Abstract
Aminopeptidase N (APN) is a member of the highly conserved M1 family of metalloproteases, and is considered to be a valuable target for the treatment of a variety of diseases, e.g., cancer, malaria, and coccidiosis. In this study, we identified an APN gene (TgAPN2) in the Toxoplasma gondii genome, and performed a biochemical characterization of the recombinant TgAPN2 (rTgAPN2) protein. Active rTgAPN2 was first produced and purified in Escherichia coli. The catalytic activity of the enzyme was verified using a specific fluorescent substrate, H-Ala-MCA; the rTgAPN2 was relatively active in the absence of added metal ions. The addition of some metal ions, especially Zn2+, inhibited the activity of the recombinant enzyme. The activity of rTgAPN2 was reduced in the presence of the EDTA chelator in the absence of added metal ions. The optimum pH for enzyme activity was 8.0; the enzyme was active in the 3–10 pH range. The substrate preference of rTgAPN2 was evaluated. The enzyme showed a preference for substrates containing N-terminal Ala and Arg residues. Finally, bestatin and amastatin were shown to inhibit the activity of the enzyme. In conclusion, rTgAPN2 shared general characteristics with the M1 family of aminopeptidases but also had some unique characteristics. This provides a basis for the function of aminopeptidases and the study of drug targets.
Keywords: activity, aminopeptidase N (APN), enzyme, Toxoplasma gondii
Aminopeptidases, which catalyze the cleavage of N-terminal amino acid residues in proteins and peptides, perform vital cellular housekeeping roles, and as such have been the targets of therapeutic intervention strategies in a range of diseases [3]. Aminopeptidase N (APN) belongs to the M1 family of peptidases and depends on a single zinc ion for activity; all members of the family exhibit an aminopeptidase activity. The catalytic zinc ion is bound by two histidines and a glutamate. The histidines are part of an HEXXH motif on one helix, with the glutamate located on another antiparallel helix [23]. The catalytic mechanism is believed to involve activation of a water molecule by the zinc ion. The glutamate of the HEXXH motif is important for catalysis, and a tyrosine residue may also be involved.
In parasite, Plasmodium falciparum M1 alanyl-aminopeptidase (PfA-M1) is an enzyme involved in the terminal stages of hemoglobin digestion and the generation of an amino acid pool within the parasite. PfA-M1 is essential for protein synthesis and metabolism of the parasite. The enzyme has been validated as a potential drug target since inhibitors of the enzyme block parasite growth in vitro and in vivo [16]. APN of another protozoan, Eimeria tenella, shares similarity with PfA-M1; specific bestatin-derived APN inhibitors abrogate parasite development, rendering APN a valuable target for anticoccidiosis drugs [9]. Cryptosporidium parvum aminopeptidase N was found to be highly conserved when compared with other Apicomplexan aminopeptidases. The aminopeptidase was expressed in infective sporozoites and during the infection of human HCT-8 enterocytes [18]. As immunogenic antigens, two M1 type aminopeptidases of Trypanosoma congolense were identified by immunoprecipitation from infected trypan tolerant cattle. The aminopeptidases showed a distinct substrate preference for H-Ala-AMC, an optimum pH of 8.0, inhibition by bestatin and amastatin, and cytoplasmic localization. Down-regulation of both APs by RNAi resulted in a slightly reduced growth rate in procyclic parasites in vitro [20].
Toxoplasma gondii is a model protozoan [27]. T. gondii can also be used as a research model for malignant tumors according to the previous report [15], since the RH strain of T. gondii behaves as a protozoan equivalent of cancer. In human, APN plays an important role in many physiological or pathological processes, such as pain, regulation of blood pressure, tumor angiogenesis and metastasis [19, 28]. In contrast with normal cells, APN is overexpressed on the surface of tumor cells and degrades the extracellular matrix, thereby promoting tumor cell infiltration and metastasis [12]. It can thus be imagined that the study of toxoplasma proteins would not only benefit the development of anti-toxoplasmosis drugs, but may also help to better understand the origin of cancer.
According to the previous report [10] and sequence alignment in the ToxoDB, there are 3 APNs in T. gondii genome that belong to the M1 metalloprotease family (TgAPN1, TgAPN2 and TgAPN3). To date, no data are available on the biochemical activity characteristics of TgAPNs. Recent analysis of the T. gondii genome led to the identification of a gene encoding a new putative APN-like protease (TgAPN2) that belongs to the M1 metalloprotease family. In this study, this is the first report on the enzymatic activity of a recombinant TgAPN2 protein toward synthetic aminopeptidase substrates. Studying the catalytic properties of the enzyme may facilitate the understanding of biochemical characteristics of the M1 family metalloproteinases, and instigate the research and development of APN-targeting anti-toxoplasmosis drugs or other drugs.
