can be used to achieve the desired editing outcome. In the absence of a repair template with identical homologous flanking ends, the HDR pathway cannot be employed. Thus, double-stranded breaks (DSBs) are re-ligated by means of the NHEJ mechanism, leaving scars in the form of insertion/deletion mutations (indels), which indirectly represent the Cas9 cleavage efficiency.
Due to its high efficacy and simple design, we have employed the CRISPR/Cas9 system in this study for USH2A gene manipulation, with the aim of repairing the most prevalent mutations present in this gene. Through this study, the locus where the mutations c.2299delG and c.2276G>T in exon 13 of USH2A are located has been edited.
Results
USH2A Exon13 Editing in HEK293 Cells
The aim of this study was to set up a CRISPR toolkit to correct the c.2299delG and c.2276G>T mutations in cells from patients harboring these mutations. Editing primary cell lines is known to be difficult. For that reason, the broadly used HEK293 cells were selected for the first trial, since they are easy to handle and transfect. However, this cell line presents a wild-type (WT) genome; to overcome this problem, a decision was made to reverse the strategy by introducing the c.2299delG and c.2276G>T mutations in these cells instead of their correction.
Four different 18-nt-long RNA guides (sgRNAs) were designed to drive Cas9 to the target exon 13 of the USH2A gene, where mutations c.2299delG and c.2276G>T are located. Two were set on the positive strand (sgRNA-1 and sgRNA-2) and two were set on the negative strand (sgRNA-3 and sgRNA-4) (Figure 1A). These sequences were cloned into the pX330 vector containing all the primary elements of the CRISPR system.14
The resultant constructs were transfected into HEK293 cells. 48 hr post-transfection, the cells were lysed to extract the genomic DNA (gDNA) and analyze the cleavage efficiency of each sgRNA by the T7-endonuclease I (T7E1) assay. Results from the T7E1 assay demonstrated that the four Cas9-sgRNA plasmids exhibited cleavage activity at both DNA concentrations used (1 μg and 3 μg) (Figure 2A). However, transfection with 3 μg DNA produced better results: sgRNA-1 had around 40% and 53% on-target cleavage efficiency at transfection concentrations of 1 μg and 3 μg, respectively. The sgRNA-2 was almost as effective, with 30% on-target activity with 1 μg product transfection and 52% with the higher dose. The sgRNA-4 showed similar results to sgRNA-1, with 39% cleavage efficiency at 1 μg transfection and 54% after the concentration increase. Finally, sgRNA-3 was the less effective construct, with a cleavage activity below 10% for both conditions. The efficiency of the sgRNA-1 (3 μg) was confirmed through Sanger sequencing of the clonal isolated amplicons (Figure 2B),22 presenting a cleavage activity of 62% based on the indel mutations detected in these sequences.
Figure 2.
CRISPR Assay on HEK293 Cells
(A) Products of the T7E1 assay resolved on a 2% agarose gel. Green arrowheads indicate cleaved bands corresponding to the expected fragments resulting from T7E1 cutting, the intensity of which is directly correlated to the indel frequency and therefore for Cas9 on-target activity. Untreated genomic DNA from the same HEK293 cells used for transfections was used as a negative control, where only the intact band can be observed. (B) Chromatograms showing the result of Sanger sequencing of PCR products obtained after amplifying genomic DNA, and subcloned into a plasmid for E. coli transformation, from cultures transfected with RGEN-1 at 3 μg. The indels shown here are produced by NHEJ repair. The indel frequency was established by comparison with the wild-type (WT) reference sequence. Red arrowheads point out the most probable cutting position on the sgRNA sequence (bold characters), 3–4 bp upstream of the PAM sequence (shown in red). NHEJ results in small deletions (bold green dashes) or in insertions (green bold characters). The chromatogram illustrates an example of the first NHEJ event with a G insertion framed in the blue background.
To introduce the c.2299delG and c.2276G>T mutations in WT HEK293 cells through the HDR major DNA repair pathway, the sgRNA-1 construct was selected, since it had shown a higher activity and is located closer to the mutations. HDR typically occurs at lower and substantially more variable frequencies than NHEJ; nevertheless, it can be used to generate precise, defined modifications at a target locus in the presence of an exogenously introduced repair template.17, 23 A repair template in the form of a single-stranded oligodeoxynucleotide (ssODN) was utilized (Figure 3A), which provides an effective and simple method for making small edits in the genome.22, 24, 25, 26 HEK293 cells were separately transfected with 3 μg sgRNA along with an ssODN carrying either the c.2299delG mutation (ssODN-2299) or the c.2276G>T mutation (ssODN-2276). In addition to the specific mutation, the designed ssODNs had a change in the protospacer adjacent motif (PAM) sequence to remove the sequence pattern, consisting of a silent mutation (a G to C transversion) that lacked splicing alterations based on computational prediction tools. This introduced a restriction site, which is useful for a restriction fragment length polymorphism (RFLP) analysis of the genomes of the edited cells.
Figure 3.
Exon 13 Is Efficiently Edited in HEK293 Cells with sgRNA-1
(A) Diagram depicting the targeted region and the two specific ssODNs used as an exogenous template for repair. The 193-nt-long ssODN-2299 is shown above, highlighting the guanine deletion corresponding to the c.2299 position. Below, the 197-nt-long ssODN-2276 presents a similar composition, but with c.2276G>T change instead of the c.2299delG. The diagram also displays the silent mutation c.2292G>C placed in both ssODN designs. This variant is located in the PAM sequence to avoid recognition of the target by the sgRNA, and, therefore, to be cleaved by Cas9. Moreover, this variant creates the new recognition site for the MlsI restriction enzyme for the edition detection. (B) RFLP results of HEK293 transfection with 3 μg sgRNA-1 together with 10 pmol of the ssODN including either the mutation c.2299delG or c.2276G>T. Green arrowheads indicate fragments cut by the restriction enzyme MlsI.
The analysis by digestion with MlsI of the PCR products of exon 13 from gDNA from targeted cells showed considerable results of HDR for both construct-template combinations. These products were resolved through RFLP analysis and subsequent image quantification through relative band intensities. When using ssODN-2299delG, HDR efficiency reached 16%, whereas co-transfection with ssODN-2276 showed 12% effectiveness (Figure 3B).
Correction of the c.2299delG Mutation in Fibroblasts
Human dermal fibroblasts (HDFs) were isolated from an USH2 patient carrying the c.2299delG mutation in homozygosis, as well as from two healthy control patients. Unfortunately, cells carrying the c.2276G>T mutation in homozygosis could not be obtained.
The CRISPR activity in control fibroblasts using the DNA constructs was undetectable (data not shown); hence, we used ribonucleoprotein (RNP) complex delivery, consisting of purified Cas9 protein coupled with the in vitro transcripted (IVT) sgRNA. A combination of 15 μg Cas9 and 20 μg sgRNA-1 was transferred to control HDFs as RNP complexes by nucleofection obtaining an indel frequency of 18% (Figure 4A). The subsequent edition attempt using co-delivery of RNPs with ssODN-2299 resulted in 5% HDR efficiency (Figure 4B).
