Abstract
The pandemic 2009 H1N1 influenza A virus emerged in humans and caused the first influenza pandemic of the 21st century. Mexican isolates, A/Mexico/4108/2009 (H1N1) (Mex4108) and A/Mexico/InDRE4478/2009 (H1N1) (Mex4487) derived from a mild case and from a cluster of severe cases, showed heterogeneity in virulence in a cynomolgus macaque model. To compare the more pathogenic differences, we generated recombinant viruses and compared their virulence in ferrets. Ferrets infected with recombinant Mex4487 displayed a slightly higher rate of viral replication and severe pneumonia in the early stage of infection. In contrast, prolonged lower virus shedding of recombinant Mex4108 than that of recombinant Mex4487 was detected in throat swabs. Thus, Mex4487 induces severe pneumonia in infected individuals, whereas Mex4108 might have wide-spreading potential with mild disease.
Keywords: ferret, pathogenesis, virus shedding
Influenza A virus infections in humans are typically associated with limited seasonal outbreaks of commonly circulating influenza virus strains (seasonal strains). A new virus strain occasionally emerges in humans, resulting in increased morbidity and mortality compared to those of seasonal influenza (pandemic strain) [29]. A novel H1N1 influenza A virus, the pandemic 2009 H1N1 influenza A virus (A(H1N1) pdm2009), caused the first human influenza pandemic of the new millennium [3, 4, 6]. In 2010, WHO announced that A(H1N1) pdm2009 had moved into the post-pandemic period [27, 28]. A(H1N1) pdm2009 has now replaced the classical seasonal H1N1 strains and is circulating globally as a current seasonal strain with a case fatality rate similar to that of classical seasonal influenza.
Human pandemic A(H1N1) pdm2009 infections appeared to be mild in general, and some infected individuals presented with symptoms atypical for seasonal influenza; however, severe illness was also reported, particularly in young, previously healthy individuals [2, 3, 21]. Several early isolates also caused severe diseases in experimentally infected animals, and pathogenicity factors were analyzed [9, 13, 16]. Interestingly, many severe cases of seasonal A(H1N1) pdm2009 were reported in Mexico during the 2013–2014 influenza season [14]. There is the possibility that antigenic change of HA is a reason for middle-aged adults being highly susceptible to seasonal A(H1N1) pdm2009 in the 2013–2014 influenza season [12]. Thus, several factors associated in virulence and human adaptation factors of viruses have identified, and it is still important to accumulate knowledge of the pathogenic potential and analyze the potential virulence factors of these influenza viruses.
In our previous study, infection of cynomolgus macaques with two genetically similar but clinically distinct human A(H1N1) pdm2009 strains, A/Mexico/4108/2009 (Mex4108) and A/Mexico/InDRE4487/2009 (Mex4487), isolated during the early phase of the pandemic [23], resulted in higher pathogenic potential of Mex4487. To investigate the potential virulence of Mexican isolates, we generated recombinant viruses between the two Mexican A(H1N1) pdm2009 isolates and evaluated their pathogenicity in the ferret model, a widely used and well-established model for studying both the pathogenicity and transmissibility of human influenza viruses [1, 26].
MATERIALS AND METHODS
Cells
Madin-Darby canine kidney (MDCK) cells were maintained in Eagle’s minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine and penicillin/streptomycin (Pen/St). Human lung carcinoma (A549) and human embryonic kidney 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 2 mM L-glutamine and Pen/St.
Viruses
Influenza viruses, Mex4108 (kindly provided by the Centers for Disease Control and Prevention, Atlanta, GA, U.S.A.) and Mex4487 (kindly provided by Public Health Agency of Canada, Winnipeg, MB, Canada), as well as the recombinant viruses, were propagated in MDCK cells with MEM containing 2% FBS and 0.35 µg/ml of TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone)-trypsin. Virus infectivity titers were determined using the 50% tissue culture infectious dose (TCID50) assay. For this assay, 10-fold dilutions of supernatants were used to infect MDCK cells. The virus-induced cytopathogenic effect (CPE) was scored at 3 days post-infection (dpi).