MATERIALS AND METHODS
Parasite strains and growth conditions
Tachyzoites of T. gondii RH strain were maintained in human foreskin fibroblast (HFFs) cells or Vero cells cultured in a Dulbecco’s minimum essential medium (DMEM; GIBCO, Invitrogen, San Diego, CA, U.S.A.) supplemented with 8% heat-inactivated fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin at 37°C in a 95% air/5% CO2 environment. To purify T. gondii tachyzoites, parasites and host cells were washed in cold phosphate-buffered saline (PBS). The final pellet was resuspended in cold PBS and passed three times, and then passed through a 27-gauge needle. The parasites were finally passed through 5.0 µm-pore filters (Millipore, Bedford, MA, U.S.A.) and stored at −80°C until use.
Sequence alignment and construction of a phylogenetic tree
The signal peptide of TgAPN2 was predicted with the SignalP algorithm, and conserved protein domains were identified using the PFAM algorithm. The amino acid sequences of the following APN family members and several selected members of the M1 zinc metallopeptidase family were aligned using the BLAST program (GenBank accession numbers are indicated in parentheses): T. gondii (XP_008881821), P. falciparum (XP_001349846.1), Neospora caninum (XP_003884424.1), and Homo sapiens (NP_001141.2). The three-dimensional protein structure of TgAPN2 was predicted with the SWISS-MODEL (https://swissmodel.expasy.org/) [5]. The phylogenetic tree was computed using the MEGA (version 5) program with a Muscle alignment and the neighbor-joining method. Bootstrap values were calculated after resampling 10,000 times. The predicted APN sequences of T. gondii (TGGT1_224350A), Cytauxzoon felis (CF002488), Eimeria acervuline (EAH_00017220), Eimeria maxima (EMWEY_00046640), Babesia bigeminy (BBBOND_0208790), and N. caninum were obtained from the ToxoDB database (http://toxodb.org/toxo/). The sequences of E. tenella, P. falciparum, and H. sapiens enzymes were obtained from the NCBI protein database.
Expression of rTgAPN2 in Escherichia coli
TgAPN2 cDNA was amplified using the following primers: pET30aAPNFwd, 5′-GCTGATATCGGATCCGAATTCAAACACCGCCTCGACTATAA-3′, and pET30aAPNRev, 5′-TTGTCGACGGAGCTCGAATTCGGCGGTCCGTTCGGGTCCTT-3′ (EcoR I sites are underlined). The PCR product with a 5′-terminal His-encoding sequence was cloned into the pET30a vector (Takara, Dalian, China). Verified plasmids were used to transform E. coli BL21 strain. Liquid culture of transformed E. coli cells (1 l) was grown at 37°C until OD600 reached 0.5; the temperature was then reduced to 22°C, and protein synthesis was induced with isopropyl-β-d-galactoside (IPTG; 1.0 mM final concentration). After centrifugation, E. coli cells were resuspended in cold PBS and lysed by ultrasonic treatment. Purification of rTgAPN2 was performed using the nickel-nitriloacetic acid (NTA)-agarose (Qiagen, Valencia, CA, U.S.A.) according to the manufacturer’s protocol. Briefly, cleared cell lysates were prepared in a buffer containing 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 1 mg/ml lysozyme, 1 mM Na3VO4, 40 mM NaF, 100 µM leupeptin, 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride, and 0.05 TIU/ml aprotinin (pH 8.0); they were combined with 1 ml of nickel-NTA-agarose, rotated for 1 hr at 4°C, and then transferred onto a chromatography column. The rTgAPN2 nickel-NTA-agarose complexes were washed three times with 50 mM imidazole wash buffer (50 mM NaH2PO4, 300 mM NaCl, and 50 mM imidazole; pH 8.0) and eluted with 250 mM imidazole elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole; pH 8.0). The His-Glutathione S-transferase (His-GST) nickel-NTA-agarose complex was purified using the same methods. His-GST and rTgAPN2 elution fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to determine the appropriate fractions for pooling. For SDS-polyacrylamide gel electrophoresis (SDS-PAGE), the samples were mixed with an equal volume of sample buffer containing 10% beta-mercaptoethanol (5% final concentration) and thenboiled for 10 min. Proteins (20 µg/lane) were run on 10% polyacrylamide slab gels and once the electrophoresis is over, SDS-PAGE gel will be stained in Coomassie Stain Solution for 1 hr. Finally, SDS-PAGE Destain Solution is used to destain Coomassie dye from the gel and the gel would be imaged. Proteins were dialyzed in a buffer containing 50 mM HEPES, 150 mM NaCl, and 10% glycerol, overnight at 4°C, and then aliquoted, and used biochemical assays. The concentration of purified rTgAPN2 was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific, Schwerte, Germany).
Enzyme activity assay
Following the purification, protein activity was evaluated. The enzymatic activity of rTgAPN2 was determined with H-Ala-(4-methyl-coumaryl-7-amide) (H-Ala-MCA) as a substrate, using an EnSpire Multimode Plate Reader (PerkinElmer) at 355–460 nm. Different concentrations of the purified protein were added to 50 mM Tris-HCl buffer (pH 8.0) and pre-heated at 37°C for 20 min, and then the substrate (0.1 mM final concentration) was added. As the substrate was released, the fluorescence increased. All substrates were completely enzymatic, the fluorescence value reached the highest, the curve reached the plateau. Reactions containing only the GST were used as a negative control.