Figure 4.
Editing of HDFs
(A) T7E1 assay resolved on a 2% agarose gel with HDF amplification products, noting the cleaved fragments corresponding to indels (shown with green arrowheads). Untreated genomic DNA from the same HDF cells used for transfections was used as a negative control, where only the intact band can be appreciated. (B) RFLP assay results of the HDF transfection with 15 μg Cas9 and 20 μg sgRNA-1 with 500 pmol of the ssODN with the wild-type sequence lacking the PAM. Green arrowheads show fragments cut by the restriction enzyme MlsI. (C) Scheme depicting the targeted region and the 194-nt ssODN-WT with the guanine restoration on the c.2299 position. The silent mutation c.2292G>C was included in the ssODN design in order to prevent Cas9 cleavage as well as the MlsI target sequence for the edition detection by RFLP.
In view of the promising results, the same process was performed on c.2299delG patient HDFs using an ssODN with a WT sequence and the PAM sequence ablated (ssODN-WT) (Figure 4C). An indel frequency of 6% and an HDR mutation correction of 2.5%, according to image quantification, were achieved (Figures 4A and 4B).
Analysis of Edition Rates Using High-Throughput Sequencing
Cas9 presents a certain capability to cleave on DNA sites differing from some nucleotides in the sgRNA sequence,27 causing collateral off-target mutations that should be taken into consideration.
The resulting potential off-target ensemble obtained from the in silico prediction tools consisted of a total of 21 off-targets (Table 1) to be screened through high-throughput sequencing, since the T7E1 assay is only sensitive to mismatches above 1%.28
Table 1.
Selection of In Silico Potential Off-Targets for sgRNA-1
| Positiona | Gene | Locus Type | Sequence | Strand | Mismatches | Bulge Size |
|---|---|---|---|---|---|---|
| chr13:36343748 | SPG20 | intronic | TTaTcCAATCCTCACTCTTGG | positive | 2 | 0 |
| chrX:31429579 | DMD | intronic | TTCTGgAATgCTCACTCTGGG | negative | 2 | 0 |
| chr1:63817856 | ROR1 | intronic | TTCTGCAtTCCTCcCTCTTGG | positive | 2 | 0 |
| chr3:60966504 | FHIT | intronic | TTCTGCAcTCCTCACgCTGGG | positive | 2 | 0 |
| chr5:168083516 | TENM2 | intronic | TTCTGCAATgCTCACaCTGGG | positive | 2 | 0 |
| chr8:98710205 | STK3 | intronic | TTCTGCAATgCTCACgCTGGG | negative | 2 | 0 |
| chr2:161421064 | TBR1 | intronic | TTCTGCAATtCTCACTCaGGG | positive | 2 | 0 |
| chr5:64594732 | RGS7BP | exonic | TTtTGCAATCCaaACTCTTGG | negative | 3 | 0 |
| chr11:64851825 | EHD1 | UTR | TTCgGCAgTCCTCAgTCTCGG | positive | 3 | 0 |
| chrX:24006521 | KLHL15 | exonic | TTCTGaAgTaaTCACTCTGGG | positive | 4 | 0 |
| chr15:81827587 | — | intergenic | TTCTGCAATCCTCAGCTCTTGG | positive | 0 | 1 |
| chr11:62715832 | HNRNPUL2-BSCL2 | intronic | TTCTGC–TCCTCACTCTGGG | negative | 0 | 2 |
| chr17:997306 | TIMM22 | exonic | TTCTGC–TCCTCACTCTTGG | negative | 0 | 2 |
| chr17:63411113 | TANC2 | intronic | TcCTGC-ATCCTCACTCTTGG | negative | 1 | 1 |
| chr5:135075878 | C5orf66 | intronic | TT-TGCAATtCTCACTCTGGG | positive | 1 | 1 |
| chr2:52242405 | LOC730100 | intronic | TTCTGCAATtC-CACTCTTGG | positive | 1 | 1 |
| chr2:159450247 | BAZ2B | intronic | TTCTGtAATCCTCACTACTCGG | positive | 1 | 1 |
| chr2:89267421 | IGKV1-33 | intronic | TTCTGCAgTCCTCAC-CTCGG | negative | 1 | 1 |
| chr2:89915107 | IGKV1D-33 | intronic | TTCTGCAgTCCTCAC-CTCGG | positive | 1 | 1 |
| chr16:7651515 | RBFOX1 | intronic | TTCTGCAAcCCTCAC-CTGGG | positive | 1 | 1 |
| chr20:10492244 | SLX4IP | intronic | TTCTGCAATCC-CACTCcAGG | negative | 1 | 1 |
Position, chromosomal coordinates according to GRCh38 (Genome Reference Consortium Human Build 38); lower-case letters, mismatches; superscripted characters, DNA bulge position; dashes, RNA bulge position; and bulge size, the presence of a bulge and the size of the loop.
Off-targets are arranged according to the recognition probability based on the mismatch-bulge display and proximity to the PAM. In the case of equal conditions, a chromosomal position numerical order is applied.
Amplicon deep sequencing of the treated cells using the Illumina MiSeq platform allowed for the validation of on-target Cas9 cleavage by indel detection, as well as the effective c.2299delG mutation correction by HDR (Figure 5). Reads inspection exposed 20.1% of the specific target cleavage and 1.7% of direct edited sequences harboring only the two directed variants included in the ssODN-WT design (c.2299insG and c.2292G>C). Furthermore, some sequences presenting only one of the two variants were detected and these were considered as partially edited.
Figure 5.
Deep Sequencing of the HDFs Transfected with RGEN-1 Plus ssODN-WT
Representation of real sequencing reads mapped against the USH2A locus, showing examples of the different editing possibilities obtained with RGEN-1: Cas9-specific action is appreciated by the small insertions (bold superscripted characters) or deletions (bold dashes) upstream of the PAM sequence, due to the NHEJ repair mechanism. Directed editing events of the exon 13 of USH2A are represented by sequences that contain the c.2299delG mutation correction, as well as the silent G>C transversion in the PAM sequence (directed changes highlighted in black background). Sequences with only one of the aimed changes are considered partially edited. RGEN-1 cleavage efficiency is estimated as the number of all sequences that are different from the reference sequence of the patient (20.1%; i.e., the sum of the edited, partially edited, and indel carrying sequences). Reads identical to the reference sequence correspond to intact cells where Cas9 had no cleavage activity.
On the other hand, no detectable off-target activity was identified for almost all potential loci, since no significant number of reads with indels on the potential cutting area was tracked. Unfortunately, sequences from one of the tested loci (SLX4IP locus) were not captured by the sequencing platform; therefore, no data could be obtained from this region. Nevertheless, this region presents a minor site recognition probability according to the ranking list (Table 1) and since none of the other targets showed cleavage activity, it seems unlikely that this locus should behave otherwise.