Generation of recombinant viruses
Genomic RNA of Mex4108 and Mex4487 was extracted from virus stocks and used to amplify the eight gene segments by reverse transcription-polymerase chain reaction (RT-PCR). Each PCR product was individually cloned into a polI-promoter plasmid (ppolI) [18]. The open reading frames coding for components of the influenza virus RNP complex, polymerase basic protein 2 (PB2), polymerase basic protein 1 (PB1), polymerase acidic protein (PA) and nucleoprotein (NP), were cloned into the expression plasmid pCAGGs (helper plasmids). All plasmids were sequence confirmed prior to use. To generate recombinant viruses, different combinations of the eight ppolI plasmids from either Mex4108 or Mex4487 were transfected together with the four helper plasmids into 293T cells. After 30 hr of incubation, transfection supernatants were removed, and OPTI-MEM with TPCK trypsin was added. At 48 hr post-transfection (hpt), the supernatant was collected, and the rescued virus was subsequently propagated in MDCK cells. All rescued recombinant viruses were sequence confirmed, and virus titers were determined by a standard TCID50 assay in MDCK cells (Table 1 ). Recombinant viruses, rgMex4108 and rgMex4487, rescued with titers similar to those of the original isolates.
Table 1. Recombinant viruses generated by a reverse genetics system.
In vitro growth kinetics of recombinant viruses
Confluent monolayers of A549 and MDCK cells were inoculated with rgMex4108, rgMex4487, rgM4108/M4487-HA.NP.M, rgM4108/M4487-PA.PB2, rgM4108/M4487-PB2 or rgM4108/M4487-PA at a multiplicity of infection (MOI) of 0.1 (A549 cells) or 0.001 (MDCK cells). Virus was allowed to adsorb for 1 hr, then unbound viruses were washed away, and DMEM or MEM with TPCK-trypsin was added. At determined time points, the supernatants were collected from 3 wells per virus. Virus titers were determined as TCID50 on MDCK cells.
Animal study
Groups of 12 ferrets (females, 4–12 months, weight range: 630 to 990 g) were inoculated with rgMex4487 or rgMex4108 (106 TCID50/ferret) via the intranasal or intratracheal route, and the animals were monitored daily for body weight and signs of disease for 14 days. Four animals from each group were euthanized at 3 or 6 dpi, and tissue samples were collected for virology and histopathological evaluation. Tissues were placed in cassettes and fixed in 10% Neutral Buffered Formalin x2 changes, for a minimum of 7 days. Cassettes were processed with a Sakura Tissue-Tek VIP-6, on a 12 hr automated schedule, using a graded series of ethanol, xylene and Ultraffin-X paraffin. Embedded tissues are sectioned at 5 µm and dried overnight at 42°C prior to staining. Tissues were stained by hematoxylin and eosin (H&E) stain and scored as: 0=no, 1=minimal, 2=mild, 3=moderate, 4=marked and 5=severe lesion. Infectivity of the virus was determined as TCID50. All animal experiments were approved by the Institutional Animal Care and Use Committee of Rocky Mountain Laboratories and performed following the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care, International (AAALAC) by certified staff in an AAALAC-approved facility.
Biosafety
All infectious in vitro and in vivo studies were performed in high biocontainment at the Integrated Research Facility (IRF) of Rocky Mountain Laboratories (RML), Division of Intramural Research (DIR), National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). Sample inactivation and removal from the containment facility was performed according to standard operating protocols approved by the local Institutional Biosafety Committee.
Statistical analysis
Statistical analyses were performed using a two-tailed Student’s t-test.