The effect of pH on enzyme activity
The pH of 50 mM Tris-HCl buffer was adjusted to different values. The specified amount of purified protein was added to the different pH buffers and pre-heated at 37°C for 20 min, and then the substrate H-Ala-MCA (0.1 mM final concentration) was added. The initial reaction velocities at different pH values were then measured and calculated.
The effect of metal ions on enzyme activity
The cation sensitivity was investigated by assaying the activity of rTgAPN2 after pre-incubation at 37°C for 30 min in 50 mM Tris-HCl (pH 8.0) supplemented with a metal chloride (Co2+, Mn2+, Fe2+, Ni2+, Mg2+, Ca2+, Cu2+, or Zn2+; Sigma-Aldrich); the substrate was H-Ala-MCA (0.1 mM final concentration). The initial reaction velocities at different metal ions were then measured and calculated. To test the effect of metal ion chelation by EDTA on enzyme activity, the enzyme was pre-incubated with different concentrations of EDTA (0.5, 1, or 10 mM) for 30 min at 37°C before the substrate (H-Ala-MCA) was added.
The rTgAPN2 substrate fingerprint and determination of kinetic parameters
The substrate preference and Km, kcat, and kcat/Km (Km, the Michaelis constant; kcat, the catalytic constant; kcat/Km, the second-order rate constant) values were determined as previously described [21]. Briefly, rTgAPN2 substrate specificity was assayed using a library of MCA substrates. rTgAPN2 activity was assayed in 50 mM Tris-HCl buffer (pH 8.0) at 37°C. The enzyme and substrate concentrations were 0.1 mM. The peptidase activity was monitored at 355–460 nm, and each kinetic assay was repeated three times. The average values and standard deviations (SD) were calculated.
Inhibition assay
To determine the inhibitory effect of bestatin and amastatin on peptidase activity, rTgAPN2 was pre-incubated with bestatin or amastatin for 30 min at 37°C before the addition of the fluorogenic substrate H-Ala-MCA. Relative enzyme inhibition was assessed with different concentrations of bestatin or amastatin.
RESULTS
Identification and sequence analysis of the TgAPN2 gene
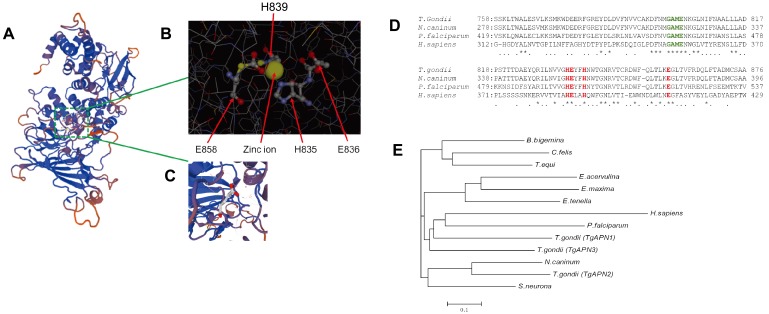
The full-length TgAPN2 cDNA contains an open reading frame of 3,659 bp; the gene resides on chromosome X. An “HEXXH” motif (residues 529–922) and a DUF3485 sequence motif (residues 997–1217) were identified indeduced protein sequence using the Pfam algorithm in the SMART program (http://smart.embl-heidelberg.de/) and SWISS-MODEL program, respectively (Fig. 1A). In these enzymes HEXXH, where the Glu residues act to coordinate the catalytic zinc ion and the first glutamic acid residue is proposed to be one of the putative nucleophiles participating in the enzymatic reaction (Fig. 1B and 1C). Multiple-sequence alignment of the active sites of the M1 protease family members revealed that TgAPN2 contains the functional domains of the M1 protease family, “GAME”, and HEXXH (Fig. 1D); the generated recombinant protein contained these two domains as well (Fig. S1).
Fig. 1.
Structural prediction and evolutionary tree of TgAPN2. (A) The spatial protein structure was predicted using the SWISS-MODEL. (B) Residues H835, E835, H839 and E858 constitute the “HEXXH” domain of M1 aminopeptidases. Ligand: zinc ion. (C) Glutamic acid (1×). (D) Multiple-sequence alignment of the active sites (boxed region) and flanking amino acid sequences of proteins from the M1 protease family, including T. gondii APN, P. falciparum A-M1, N. caninum APN, and H. sapiens APN. TgAPN2 contains the “GAME” and “HEXXH” functional domains of the M1 protease family. (E) Evolutionary tree of TgAPN2. The predicted sequences of enzymes from T. gondii, C. felis, E. acervuline, E. maxima, B. bigeminy, and N. caninum APN1 enzymes were obtained from the ToxoDB database (http://toxodb.org/toxo/). Sequences of the E. tenella N1, P. falciparum, and H. sapiens enzymes were obtained from the NCBI protein database (accession numbers are given in parentheses). TgAPN2 was similar to APN from N. caninum (44.9% identity) and S. neurona (34.5% identity); the homology between TgAPN2 and PfA-M1 was relatively low, only 20%.