Discussion
USH is a disorder that heavily impairs the social and cognitive development of patients, since nowadays in developed countries communication is based on media. This is a major handicap for people with hearing and vision problems, interfering with their educational and intellectual development and eventually leading to a hampering of professional integration. To date, there is no medical treatment except hearing aids or cochlear implants for audiological disabilities, so new therapies are being explored to cure or alleviate RP symptoms. Gene replacement therapies could be a potential treatment if it were not for the large size of USH2A. The limited cargo capacity of the most widely used viral vectors such as adeno-associated viruses (AAVs) could be bypassed using helper-dependent adenoviruses (Hd-Ads) or herpes simplex viruses (HSVs) as gene carriers that are able to accommodate larger sequences, with respective cargo limits up to 36 kb and 150 kb.29, 30, 31 However, these viral vectors have particular restrictions that directly concern this case study. Hd-Ad and HSV have been reported as transducing primarily the retinal pigment epithelium (RPE) but in a very limited way or not at all the photoreceptors (PR),32, 33 apart from some Ad engineered vectors without a persistent transgene expression that require systematic infections with the construct.34 In addition, the large size of these vectors makes trespassing through the inner plexiform layer very difficult, not permitting intravitreal administration but only the more invasive subretinal injection.
A genome editing therapeutic approach could be a suitable alternative to safely correct the specific mutations of each patient. Various genome editing technologies have emerged in recent years, including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the CRISPR/Cas9 nuclease system. The first two technologies use a strategy of tethering endonuclease catalytic domains to modular DNA binding proteins for inducing targeted DNA DSBs at specific genomic loci.35 By contrast, Cas9 is a nuclease guided by small RNAs through Watson-Crick base pairing with target DNA, representing a system that is markedly easier to design, highly specific, and efficient for a variety of cell types and organisms.36 Using a direct mutation repair strategy like engineered nucleases represents a new panorama for future therapies. In this study, the most prevalent mutation involved in USH, c.2299delG, has been edited. This mutation is responsible for USH in a considerable number of patients.
This study has demonstrated successful targeting of exon 13 of USH2A in HEK293 cells with CRISPR, allowing for the editing of the locus to introduce the c.2299delG and c.2276G>T mutations. However, the approach utilizing DNA constructs was unsuccessfully transferred to control HDFs, most probably due to the hard-to-transfect feature of the fibroblasts, in addition to the fact that the isolated cells are not neonatal but are from adult donors. This issue was addressed by transferring the RGEN designs from plasmids to pre-assembled Cas9-sgRNA (RNPs) cell delivery, a method that offers a host-transcriptome-free option. This method has previously been reported as an efficient alternative that also provides the advantage of a reduction in off-target events because of the short lifespan of these RNP complexes, compared to the continuous expression of the elements providing plasmid transfection.37, 38, 39, 40
In this project, high indel frequencies were achieved with 3 of the 4 designed sgRNAs in HEK293 cells. sgRNA-3 is the only construct showing low cleavage activity, which may be due to the locus accessibility. Structural or epigenetic arrangements such as methylation patterns have previously been reported to hinder Cas9-sgRNA attachment to the DNA.41, 42 Other studies suggest that sgRNAs targeting the active strand show a higher cleavage efficiency compared to those targeting the inactive strand.43 Nevertheless, if we accept this statement, sgRNA-4 should have shown a lower NHEJ efficiency instead of rates similar to sgRNA-1 and sgRNA-2. We suspect that the sgRNA-3 failure could be affected by any of these phenomena or by other still unknown aspects regarding the CRISPR/Cas9 system.
Substantial editing rates were achieved on HEK293 and more limited percentages on HDFs. Editing differences between the fibroblasts of the control (18% indel and 5% HDR) and USH patients (6% indel and 2.5% HDR) may be due to a survival detriment associated with the c.2299delG mutation, but other explanations cannot be ruled out. The discrepancies between the RFLP and the deep-sequencing rates have led to further analyses being performed with the next-generation sequencing (NGS) data, resulting in interesting findings. A small number of sequences presented either c.2292G>C silent change or c.2299G insertion in equal proportions (0.8% each), suggesting that HDR is rendered in some cases with one end of the template or the other. Partially edited fractions with only the PAM change would be recognized in the RFLP assay, since the MlsI target sequence would be generated as well. Therefore, when we compute this rate (0.8%) along with the absolute edited cells registered by deep sequencing (1.7%), the percentage approximates to the results obtained by RFLP (2.5%). Similarly, the partially edited cells for only the c.2299delG repair (0.8%) could be taken into account along with the whole edited sequences number (1.7%), inferring a 2.5% rate of mutation correction (Figure 5). These data, therefore, show evidence of in vitro genomic editing of the pathological mutation c.2299delG in fibroblasts from patients. It has to be noted that HDFs are considered to be refractory to transfection, so that the exogenous DNA is rapidly excluded from the nucleus hindering HDR.44 In addition, HDFs from adult skin biopsies are less pliant than those used in other studies from newborn cell lines, where authors achieve similar or lower editing rates.37, 39 For that reason, we consider that our knock-in results are high compared with those described in the literature, giving grounds for optimism for future therapeutic implementation of this technology in USH.
Concerning the cleavage frequency differences observed in the HDFs of the USH patient (6% in the T7E1 assay versus 20.1% in the deep-sequencing results; Figures 4A and 5, respectively), these are thought to be due to the correction equation used to calculate the indel percentage based on the T7E1 assay (see the Materials and Methods), which may be excessively conservative. If we do not use this formula, the cleavage efficiency on HDFs increases from 6% to 12%, resembling more the 20.1% count based on deep-sequencing data.
The retina has emerged as an increasingly promising tissue for genome editing-based and cell transplantation therapies, owing to its unique features as a surgically accessible, immune privileged organ that can be non-invasively imaged. The development of site-specific CRISPR-based genome editing approaches to correct the genetic mutation causing blindness will provide potential strategies for future gene therapy in patients.
There are a number of researchers carrying out preclinical studies using induced pluripotent stem cells (iPSCs) or photoreceptor cells for transplantation in the retina,45, 46 but these approaches still have limitations. In the case of autologous transplants, the replacement cells still carry the mutations responsible for the pathology and are thus a short-term solution. Otherwise, the use of donor cells carries the usual risk of tissue rejection.45, 46 Another alternative strategy would be to use the CRISPR/Cas9 system on the iPSCs prior to transplantation in order to deliver repaired self-donated cells into the retina.47, 48, 49 However, producing and maintaining iPSCs from fibroblasts is expensive and time-consuming; therefore, attempting gene targeting in these cells may not be affordable for laboratories lacking the necessary expertise. With reference to this issue, several groups have claimed successful iPSC-derived photoreceptor progenitor (PhRP) transplantations in animal models, which form functional synapses with the host bipolar cells and partially restore visual function based on electrophysiology and anatomical diagnosis.50, 51, 52 In contrast, feasible visual recovery on human translation still remains unproven and many questions need to be answered before these techniques can be transferred to clinical trials for RP patients.