RESULTS
Growth kinetics of recombinant viruses
A comparison of the two Mexican isolates, rgMex4108 and rgMex4487, revealed a difference in seven amino acids affecting five proteins [PB2 (amino acid position (aa) 82) and PA (aa275, aa581), HA (aa444), NP (aa100, aa373) and M2 protein (aa82)] [23]. We generated rgMex4108, rgMex4487 and other 4 recombinant viruses of which two were single-gene replacements (PB2, PA), a dual-gene replacement of PB2 and PA and a triple-gene replacement of HA, NP and M. The titers of the all rescued recombinant viruses ranged from 106.0 to 107.3 TCID50/ml (Table 1). In vitro growth kinetics was performed in A549 cells infected with a MOI of 0.1. As shown in Fig. 1A, rgMex4487 showed approximately 1 log higher replication compared to rgMex4108 at 12 hr post-inoculation (hpi). Mex4108 backbone viruses of which PA and/or PB2 genes are from Mex4487 (white symbols) tended to higher replication than rgMex4108 or rgM4108/M4487-HA.NP.M (black symbols), however, no significant difference was observed. We also evaluated the growth kinetics in MDCK cells that was used for virus propagation. All recombinant viruses replicated similarly over a time course of 72 hr with no significant differences in titers and reached highest virus titer at 48 hpi as similar in Table 1 (Fig. 1B).
Fig. 1.
In vitro growth kinetics of recombinant viruses. Confluent A549 (A) or MDCK (B) cells were infected with recombinant viruses, rgMex4108, rgMex4487, M4108/M4487-PB2.PA, M4108/M4487-HA.NP.M, M4108/M4487-PB2 or M4108/M4487-PA at a MOI of 0.1 (A549) or 0.001 (MDCK), respectively. Infected cells were incubated at 37°C, and supernatants were collected at the indicated times. Virus titer of supernatants was expressed as TCID50 with standard deviations (S.D.).
Pathogenicity of recombinant viruses in ferrets
Ferrets were inoculated with rgMex4487 or rgMex4108 via the intranasal (Fig. 2A and 2B) or intratracheal (Fig. 2C and 2D) route. Both groups developed only mild clinical symptoms, and all of the ferrets survived except for two ferrets inoculated with rgMex4108 via the intratracheal route. Those two ferrets died at 2 and 9 dpi with suspected secondary bacterial infection (Supplemental Fig. 1).
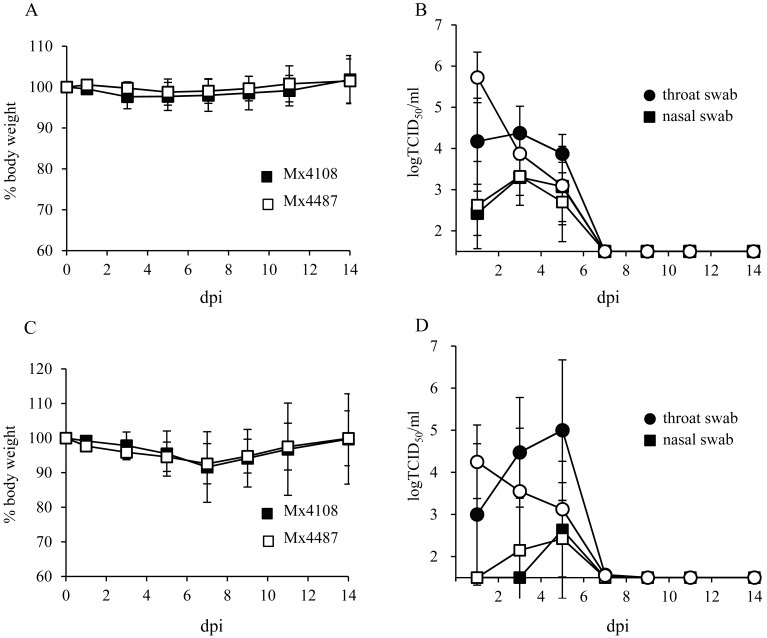
Fig. 2.