These results indicated that the amino acid sequence of TgAPN2 was similar to homohexameric APN proteins, such as APN from N. caninum (44.9% identity) or Sarcocystisneurona (34.5% identity); however, the homology between TgAPN2 and PfA-M1 was relatively low, only 20% (Fig. 1E).
Expression of rTgAPN2
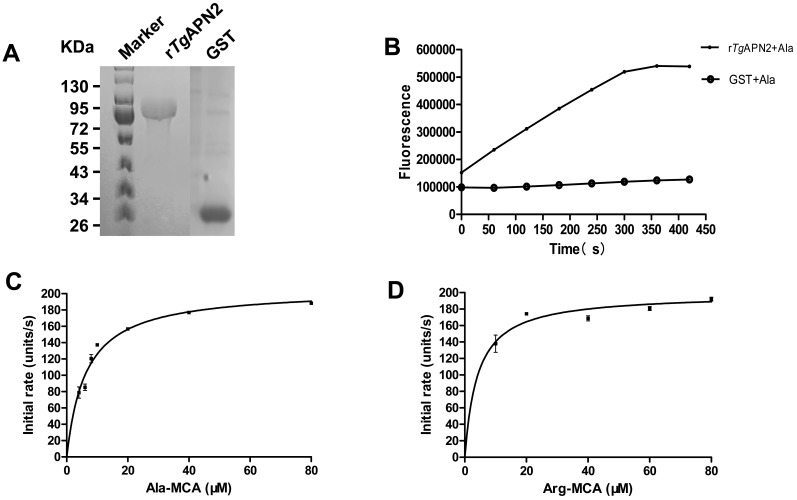
rTgAPN2 protein containing an N-terminal His-tag was expressed as expected. The molecular mass of rTgAPN2 was 85 kDa, as assessed by SDS-PAGE. His-GST protein was used as a control (Fig. 2A).
Fig. 2.
The expression of rTgAPN2 and verification of its activity. (A) SDS-PAGE analysis of the purified rTgAPN2. The molecular mass of rTgAPN2 with a His-tag was 85 kDa. GST was used as a control. (B) The catalytic activity of rTgAPN2. The activity of purified rTgAPN2 was evaluated using a specific fluorescent substrate H-Ala-MCA. GST was used as a negative control. The plateau stage indicates a complete release of the substrate. (C) & (D) Michaelis-Menten enzyme kinetics. Enzymatic activity of purified rTgAPN2. An initial assay with different concentrations of the fluorogenic peptide substrates H-Ala-MCA and H-Arg-MCA is shown. One unit of activity is defined as the amount of MCA (pM) released per mg of recombinant protein. Data points indicate the mean activity ± SD (n=3).
The activity of rTgAPN2
Previous studies demonstrated that APNs have a strict preference for Ala as the N-terminal amino acid residue. The activity of purified rTgAPN2 was therefore tested using a specific fluorescent substrate, H-Ala-MCA. The purified protein exhibited a relatively catalytic activity in 50 mM Tris-HCl (pH 8.0). The negative control was GST protein (Fig. 2B). Under the same conditions, the reaction catalyzed by rTgAPN2 exhibited the Michaelis-Menten kinetics with substrates H-Ala-MCA and H-Arg-MCA (Fig. 2C and 2D).
The metal ion dependence of enzyme activity
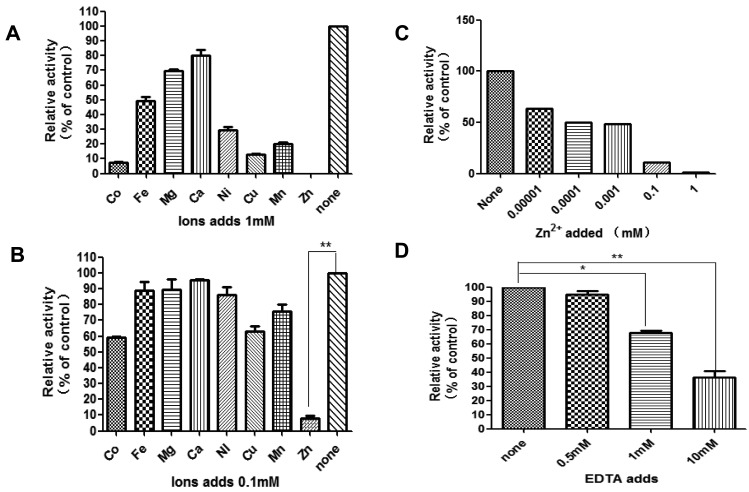
The effect of divalent metal ions on enzyme activity was next evaluated. H-Ala-MCA was used as a standard substrate to examine the protein’s dependence on metal ions; the enzyme activity was inhibited by the addition of the bivalent metal cations Co2+, Mn2+, Fe2+, Ni2+, Mg2+, Ca2+, Cu2+ and Zn2+ (Fig. 3A). To test whether a higher concentration of metal ions would further inhibit the activity of rTgAPN2, different concentration of Zn2+ ions were used in the reactions. The experiment revealed that the activity of the enzyme somewhat recovered with a decreasing concentration of metal ions; however, low Zn2+ concentration still significantly (P<0.01) inhibited the activity of the enzyme (Fig. 3B and 3C). EDTA competitively chelates divalent metal ions and inhibits the activity of some metal ion-dependent enzymes, e.g., human-APN (CD13) [14] and EtAPN [7]. We speculated that the rTgAPN2 might have been activated by metal ions during production in the heterologous (prokaryotic) system. Therefore, the ability of EDTA to chelate the metal ions and inhibit rTgAPN2 activity was next examined. Different concentrations of EDTA were added to the reaction mixture; 1 mM EDTA significantly inhibited the activity of the recombinant protein compared with the untreated group (P<0.05) (Fig. 3D).