An ultimate option would be the in vivo photoreceptor targeting with CRISPR and, for that purpose, different methods are currently being tested such as AAV-based delivery strategies or even other carriers for RNPs like nanoparticles, cell-penetrating peptides, exosomes, or liposomes.53, 54, 55, 56, 57, 58, 59, 60, 61 Utilizing these non-viral vehicles for delivery would likely lead to a less toxic approach, also allowing for the benefit of preserving the previously designed RNP protocol from the present study.
The HDR rate achieved in this study with HDFs is still too low to be considered for an in vivo procedure. Other strategies that should be investigated for increasing HDR efficiencies have been developed in recent years, such as the use of chemically modified or asymmetric ssODNs or the addition of NHEJ inhibitors to the culture medium.62, 63, 64 Another controversial issue worth considering is the fact that post-mitotic cells are considered as lacking the HDR-mediated repair mechanism,22 yet recent studies have detected HDR in retinal pigment epithelial cells and developed adult photoreceptors.8 Nevertheless, even if HDR is considered as non-active, it has also been demonstrated that non-dividing cells do have a repair mechanism in the form of transcription-coupled homologous recombination,65 enabling a repair pathway through an RNA template instead of ssODNs. The only major hindrance in using this technique would be the remote chance of introducing a mutation at potential off-target sites, thereby resulting in oncological consequences. However, the design of this project indicated no evidence for off-target activity from the final treated cells. In addition, some studies relating to this dilemma have recently been published, offering improved Cas9 engineered versions with an off-target reduction, such as eSpCas9(1.1) or SpCas9-HF1.66, 67 NHEJ repair would still outline HDR, but since USH is an autosomal recessive disease and both gene alleles are already nonfunctional, the possible non-repaired indel scar would have a neutral impact on the photoreceptor phenotype of the patients.
Some attempts to correct the underlying genetic defect of USH, such as the use of antisense oligonucleotides (AONs) or cDNA supplementation,68, 69 are ongoing. Furthermore, a study was recently published where the visual function of an RP animal model seemed to be restored after correcting the genetic cause by CRISPR/Cas9 techniques.70 In conclusion, the present study demonstrates for the first time that correcting the two most prevalent mutations of USH is feasible using the CRISPR/Cas9 system and homologous recombination, thereby offering future promise for repairing these and other mutations in cells from patients.
Materials and Methods
This study was approved by the University Hospital La Fe Research Ethics Committee.
Design of the sgRNAs
Four different sgRNAs (more specifically, the crRNA domain) were designed according to the following criteria: close location to the mutation(s); following PAM at the 3′ end; and 18-nt length, since it has been demonstrated that a 2-nt shortening in the sequence length brings higher specificity rather than the initially proposed 20-nt design.71
Plasmid Constructions
The sgRNA consists of two RNA domains: crispr RNA (crRNA), which is specifically designed to be complementary to the target locus, and the constant trans-activating crRNA (tracrRNA), which is required for coupling with the Cas9 nuclease.
In order to create each Cas9-sgRNA construct, crRNAs were cloned in pX330-U6-Chimeric_BB-CBh-hSpCas9 (a gift from Feng Zhang, plasmid no. 42230; Addgene). The two opposite BbsI restriction sites were used to insert the guide under the control of a U6 promoter and were then linked to the tracrRNA sequence. For this purpose, self-complementary oligonucleotides (Integrated DNA Technologies, IA, USA) with the corresponding crRNA sequences were annealed by gradual cooling with prior denaturalization at 94°C. The duplex oligonucleotides with the crRNA sequences also presented cohesive ends with the 3′ overhangs left after pX330 incubation with the BbsI restriction enzyme, serving for the ligation of the insert-plasmid with T4 DNA ligase (EL0014; Thermo Fisher Scientific, Waltham, MA, USA). Top 10 Escherichia coli electrocompetent cells were transformed by electroporation with each of the plasmid constructs for their plate selection and amplification in liquid culture. The vectors were purified using a HiSpeed Plasmid Midi Kit (no. 12643; QIAGEN, Hilden, Germany) and were Sanger sequenced to verify the correct cloning of the specific crRNA inserts.
HEK293 Cell Culture and Transfections
HEK293 cells were maintained at 37°C and 5% CO2 in DMEM high glucose without L-glutamine without sodium pyruvate medium (Biowest SAS, Nuaillé, France) supplemented with 10% fetal bovine serum (FBS) (Biowest) and 1% penicillin-streptomycin (PS) solution 100× (Biowest).
For the functional validation of the RGEN activity, 5 × 105 HEK293 cells were separately transfected with each of the plasmid constructs at 1 μg and 3 μg concentrations using Lipofectamine 3000 (no. L3000001; Invitrogen, Carlsbad, CA, USA).
For the directed edition, 3 μg RGEN was delivered to 5 × 105 HEK293 cells along with 1 μL of the pertinent ssODN at 10 μM (10 pmol). Parallel GFP transfections were carried out in all transfection assays as positive controls. All experiments were assayed in duplicate.
T7-Endonuclease 1 Assay
The widely used T7-endonucelase I assay targets and digests heteroduplexes formed by hybridization of mutant WT strands resulting in two smaller fragments,72 and this method was performed to assess sgRNA-specific activity.
After transfection, cells were incubated for 48 hr. The cells were then pelleted and resuspended in 10 μL PBS. For lysis, 20 μL lysis buffer (0.3 mM Tris/HCl, 0.6 mM CaCl2, 1.5% glycerol, 0.675% Tween 20, 0.3 μg/μL proteinase K, and 0.954% H2O) were added and the suspension was treated at 65°C for 30 min, 90°C for 10 min, and then cooled for 4°C. Finally, 30 μL H2O was added to the lysed product. The target locus was amplified for 35 cycles with specific forward (5′-GGCATTGCTTGTGAGAAAACAC-3′) and reverse (5′-CAGATGTGTGAGTGTGATTCCT-3′) primers targeting exon 13 of the USH2A locus with KAPA HiFi HotStart DNA Polymerase (no. KK2501; Kapa Biosystems, Wilmington, MA, USA). These PCR products were gel purified using the E.Z.N.A. Gel Extraction Kit (no. D2500-02; Omega Bio-tek, Norcross, GA, USA). For heteroduplex formation, 200 ng purified DNA amplification was denatured and then reannealed using the following program: 95°C for 5 min, ramp down to 85°C at −2°C/s, and ramp down to 25°C at −0.1°C/s. At this step, 1 μL T7 endonuclease I (no. M0302S; New England Biolabs, Ipswich, MA) was added to the mix and incubated for 15 min at 37°C. The reaction was stopped by adding 2.5 μL of 0.20 M EDTA. The digested product was immediately loaded on a 2% agarose gel and stained with SYBR Safe DNA Gel Stain (no. S33102; Invitrogen). Indel frequencies were estimated as previously described by calculating band intensities with ImageJ (NIH) software and applying the following equation:
where is the fraction cleaved, corresponding to the sum of intensities of the cleaved bands divided by the sum of total band intensities.72, 73, 74
Design of the ssODNs
Single-stranded ODNs for the HDR were designed with 90-nt-long homology arms, including the specific change (as for the mutation of interest) in the core. Further changes with silent effect were incorporated in the template to prevent Cas9 from reiterated cleavage after HDR, by means of the PAM sequence removal exchanging the last guanine of the sequence for a cytosine nucleotide to escape Cas9 recognition. Several in silico web tools (Human Splicing Finder,75 NetGene2,76 and MaxEnt77) were used to test the possible splicing alterations due to this synonymous nucleotide change, resulting in zero predicted effects. The introduced variant also involved a new recognition site for the specific restriction enzyme MlsI (no. FD1214; Thermo Fisher Scientific), which in turn aided RFLP analysis for the edition testing. These long ssODNs were synthesized as Ultramer Oligonucleotides (Integrated DNA Technologies).