Comparison of pathogenicities of recombinant viruses. Ferrets were infected with a dose of 106 TCID50 of either rgMex4108 or rgMex4487 via the intranasal (A, B) or intratracheal (C, D) route. (A, C) Body weight loss of ferrets was monitored until 14 dpi. (B, D) Virus shedding in swabs was monitored until 14 dpi. Black symbols indicate Mex4108, and white symbols indicate Mex4487.
By the intranasal inoculation route, no weight loss was induced in either of the groups (Fig. 2A). Virus shedding in nasal and throat swabs was monitored every other day until 14 dpi. rgMex4487 reached the highest titer of 5 × 105 TCID50/ml at 1 dpi from throat swabs, and then, the virus titer decreased (Fig. 2B). In contrast, the titer of rgMex4108 in throat swabs remained at 104 TCID50/ml until 5 dpi. The virus titers in nasal swab were similar in the two groups. Ferrets inoculated with viruses via the intratracheal route exhibited mild weight loss until 7 dpi in both groups and subsequently recovered (Fig. 2C). As was observed in the groups with intranasal inoculation, the highest virus titer of rgMex4487 was detected at 1 dpi from throat swabs (Fig. 2D). The virus titer of rgMex4108 in throat swabs increased and reached 105 TCID50 at 5 dpi, but no significant difference was observed in all time points. The virus titers in nasal swab were similar in the two groups.
Histopathological differences of recombinant viruses
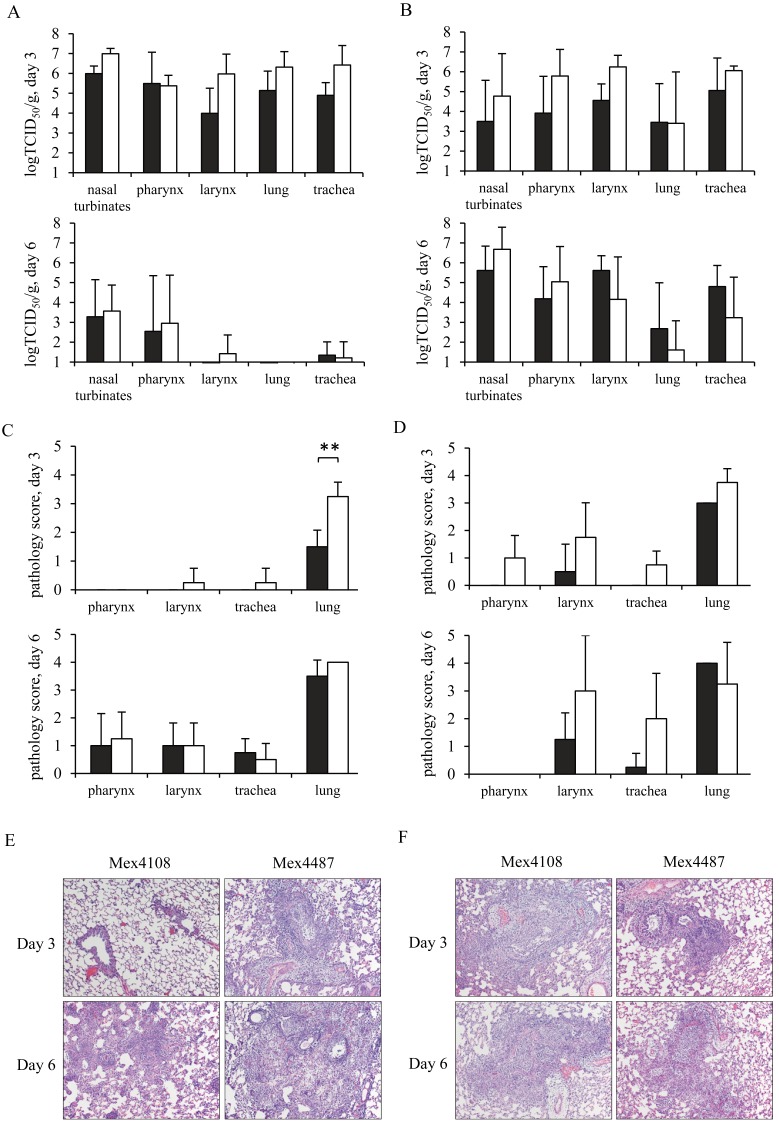
We next compared the viral replication and histopathological changes in respiratory tissues. Four animals from each group were euthanized at 3 or 6 dpi, and tissue samples were collected. By the intranasal inoculation, the highest rate of virus replication was detected in all respiratory tissues at 3 dpi (Fig. 3A). Although virus titers of rgMex4487 were slightly higher than those of rgMex4108 at 3 dpi in most of the respiratory tissues tested, no significant differences were observed. Virus replication decreased by 6 dpi in all respiratory tissues. Ferrets inoculated with rgMex4487 had developed severe inflammation and pneumonia on day 3 (Fig. 3C and 3E). Mild inflammation and pneumonia were still observed in all respiratory tissues at 6 dpi, even though the virus replication on day 6 was lower than on day 3. Ferrets inoculated with viruses via the intratracheal route showed more severe pneumonia at 3 dpi compared with ferrets inoculated with viruses via the intranasal route (Fig. 3D and 3F). The high rate of virus replication was detected in respiratory tissues at 3 dpi and continued through 6 dpi (Fig. 3B). Although moderate pneumonia was detected in both groups until day 6, rgMex4108 seemed to replicate well in respiratory tissues without severe inflammation, as compared to rgMex4487. All four groups of ferrets developed multifocal, moderate to marked bronchointerstitial pneumonia; however, ferrets inoculated via the intratracheal route tended to have more severe lesions. Pulmonary changes are predominately centered on terminal bronchioles, but often extend into adjacent bronchi and surrounding alveoli (Fig. 3E and 3F).