Fig. 3.
Restoration of rTgAPN2 activity by divalent metal ions. rTgAPN2 was pre-incubated at 37°C for 30 min in 50 mM Tris-HCl supplemented with the specified metal chloride, and then the substrate (H-Ala-MCA, 0.1 mM final concentration) was added. (A) Enzyme activity in the presence of 1 µM metal ions. (B) Enzyme activity in the presence of 0.1 µM metal ions. (C) Different concentration of Zn2+ ions. (D) Enzyme activity in the presence of EDTA. The differences between samples were evaluated by Student’s t-test (n=3). *P<0.05, **P<0.01.
The pH dependence of rTgAPN2 activity
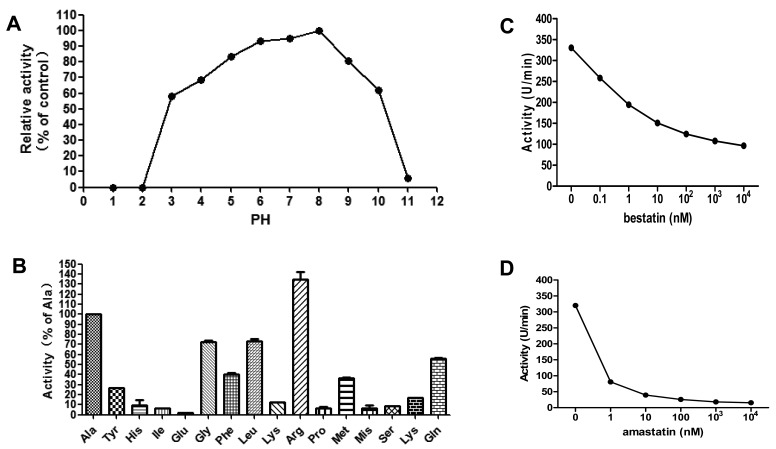
Next, the catalytic activity of the recombinant protein was evaluated at different pH values. The activity of rTgAPN2 was tested in 50 mM Tris-HCl buffers with the pH adjusted from 1 to11; the optimum enzyme activity was observed at pH 8.0. The enzyme also retained some activity at pH 3.0 (Fig. 4A). This indicated that rTgAPN2 is active over a wide range of pH values.
Fig. 4.
(A) pH dependence of rTgAPN2. Optimum enzyme activity was observed at pH 8.0. (B) Substrate specificity of rTgAPN2. One unit of activity is defined as the amount of MCA (pM) released per mg of recombinant protein. Relative rTgAPN2 activity is expressed as a percentage of activity with H-Ala-MCA. (C) The activity of rTgAPN2 is inhibited by bestatin and amastatin. rTgAPN2 was incubated with different concentrations of either bestatin or amastatin for 30 min before the addition of H-Ala-MCA. The residual enzyme activity was recorded and expressed as a percentage of the activity of enzyme incubated in the absence of inhibitors. The inhibitory effect of bestatin and amastatin on the aminopeptidase activity was dose-dependent.
Substrate specificity and enzyme kinetics of rTgAPN2
To characterize the enzymatic properties of rTgAPN2, its activity was measured in fluorescence assays with various peptidase substrates (Fig. 4B). The initial reaction rate of rTgAPN2 with H-Ala-MCA was first calculated. Then, an rTgAPN2 fingerprint for MCA substrate specificity in comparison with H-Ala-MCA was obtained using a library of synthetic substrates. The preferred amino acids were Ala and Arg. rTgAPN2 also accommodated Gly, Leu, Phe, and Pro, while H-Lys-MCA and H-Trp-MCA substrates were cleaved at lower rates (Figs. 4B and S2). The kinetic parameters of rTgAPN2 were determined using a panel of selected substrates (Table 1).
Table 1. Kinetic parameters for the hydrolysis of peptide substrates by rTgAPN2.
| Substrates | kcat (s−1)a) | Km (µM)a) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| Arg-MCA | 6.73 ± 0.001 | 2.15 ± 0.458 | 3,367.30 |
| Ala-MCA | 6.02 ± 0.002 | 6.34 ± 0.643 | 1,004.90 |
| Leu-MCA | 2.73 ± 0.037 | 4.35 ± 1.259 | 683.40 |
| Gly-MCA | 1.54 ± 0.009 | 7.80 ± 1.127 | 220.40 |
| Met-MCA | 0.36 ± 0.170 | 62.08 ± 5.190 | 57.80 |
| Phe-MCA | 1.81 ± 0.183 | 21.56 ± 6.231 | 56.26 |
| Pro-MCA | 0.31 ± 0.140 | 60.53 ± 10.860 | 5.21 |
a) Data represent means ± SD from three independent experiments.