RFLP Analysis for Edition Testing
gDNA extraction, target amplification, and purification were performed identically to the T7E1 assay. Digestion with corresponding restriction enzymes was fulfilled according to the manufacturer’s instructions using 300 ng purified product and was subsequently resolved on a 4%–20% high-resolution Tris-borate-EDTA (TBE) polyacrylamide gel (no. 4561093S; Bio-Rad, Hercules, CA, USA), following staining with SYBR Safe DNA Gel Stain.
Clonal Amplicon Sanger Sequencing
A PCR product obtained from the target locus was cloned using a kit for sequencing purposes (TOPO TA Cloning Kit, no. K457502; Thermo Fisher Scientific) and introduced in E. coli.
Sanger sequencing was used to sequence 45 individual colonies to reveal the clonal genotype and thus the general indel frequency.
HDF Cell Culture and Transfections
Fibroblasts from WT controls and USH2 patients harboring the c.2299delG homozygotic mutation were isolated from skin biopsies and were maintained at 37°C and 5% CO2 in Biowest DMEM high glucose without L-glutamine, without sodium pyruvate medium supplemented with 10% FBS, 1% Biowest PS solution 100×, and 1% L-glutamine (no. X0550; Biowest).
Plasmid delivery trials were performed with several compounds: namely, Lipofectamine LTX (Thermo Fisher Scientific), Lipofectamine 3000 (Thermo Fisher Scientific), NanoJuice (Merck Millipore), polyethylenimine (Polysciences), Viromer (Lipocalyx), and Nucleofector 4D (Lonza), parallel to pmaxGFP transfection as a positive control. Neither the control GFP vector nor the CRISPR constructs showed significant results; the former achieved less than a 10% transfection efficiency with all the transfection products, except for Lipofectamine 3000, which showed no GFP-positive cells at all (data not shown). For that reason, the Cas9-sgRNA design was shifted from DNA usage to the RNP strategy, according to protocols from previous studies.37
For the RNP complexes, 15 μg purified Cas9 endonuclease (no. M0646T; New England Biolabs) was blended with 20 μg IVT synthesized sgRNA (PNA Bio, CA, USA) by 10-min incubation at room temperature with 1× Cas9 Nuclease Reaction Buffer (New England Biolabs).
For HDF targeting, nucleofection assays were performed using program DT-130 on the 4D-Nucleofector System (Lonza, Basel, Switzerland). Premixed RNPs were added to 100 μL P2 nucleofection solution where 2 × 105 HDFs had been previously resuspended for the nucleofection. The same concentration was used for the directed edition, together with 5 μL ssODN-WT at 100 μM (500 pmol) resuspended in the nucleofection solution. All experiments were assayed in duplicate.
Analysis of Potential Off-Targets
For the off-target analysis, two free-access bioinformatic prediction tools were used: Cas-OFFinder78 from RGEN tools and the WTSI Genome Editing (WGE) tool from the Wellcome Trust Sanger Institute.79 These were used to search for algorithms for highly similar sgRNA sequences throughout the genome. Algorithm parameters were set for a maximum of 2 mismatches, 2 bulges, or the combination of 1 mismatch and 1 bulge. The bulges appear when insertions or deletions are present in the DNA compared to the sgRNA sequence, forming a DNA bulge in the case of an insertion and an RNA bulge in the case of a deletion. The outcome of 45 potential off-targets was filtered by selecting all bulge-involving loci and those with mismatches with a higher impact, meaning they were located in genes. Two possible additional exonic off-targets were included despite the higher number of mismatches, in view of the risky repercussion. Putative off-target sites were ranked according to previous studies, where the quantity and PAM proximity of the dissimilarities have been proven to be a key factor for site recognition probability.27, 80, 81
High-Throughput Sequence Analysis
Deep sequencing with a 500-cycle v2 Nano Reagent Kit (Illumina, San Diego, CA, USA) on a MiSeq platform (Illumina) was performed in order to precisely determine sgRNA-1 on-target activity in HDFs, either NHEJ events or efficiently edited cells by HDR, and potential off-target issues. For this purpose, in silico predicted off-target loci and on-target regions from sgRNA-1 treated cell gDNA were amplified. Library preparation was fulfilled by a two-step PCR. The initial PCR was performed with specific locus primers with flanking TruSeq partial sequence adapters (Table 2). For the second PCR, primers with TruSeq adaptor sequences overlapping with PCR1 primers were used. The PCR products were pooled in equimolar amounts following the manufacturer’s protocol. The sequencing yield exposed overall coverage >50,000 for each target.
Table 2.