Fig. 3.
Comparison of pathological differences in ferrets. Ferrets were infected with a dose of 106 TCID50 of either rgMex4108 or rgMex4487 via the intranasal (A, C, E) or intratracheal (B, D, F) route. Four ferrets were euthanized at 3 or 6 dpi. (A, B) Virus titers of respiratory tissues are shown as TCID50 with S.D. (C, D) Histopathological data are shown as the average of pathological scores with S.D. *P<0.05. (E, F) Histpathological sections of lung tissues were stained with H&E stain, original magnification ×100. Black symbols indicate Mex4108, and white symbols indicate Mex4487.
DISCUSSION
Since the pandemic of 2009, A(H1N1) pdm2009 circulating worldwide as seasonal flu has caused mild respiratory illness in infected individuals. A(H1N1) pdm2009 has occasionally caused severe disease and related deaths depending on the virus or the patient’s condition [2, 4, 14, 19, 21]. In our previous study, we characterized two Mexican isolates, Mex4108 and Mex4487, derived from a mild case and from a cluster of severe cases, respectively. They also produced heterogeneity in virulence in a cynomolgus macaque model [23]. However, in this study, experimental infection with both rgMex4108 and rgMex4487 caused only mild and undistinguishable respiratory diseases in ferrets. Ferrets inoculated with original isolates also showed mild symptoms, as was observed by the recombinant viruses (data not shown). In this experiment, we compared both intranasal and intratracheal inoculation routes, and ferrets inoculated via the intratracheal route showed slightly reduced body weight by day 7 post-infection and tended to have more severe lesions by histopathological analysis. However, ferrets showed similar mild symptom in all groups. We did not find the significant clinical differences between the two isolates that were observed in the cynomolgus macaque model. The ferret is a common animal model for an influenza virus study. However, animal models do not always seem to mimic symptoms that were observed in human cases. To evaluate the pathogenicity difference of two Mexican isolates, we may need to use the cynomolgus macaque model, since cynomolgus macaque demonstrated similar clinical differences reflecting human cases in the previous study. On the other hand, it has been reported that Mex4487-infected ferrets showed severe clinical signs and that approximately 50% of the animals succumbed to infection within 9 dpi [11, 15]. This difference indicates that the outcome of disease might be affected by rearing environments of infected hosts as well as technical differences of experiments. However, rgMex4108 yielded a higher and more prolonged virus shedding in throat swabs, and a higher virus replication was detected until 5 dpi in throat swab. Although there is also no significant difference between viruses in our ferret model, these results suggested that Mex4108 may have the potential for long and wide spreading of viruses. To compare the transmissibility of viruses, contact transmission experiments are required.