The inhibitors of rTgAPN2
Inhibition assays were then performed with the purified rTgAPN2 protein. The activity of rTgAPN2 was reduced in a dose-dependent manner in the presence of bestatin or amastatin. When the same concentrations of inhibitors were tested, the inhibitory effect of amastatin appeared to be slightly stronger than the inhibitory effect of bestatin (Fig. 4C and 4D).
DISCUSSION
APN is a member of the M1 family of metalloproteases, which hydrolyzes N-terminal neutral (e.g., leucine and alanine) or basic (e.g., arginine and lysine) amino acid residues in oligopeptides or polypeptides. Such substrate diversity indicates a wide range of biological functions of APNs.
In this study, a novel T. gondii aminopeptidase belonging to the M1 metalloproteinase family was produced and purified in E. coli, and its biochemical characteristics were evaluated in a series of experiments. Although it was difficult to produce the full-length protein in E. coli, the enzymaticallyactive region of TgAPN2 was analyzed and successfully produced as a recombinant protein; the activity of the recombinant protein was then verified.
Based on protein sequence analysis, TgAPN2 contains a highly-conserved ion-binding motif and a catalytic motif [1]. These two motifs are characteristic for all M1 family aminopeptidases. Most APNs are thought to be zinc-dependent metalloproteinases [2, 25]. We previously produced a family of metalloproteinases in E. coli, demonstrating that these proteases were metal-dependent [11, 29]; i.e., recombinant aspartyl aminopeptidase (rTgAAP) and leucyl aminopeptidase (rTgLAP) of the T. gondii had some activity in the absence of added metal ions, and the addition of metal ions improved their activity. As shown in the current study, TgAPN2 is somewhat different from rTgAAP and rTgLAP. Purified rTgAPN2, produced in E. coli, did not require the addition of metal ions for activity and the addition of low concentration of zinc ions inhibited its activity. Some Zn2+ metalloproteases acquire zinc ions during the process of production in prokaryotic systems, and a subsequent addition of zinc ions significantly inhibits their activity [13]. We hypothesize that rTgAPN2 may also bind Zn2+ metal ions during prokaryotic expression and purification. If that were the case, the enzymatic activity would be decreased after the addition of a metal chelator, EDTA. Indeed, EDTA inhibited the activity of rTgAPN2. Thus, TgAPN2 is likely to be a Zn2+ metalloprotease from the M1 family. Protein expression in eukaryotic systems, e.g., yeast, might yield different results. We are planning to explore the activity in follow-up experiments.
PfA-M1 is essential for protein metabolism and plays an important role in the digestion of host hemoglobin into free amino acids [16]. PfA-M1 exists in three soluble forms, p120, p96 and p68, localized in different parts of the Plasmodium cell. Protein p68 is marginally delivered into the food vacuole at trophozoite stages, but is not in a direct contact with the parasite cytoplasm [4]. In fact, the intravacuolar pH of the food vacuole can decline from 7 to 3 [8], and studies show that the optimal pH for Plasmodium falciparum APN activity is 7.4. We found that rTgAPN2 was active in the 3–10 pH range, suggesting that M1 family aminopeptidases share some common features.
PfA-M1 is used as an exciting target for novel anti-malarial drugs [24]. Unlike P. falciparum, there are three APN-encoding genes in T. gondii genome. These three genes that belong to the M1 family aminopeptidases are located on different chromosomes of T. gondii, named APN1, APN2 and APN3 according to the previous report [10], respectively. So, they may or not have a common effect. Therefore, to develop anti-Toxoplasma drugs, it is important to understand the functions of each TgAPN. The current study demonstrated that the short-segmented rTgAPN2 protein retained appreciable enzymatic activity, with a broad substrate specificity for unnatural amino acids. Ala is considered to be the most suitable substrate for the M1 family aminopeptidases in P. falciparum [6]. Here, similar results were obtained with the T. gondii enzyme, with rTgAPN2 being highly active toward H-Ala-MCA; however, the activity of rTgAPN2 was even higher with H-Arg-MCA. By contrast, the activity of E. tenella aminopeptidase with H-Ala-MCA and H-Arg-MCA is substantially the same [7]. This suggests that the substrate preference of TgAPN2 may have its own characteristics.