Locus-Specific Primer Sequences for MiSeq Deep Sequencing
| Locus Name | 5′-3′ Forward Primera | 5′-3′ Reverse Primerb |
|---|---|---|
| USH2A (on-target) | AGGGCTTAGGTGTGATCATTGC | TAGCATTACAGACAGTCCCAGG |
| STK3 | TCATGCTCAGTGCACTGTAC | CTCTGTTCACAGATGATATG |
| FHIT | CTTCTCTTGACTAGGAAAGG | CCACCATTTAAAAGCCTCCC |
| TENM2 | TTGCACTTCCCAGATGAGGTG | GGATATGAACTTTCTCCAACAC |
| ROR1 | CAGAATTCCGATTTCTTGTCTC | GCCCACCATTATAACATTTCAG |
| SPG20 | GCAAAGATATAATGGACATGG | TGACAGATAATCTTCTGATCC |
| TBR1 | TGCTGGTGCCCTTTTCTTTAGG | TGTCTCAGGCGCTCAATGTAAG |
| DMD | AGAGGAACACACCAAATCTGG | AGCCACGTATTTGTCTTTCC |
| RGS7BP | CAGTGTTCTCAATGTCTGTGG | TGACTCAAAGCATAGCCAACC |
| EHD1 | CCTCTTAGTGTGTGCTTTGG | AGTGGATGCTTCAGTTGCTG |
| KLHL15 | ACATAAATTGCAGGACATGGC | AGGATTCTGTTCCTCCATCC |
| Intergenic | GTGCATACAAGAGTGGCTTG | ACCCTTCTGGATGAAAGCTG |
| TIM22 | TGGTCTTCTCGGCAGAGATC | AAGCTCCGCTGCAGTACAGC |
| HNRNPUL2 | CTCCATTTCTCTTCCAGTGG | TGCGACACATCTTAGCTCG |
| C5orf66 | AGTCTCTGAGAGGCTTTGGATG | ACAGCTTGGGTGTTCAGAAACC |
| RBFOX1 | CTAAGTGCACTACATGGTCACC | ACTGCCAAGAAGAGCTTCTTCC |
| LOC730100 | AAAGATTGACCTCAGAGCC | GTAAATATGCTGCAGAGAC |
| IGKV1-33 | TACAACTGCCAATCATGTGGTC | AGAAACCCTATGCCTTCTCTGG |
| IGKV1D-33 | ATAGAGAAGGTACAACTGCC | CAGAAACCCTATGCCTTCTC |
| BAZ2B | TGCTAAGGTAATCAAAGTG | CTTTGAGAGAGTTTCACTC |
| TANC2 | TGTGAGAATCCTCCACTTTGG | ATGAGGTAGCAGTGAACTGTG |
| SLX4IP | GGAAATTCTCATATGGCTGG | GCTATTCCCAACAGTTTGTG |
Forward primers have appended the last 5′-3′ 34 bases of the TruSeq adaptor sequence: TACACTCTTTCCCTACACGACGCTCTTCCGATCT.
Reverse primers have appended the first 3′-5′ 34 bases of the TruSeq adaptor sequence: GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT.
For the data analysis, CRISPResso82 was used applying the following parameters: adaptor and 50-bp trimming, mean average quality 30 phred score, minimum (100) and maximum (400) paired end reads overlap, and ignoring substitutions and hiding mutations outside the NHEJ region. The on-target locus was studied for cleavage and edition quantification, considering all the reads with only the two introduced variants (c.2299insG and c.2292G>C) as edited sequences. The CRISPR system is supposed to cleave virtually 3–4 bp upstream of any preceding PAM sequence,83 yet the previous clonal amplicon Sanger sequencing on HEK293 cells showed evidence repair scars that appeared displaced some bases from the canonic cutting spot (Figure 2B). Therefore, a 10-bp window for the survey was established, straying from the canonical cleavage point. A parallel custom data analysis was performed and the results were equivalent to the outcome from the CRISPResso tool. The number of cleaved sequences was calculated by the sum of reads with indels enclosed in this frame and the number of edited cells that had been cleaved in the first instance.
Author Contributions
Conceptualization, C.F.-G. and E.A.; Methodology, C.F.-G., G.G.-G., and E.A.; Investigation, C.F.-G., G.G.-G., E.G.-R., and M.D.S.; Writing – Original Draft, C.F.-G.; Writing – Review & Editing, G.G.-G, T.J., R.P.V.-M., J.M.M., and E.A.; Resources, R.V.P.-M., C.A., and E.A.; Supervision, T.J., R.P.V.M., J.M.M., and E.A.; Funding Acquisition, J.M.M. and E.A.
Acknowledgments
We thank the patients involved in this study and the patient associations Retina CV and FARPE. We thank the CIBERER biobank (Valencia, Spain) for sharing the skin biopsy-derived HDFs from one of the control patients, and we also thank dermatologist Montserrat Évole for the medical procedure. We also thank Paul E. Gutteridge for the grammatical revision of the English. We acknowledge Sheila Zúñiga at the Medical Research Institute Hospital La Fe and Galo A. Goig from the Genomics and Health Unit (Fundación para el Fomento de la Investigación Sanitaria y Biomédica de la Comunitat Valenciana [FISABIO]) for help with the bioinformatics analysis. We are grateful to Dr. Juan Bueren (Centro de Investigaciones Energéticas, Medioambientales y Tecnológicas [CIEMAT]) and Dr. Lluís Montoliu (Centro Nacional de Biotecnología-Centro Superior de Investigaciones Científicas [CNB-CSIC]) for their technical support. This work was financially supported by the Institute of Health Carlos III and FEDER funds (ISCIII; grants PI13/00638, PI16/00425, PI16/00539, and PIE13/00046), Fundación ONCE (grant 2015/0398), XVIII Fundaluce-FARPE, and “Telemaratón: Todos Somos Raros, Todos Somos Únicos” (grant IP58). C.F.-G. is a recipient of a fellowship (grant IFI14/00021) from the ISCIII. R.P.V.-M. is a Miguel Servet researcher (grant CP11/00090 funded by ISCIII, Madrid, Spain). The funds from the ISCIII are partially supported by the European Regional Development Fund. R.-P.V.M. is also a Marie Curie fellow (grant CIG322034 from the European Commission).
References
- 1.Millán J.M., Aller E., Jaijo T., Blanco-Kelly F., Gimenez-Pardo A., Ayuso C. An update on the genetics of Usher syndrome. J. Ophthalmol. 2011;2011:417217. doi: 10.1155/2011/417217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathur P., Yang J. Usher syndrome: hearing loss, retinal degeneration and associated abnormalities. Biochim. Biophys. Acta. 2015;1852:406–420. doi: 10.1016/j.bbadis.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnet C., Grati M., Marlin S., Levilliers J., Hardelin J.P., Parodi M., Niasme-Grare M., Zelenika D., Délépine M., Feldmann D. Complete exon sequencing of all known Usher syndrome genes greatly improves molecular diagnosis. Orphanet J. Rare Dis. 2011;6:21. doi: 10.1186/1750-1172-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenarduzzi S., Vozzi D., Morgan A., Rubinato E., D’Eustacchio A., Osland T.M., Rossi C., Graziano C., Castorina P., Ambrosetti U. Usher syndrome: an effective sequencing approach to establish a genetic and clinical diagnosis. Hear. Res. 2015;320:18–23. doi: 10.1016/j.heares.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Hartong D.T., Berson E.L., Dryja T.P. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 6.Baux D., Blanchet C., Hamel C., Meunier I., Larrieu L., Faugère V., Vaché C., Castorina P., Puech B., Bonneau D. Enrichment of LOVD-USHbases with 152 USH2A genotypes defines an extensive mutational spectrum and highlights missense hotspots. Hum. Mutat. 2014;35:1179–1186. doi: 10.1002/humu.22608. [DOI] [PubMed] [Google Scholar]
- 7.Aller E., Nájera C., Millán J.M., Oltra J.S., Pérez-Garrigues H., Vilela C., Navea A., Beneyto M. Genetic analysis of 2299delG and C759F mutations (USH2A) in patients with visual and/or auditory impairments. Eur. J. Hum. Genet. 2004;12:407–410. doi: 10.1038/sj.ejhg.5201138. [DOI] [PubMed] [Google Scholar]
- 8.Williams D.S., Chadha A., Hazim R., Gibbs D. Gene therapy approaches for prevention of retinal degeneration in Usher syndrome. Gene Ther. 2017;24:68–71. doi: 10.1038/gt.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabebordbar M., Zhu K., Cheng J.K.W., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox D.B.T., Platt R.J., Zhang F. Therapeutic genome editing: prospects and challenges. Nat. Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai W.-J., Zhu L.-Y., Yan Z.-Y., Xu Y., Wang Q.-L., Lu X.-J. CRISPR-Cas9 for in vivo gene therapy: promise and hurdles. Mol. Ther. Nucleic Acids. 2016;5:e349. doi: 10.1038/mtna.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 18.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnes D.E. Non-homologous end joining as a mechanism of DNA repair. Curr. Biol. 2001;11:R455–R457. doi: 10.1016/s0960-9822(01)00279-2. [DOI] [PubMed] [Google Scholar]
- 21.van den Bosch M., Lohman P.H.M., Pastink A. DNA double-strand break repair by homologous recombination. Biol. Chem. 2002;383:873–892. doi: 10.1515/BC.2002.095. [DOI] [PubMed] [Google Scholar]
- 22.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong C., Bongiorno P., Martins A., Stephanou N.C., Zhu H., Shuman S., Glickman M.S. Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat. Struct. Mol. Biol. 2005;12:304–312. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- 24.Chen F., Pruett-Miller S.M., Huang Y., Gjoka M., Duda K., Taunton J., Collingwood T.N., Frodin M., Davis G.D. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storici F., Snipe J.R., Chan G.K., Gordenin D.A., Resnick M.A. Conservative repair of a chromosomal double-strand break by single-strand DNA through two steps of annealing. Mol. Cell. Biol. 2006;26:7645–7657. doi: 10.1128/MCB.00672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radecke S., Radecke F., Cathomen T., Schwarz K. Zinc-finger nuclease-induced gene repair with oligodeoxynucleotides: wanted and unwanted target locus modifications. Mol. Ther. 2010;18:743–753. doi: 10.1038/mt.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Y., Cradick T.J., Brown M.T., Deshmukh H., Ranjan P., Sarode N., Wile B.M., Vertino P.M., Stewart F.J., Bao G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H.J., Lee H.J., Kim H., Cho S.W., Kim J.-S. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parks R.J., Chen L., Anton M., Sankar U., Rudnicki M.A., Graham F.L. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc. Natl. Acad. Sci. USA. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melendez M.E., Fraefel C., Epstein A.L. Herpes simplex virus type 1 (HSV-1)-derived amplicon vectors. Methods Mol. Biol. 2014;1144:81–98. doi: 10.1007/978-1-4939-0428-0_6. [DOI] [PubMed] [Google Scholar]
- 31.Hibbitt O.C., Wade-Martins R. Delivery of large genomic DNA inserts >100 kb using HSV-1 amplicons. Curr. Gene Ther. 2006;6:325–336. doi: 10.2174/156652306777592054. [DOI] [PubMed] [Google Scholar]
- 32.Kumar-Singh R. Barriers for retinal gene therapy: separating fact from fiction. Vision Res. 2008;48:1671–1680. doi: 10.1016/j.visres.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraefel C., Mendes-Madeira A., Mabon O., Lefebvre A., Le Meur G., Ackermann M., Moullier P., Rolling F. In vivo gene transfer to the rat retina using herpes simplex virus type 1 (HSV-1)-based amplicon vectors. Gene Ther. 2005;12:1283–1288. doi: 10.1038/sj.gt.3302553. [DOI] [PubMed] [Google Scholar]
- 34.Cashman S.M., McCullough L., Kumar-Singh R. Improved retinal transduction in vivo and photoreceptor-specific transgene expression using adenovirus vectors with modified penton base. Mol. Ther. 2007;15:1640–1646. doi: 10.1038/sj.mt.6300203. [DOI] [PubMed] [Google Scholar]
- 35.Mortensen R. Overview of gene targeting by homologous recombination. Curr. Protoc. Neurosci. 2007;Chapter 4 doi: 10.1002/0471142301.ns0429s40. Unit 4.29. [DOI] [PubMed] [Google Scholar]
- 36.Burgess D.J. Technology: a CRISPR genome-editing tool. Nat. Rev. Genet. 2013;14:80. doi: 10.1038/nrg3409. [DOI] [PubMed] [Google Scholar]
- 37.Kim S., Kim D., Cho S.W., Kim J., Kim J.-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kouranova E., Forbes K., Zhao G., Warren J., Bartels A., Wu Y., Cui X. CRISPRs for optimal targeting: delivery of CRISPR components as DNA, RNA, and protein into cultured cells and single-cell embryos. Hum. Gene Ther. 2016;27:464–475. doi: 10.1089/hum.2016.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang X., Potter J., Kumar S., Zou Y., Quintanilla R., Sridharan M., Carte J., Chen W., Roark N., Ranganathan S. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 41.Vojta A., Dobrinić P., Tadić V., Bočkor L., Korać P., Julg B., Klasić M., Zoldoš V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X., Rinsma M., Janssen J.M., Liu J., Maggio I., Gonçalves M.A.F.V. Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res. 2016;44:6482–6492. doi: 10.1093/nar/gkw524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song F., Stieger K. Optimizing the DNA donor template for homology-directed repair of double-strand breaks. Mol. Ther. Nucleic Acids. 2017;7:53–60. doi: 10.1016/j.omtn.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coonrod A., Li F.Q., Horwitz M. On the mechanism of DNA transfection: efficient gene transfer without viruses. Gene Ther. 1997;4:1313–1321. doi: 10.1038/sj.gt.3300536. [DOI] [PubMed] [Google Scholar]