The involvement of aminopeptidase activities during Plasmodium species development was initially illustrated using bestatin [17], a well-known broad-spectrum inhibitor of aminopeptidases. And E. tenella aminopeptidase N 1 (EtAPN1) is likely the most significant target of bestatin in E. tenella sporozoites, but for rEtAPN1, the inhibitory effect of amastatin was higher than that of bestatin [10]. This phenomenon is also reflected by our experimental results: the inhibitory effect of bestatin on rTgAPN2 was dose-dependent, bestatin did not completely inhibit rTgAPN2, and the inhibitory effect of amastatin was to some extent greater than that of bestatin. For human Aminopeptidase N (APN/CD13), An IC50 of 9.0 mM was determined for amastatin which is higher than that of bestatin (IC50=16.9 mM) [22, 26].
We propose that TgAPN2 is also involved in parasite development and plays a role similar to that of PfA-M1. But it is noteworthy that Plasmodium and Cryptosporidium, in contrast to Toxoplasma and Eimeria, have only one aminopeptidase enzyme of the M1 family. Complementary phylogenetic analysis should be carried out to determine if gene duplication occurred in these parasites. Given the above, to understand the biochemical characteristics of TgAPN2, to be more targeted to design anti-Toxoplasma drug target.
Supplementary Material
Acknowledgments
This work was supported by a grant from Natural Science Foundation of Heilongjiang Province of China (no. C2015063 and C2015065) to Honglin Jia and Jun Zheng.
REFERENCES
- 1.Addlagatta A., Gay L., Matthews B. W.2006. Structure of aminopeptidase N from Escherichia coli suggests a compartmentalized, gated active site. Proc. Natl. Acad. Sci. U.S.A. 103: 13339–13344. doi: 10.1073/pnas.0606167103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albiston A. L., Ye S., Chai S. Y.2004. Membrane bound members of the M1 family: more than aminopeptidases. Protein Pept. Lett. 11: 491–500. doi: 10.2174/0929866043406643 [DOI] [PubMed] [Google Scholar]
- 3.Arfin S. M., Kendall R. L., Hall L., Weaver L. H., Stewart A. E., Matthews B. W., Bradshaw R. A.1995. Eukaryotic methionyl aminopeptidases: two classes of cobalt-dependent enzymes. Proc. Natl. Acad. Sci. U.S.A. 92: 7714–7718. doi: 10.1073/pnas.92.17.7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azimzadeh O., Sow C., Gèze M., Nyalwidhe J., Florent I.2010. Plasmodium falciparum PfA-M1 aminopeptidase is trafficked via the parasitophorousvacuole and marginally delivered to the food vacuole. Malar. J. 9: 189. doi: 10.1186/1475-2875-9-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., Schwede T.2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42: W252-8. doi: 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalal S., Klemba M.2007. Roles for two aminopeptidases in vacuolar hemoglobin catabolism in Plasmodium falciparum. J. Biol. Chem. 282: 35978–35987. doi: 10.1074/jbc.M703643200 [DOI] [PubMed] [Google Scholar]
- 7.Drag M., Bogyo M., Ellman J. A., Salvesen G. S.2010. Aminopeptidase fingerprints, an integrated approach for identification of good substrates and optimal inhibitors. J. Biol. Chem. 285: 3310–3318. doi: 10.1074/jbc.M109.060418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fok A. K., Lee Y., Allen R. D.1982. The correlation of digestive vacuole pH and size with the digestive cycle in Paramecium caudatum. J. Protozool. 29: 409–414. doi: 10.1111/j.1550-7408.1982.tb05423.x [DOI] [Google Scholar]
- 9.Frankel M. B., Knoll L. J.2009. The ins and outs of nuclear trafficking: unusual aspects in apicomplexan parasites. DNA Cell Biol. 28: 277–284. doi: 10.1089/dna.2009.0853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gras S., Byzia A., Gilbert F. B., McGowan S., Drag M., Silvestre A., Niepceron A., Lecaille F., Lalmanach G., Brossier F.2014. Aminopeptidase N1 (EtAPN1), an M1 metalloprotease of the apicomplexan parasite Eimeria tenella, participates in parasite development. Eukaryot. Cell 13: 884–895. doi: 10.1128/EC.00062-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia H., Nishikawa Y., Luo Y., Yamagishi J., Sugimoto C., Xuan X.2010. Characterization of a leucine aminopeptidase from Toxoplasma gondii. Mol. Biochem. Parasitol. 170: 1–6. doi: 10.1016/j.molbiopara.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 12.Kanayama N., Kajiwara Y., Goto J., el Maradny E., Maehara K., Andou K., Terao T.1995. Inactivation of interleukin-8 by aminopeptidase N (CD13). J. Leukoc. Biol. 57: 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K. I., Baek S. H., Hong Y. M., Kang M. S., Ha D. B., Goldberg A. L., Chung C. H.1995. Purification and characterization of protease Ci, a cytoplasmic metalloendoprotease in Escherichia coli. J. Biol. Chem. 270: 29799–29805. doi: 10.1074/jbc.270.50.29799 [DOI] [PubMed] [Google Scholar]