- 45.Pearson R.A., Gonzalez-Cordero A., West E.L., Ribeiro J.R., Aghaizu N., Goh D., Sampson R.D., Georgiadis A., Waldron P.V., Duran Y. Donor and host photoreceptors engage in material transfer following transplantation of post-mitotic photoreceptor precursors. Nat. Commun. 2016;7:13029. doi: 10.1038/ncomms13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez-Cordero A., West E.L., Pearson R.A., Duran Y., Carvalho L.S., Chu C.J., Naeem A., Blackford S.J.I., Georgiadis A., Lakowski J. Photoreceptor precursors derived from three-dimensional embryonic stem cell cultures integrate and mature within adult degenerate retina. Nat. Biotechnol. 2013;31:741–747. doi: 10.1038/nbt.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ou Z., Niu X., He W., Chen Y., Song B., Xian Y., Fan D., Tang D., Sun X. The combination of CRISPR/Cas9 and iPSC technologies in the gene therapy of human β-thalassemia in mice. Sci. Rep. 2016;6:32463. doi: 10.1038/srep32463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grobarczyk B., Franco B., Hanon K., Malgrange B. Generation of isogenic human iPS cell line precisely corrected by genome editing using the CRISPR/Cas9 system. Stem Cell Rev. 2015;11:774–787. doi: 10.1007/s12015-015-9600-1. [DOI] [PubMed] [Google Scholar]
- 49.Li H.L., Gee P., Ishida K., Hotta A. Efficient genomic correction methods in human iPS cells using CRISPR-Cas9 system. Methods. 2016;101:27–35. doi: 10.1016/j.ymeth.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 50.Barnea-Cramer A.O., Wang W., Lu S.-J., Singh M.S., Luo C., Huo H., McClements M.E., Barnard A.R., MacLaren R.E., Lanza R. Function of human pluripotent stem cell-derived photoreceptor progenitors in blind mice. Sci. Rep. 2016;6:29784. doi: 10.1038/srep29784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tucker B.A., Mullins R.F., Streb L.M., Anfinson K., Eyestone M.E., Kaalberg E., Riker M.J., Drack A.V., Braun T.A., Stone E.M. Patient-specific iPSC-derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. eLife. 2013;2:e00824. doi: 10.7554/eLife.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Homma K., Okamoto S., Mandai M., Gotoh N., Rajasimha H.K., Chang Y.-S., Chen S., Li W., Cogliati T., Swaroop A., Takahashi M. Developing rods transplanted into the degenerating retina of Crx-knockout mice exhibit neural activity similar to native photoreceptors. Stem Cells. 2013;31:1149–1159. doi: 10.1002/stem.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang D., Chen Y.-S., Rupenthal I.D. Hyaluronic acid coated albumin nanoparticles for targeted peptide delivery to the retina. Mol. Pharm. 2017;14:533–545. doi: 10.1021/acs.molpharmaceut.6b01029. [DOI] [PubMed] [Google Scholar]
- 55.Li J., Wang Y., Zhu Y., Oupický D. Recent advances in delivery of drug-nucleic acid combinations for cancer treatment. J. Control. Release. 2013;172:589–600. doi: 10.1016/j.jconrel.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han C., Sun X., Liu L., Jiang H., Shen Y., Xu X., Li J., Zhang G., Huang J., Lin Z. Exosomes and their therapeutic potentials of stem cells. Stem Cells Int. 2016;2016:7653489. doi: 10.1155/2016/7653489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee J., Goh U., Lee H.J., Kim J., Jeong M., Park J.H. Effective retinal penetration of lipophilic and lipid-conjugated hydrophilic agents delivered by engineered liposomes. Mol. Pharm. 2017;14:423–430. doi: 10.1021/acs.molpharmaceut.6b00864. [DOI] [PubMed] [Google Scholar]
- 58.Mishra G.P., Bagui M., Tamboli V., Mitra A.K. Recent applications of liposomes in ophthalmic drug delivery. J. Drug Deliv. 2011;2011:863734. doi: 10.1155/2011/863734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., Rajala A., Cao B., Ranjo-Bishop M., Agbaga M.-P., Mao C., Rajala R.V. Cell-specific promoters enable lipid-based nanoparticles to deliver genes to specific cells of the retina in vivo. Theranostics. 2016;6:1514–1527. doi: 10.7150/thno.15230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao L., Chen G., Li J., Fu Y., Mavlyutov T.A., Yao A., Nickells R.W., Gong S., Guo L.W. An intraocular drug delivery system using targeted nanocarriers attenuates retinal ganglion cell degeneration. J. Control. Release. 2017;247:153–166. doi: 10.1016/j.jconrel.2016.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang K., Xu Z.P., Lu J., Tang Z.Y., Zhao H.J., Good D.A., Wei M.Q. Potential for layered double hydroxides-based, innovative drug delivery systems. Int. J. Mol. Sci. 2014;15:7409–7428. doi: 10.3390/ijms15057409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Renaud J.-B., Boix C., Charpentier M., De Cian A., Cochennec J., Duvernois-Berthet E., Perrouault L., Tesson L., Edouard J., Thinard R. Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR-Cas9 nucleases. Cell Rep. 2016;14:2263–2272. doi: 10.1016/j.celrep.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 63.Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 64.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei L., Levine A.S., Lan L. Transcription-coupled homologous recombination after oxidative damage. DNA Repair (Amst.) 2016;44:76–80. doi: 10.1016/j.dnarep.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 66.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slijkerman R.W., Vaché C., Dona M., García-García G., Claustres M., Hetterschijt L., Peters T.A., Hartel B.P., Pennings R.J., Millan J.M. Antisense oligonucleotide-based splice correction for USH2A-associated retinal degeneration caused by a frequent deep-intronic mutation. Mol. Ther. Nucleic Acids. 2016;5:e381. doi: 10.1038/mtna.2016.89. [DOI] [PubMed] [Google Scholar]
- 69.Lopes V.S., Boye S.E., Louie C.M., Boye S., Dyka F., Chiodo V., Fofo H., Hauswirth W.W., Williams D.S. Retinal gene therapy with a large MYO7A cDNA using adeno-associated virus. Gene Ther. 2013;20:824–833. doi: 10.1038/gt.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J., Hatanaka F., Yamamoto M., Araoka T., Li Z. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 73.Miller J.C., Holmes M.C., Wang J., Guschin D.Y., Lee Y.-L., Rupniewski I., Beausejour C.M., Waite A.J., Wang N.S., Kim K.A. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 74.Reyon D., Tsai S.Q., Khayter C., Foden J.A., Sander J.D., Joung J.K. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Desmet F.-O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brunak S., Engelbrecht J., Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- 77.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 78.Bae S., Park J., Kim J.-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hodgkins A., Farne A., Perera S., Grego T., Parry-Smith D.J., Skarnes W.C., Iyer V. WGE: a CRISPR database for genome engineering. Bioinformatics. 2015;31:3078–3080. doi: 10.1093/bioinformatics/btv308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anderson E.M., Haupt A., Schiel J.A., Chou E., Machado H.B., Strezoska Ž., Lenger S., McClelland S., Birmingham A., Vermeulen A., Smith Av. Systematic analysis of CRISPR-Cas9 mismatch tolerance reveals low levels of off-target activity. J. Biotechnol. 2015;211:56–65. doi: 10.1016/j.jbiotec.2015.06.427. [DOI] [PubMed] [Google Scholar]
- 81.Zhu H., Misel L., Graham M., Robinson M.L., Liang C. CT-Finder: a web service for CRISPR optimal target prediction and visualization. Sci. Rep. 2016;6:25516. doi: 10.1038/srep25516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Swarts D.C., Mosterd C., van Passel M.W.J., Brouns S.J.J. CRISPR interference directs strand specific spacer acquisition. PLoS ONE. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]