- 14.Larsen S. L., Pedersen L. O., Buus S., Stryhn A.1996. T cell responses affected by aminopeptidase N (CD13)-mediated trimming of major histocompatibility complex class II-bound peptides. J. Exp. Med. 184: 183–189. doi: 10.1084/jem.184.1.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lun Z. R., Lai D. H., Wen Y. Z., Zheng L. L., Shen J. L., Yang T. B., Zhou W. L., Qu L. H., Hide G., Ayala F. J.2015. Cancer in the parasitic protozoans Trypanosoma brucei and Toxoplasma gondii. Proc. Natl. Acad. Sci. U.S.A. 112: 8835–8842. doi: 10.1073/pnas.1502599112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mistry S. N., Drinkwater N., Ruggeri C., Sivaraman K. K., Loganathan S., Fletcher S., Drag M., Paiardini A., Avery V. M., Scammells P. J., McGowan S.2014. Two-pronged attack: dual inhibition of Plasmodium falciparum M1 and M17 metalloaminopeptidases by a novel series of hydroxamic acid-based inhibitors. J. Med. Chem. 57: 9168–9183. doi: 10.1021/jm501323a [DOI] [PubMed] [Google Scholar]
- 17.Nankya-Kitaka M. F., Curley G. P., Gavigan C. S., Bell A., Dalton J. P.1998. Plasmodium chabaudi chabaudi and P. falciparum: inhibition of aminopeptidase and parasite growth by bestatin and nitrobestatin. Parasitol. Res. 84: 552–558. doi: 10.1007/s004360050447 [DOI] [PubMed] [Google Scholar]
- 18.Padda R. S., Tsai A., Chappell C. L., Okhuysen P. C.2002. Molecular cloning and analysis of the Cryptosporidium parvum aminopeptidase N gene. Int. J. Parasitol. 32: 187–197. doi: 10.1016/S0020-7519(01)00317-4 [DOI] [PubMed] [Google Scholar]
- 19.Pasqualini R., Koivunen E., Kain R., Lahdenranta J., Sakamoto M., Stryhn A., Ashmun R. A., Shapiro L. H., Arap W., Ruoslahti E.2000. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 60: 722–727. [PMC free article] [PubMed] [Google Scholar]
- 20.Pillay D., Boulangé A. F., Coustou V., Baltz T., Coetzer T. H.2013. Recombinant expression and biochemical characterisation of two alanyl aminopeptidases of Trypanosoma congolense. Exp. Parasitol. 135: 675–684. doi: 10.1016/j.exppara.2013.10.005 [DOI] [PubMed] [Google Scholar]
- 21.Poreba M., McGowan S., Skinner-Adams T. S., Trenholme K. R., Gardiner D. L., Whisstock J. C., To J., Salvesen G. S., Dalton J. P., Drag M.2012. Fingerprinting the substrate specificity of M1 and M17 aminopeptidases of human malaria, Plasmodium falciparum. PLoS One 7: e31938. doi: 10.1371/journal.pone.0031938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rich D. H., Moon B. J., Harbeson S.1984. Inhibition of aminopeptidases by amastatin and bestatin derivatives. Effect of inhibitor structure on slow-binding processes. J. Med. Chem. 27: 417–422. doi: 10.1021/jm00370a001 [DOI] [PubMed] [Google Scholar]
- 23.Sjöström H., Norén O., Olsen J.2000. Structure and function of aminopeptidase N. Adv. Exp. Med. Biol. 477: 25–34. doi: 10.1007/0-306-46826-3_2 [DOI] [PubMed] [Google Scholar]
- 24.Skinner-Adams T. S., Stack C. M., Trenholme K. R., Brown C. L., Grembecka J., Lowther J., Mucha A., Drag M., Kafarski P., McGowan S., Whisstock J. C., Gardiner D. L., Dalton J. P.2010. Plasmodium falciparum neutral aminopeptidases: new targets for anti-malarials. Trends Biochem. Sci. 35: 53–61. doi: 10.1016/j.tibs.2009.08.004 [DOI] [PubMed] [Google Scholar]
- 25.Tsujimoto M., Hattori A.2005. The oxytocinase subfamily of M1 aminopeptidases. Biochim Biophys Acta 1751: 9–18. doi: 10.1016/j.bbapap.2004.09.011 [DOI] [PubMed] [Google Scholar]
- 26.Umezawa H., Aoyagi T., Suda H., Hamada M., Takeuchi T.1976. Bestatin, an inhibitor of aminopeptidase B, produced by actinomycetes. J. Antibiot. 29: 97–99. doi: 10.7164/antibiotics.29.97 [DOI] [PubMed] [Google Scholar]
- 27.Weiss L. M., Kim K.2013. Toxoplasma gondii: the Model Apicomplexan. Perspectives and Methods, 2nd ed., Academic Press, Cambridge. [Google Scholar]
- 28.Wong A. H., Zhou D., Rini J. M.2012. The X-ray crystal structure of human aminopeptidase N reveals a novel dimer and the basis for peptide processing. J. Biol. Chem. 287: 36804–36813. doi: 10.1074/jbc.M112.398842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng J., Cheng Z., Jia H., Zheng Y.2016. Characterization of aspartyl aminopeptidase from Toxoplasma gondii. Sci. Rep. 6: 34448. doi: 10.1038/srep34448 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.