A(H1N1) pdm2009 lacks amino acid mutations previously identified as human adaptation signature, like PB2-E627K and D701N, and these known virulence associated mutations in PB2 increased reporter gene expression, but did not affect virus replication and transmission [8]. Whereas, several A(H1N1) pdm2009 strains possessing amino acid substitutions in PB2 and HA showed enhanced virulence and transmission in ferret models [10, 31, 34]. However, both Mex4487 and Mex4108 do not have identified amino acid sequence that was contributed to virulence and transmission of viruses in animal models. Only 7 amino acid differences were identified in the genome of two Mexican isolates. During pandemic period, frequency of specific amino acids in A(H1N1) pdm2009 circulating in the world had evolved and selected the more efficient replication and transmissible strains as current seasonal strains [20]. We also compared the sequences of Mexican isolates with consensus sequence with A(H1N1) pdm2009 isolated in the early stage of pandemic, and five of seven amino acid differences are specific for either Mex4108 or Mex4487. Two amino acid residues in PB2-N82A and PA-L275I of Mex4487, and three amino acid residues in HA-V444I, NP-D100G and M2-S82N of Mex4108 were specific in each strain. The other two amino acid differences, PA-581L and NP-373I in Mex4108, were found as common in A(H1N1) pdm2009, especially in the early stage of pandemic [20]. Specific mutations in Mex4487 or Mex4108 might affect difference in virulence or transmissibility of the two isolates.
The results of in vitro growth kinetics of recombinant viruses suggested that PB2 or PA of Mex4487 is a potential factor for prolonged virus replication in human alveolar epithelial cells. rgMex4108 possessing PA and/or PB2 of Mex4487 showed a higher rate of replication than that of rgMex4108. PB2 and PA are components of the RNA-dependent RNA polymerase complex of influenza A virus, and mutations in PB2 [7, 32, 33] or PA [5, 22, 24] associated with host range or viral pathogenicity have been reported. To determine the functional differences of two Mexican isolates, we analyzed the polymerase activity and inhibitory effects of IFN induction of both RNP complexes derived from Mex4108 or Mex4487 using a reporter assay. However, no difference was observed in both assays tested (Supplemental Fig. 2). We evaluated the polymerase activity in human cell lines, A549 and 293, and polymerase activities were similar for any combination of the RNP complex derived from Mex4108 or Mex4487. As another function of PB2, we also compared the inhibition activity of MAVS-mediated IFN-β induction. From the IFN-β promoter-driven reporter assay, significant inhibition of MAVS-mediated IFN-β induction by both PB2 and PA was observed. However, again, no difference was observed in this activity between proteins derived from Mex4108 or Mex4487 (Supplemental Fig. 2). These results suggested that the polymerase activity and inhibitory effects of IFN induction of these proteins are not critical factors in pathological differences of Mex4108 and Mex4487 shown in human cases and a cynomolgus macaque model. Recently, several novel proteins encoded by polymerase genes and additional functions of these proteins have been reported, suggesting the presence of additional proteins and/or functions associated with pathogenesis [17, 25, 30]. To investigate the importance of PB2 or PA in virulence, the pathogenesis of other recombinant viruses switching of PA and PB2 genes should be examined in animal models. Also, the pathogenetic differences and molecular function of viruses will need to be determined.
In this study, we showed that Mex4487 tends to induce severe pneumonia in infected individuals, whereas Mex4108 has wide-spreading potential with mild disease. However, there is no significant difference in most experiments, despite two Mexican isolates showing heterogeneity in virulence in a cynomolgus macaque. It might be the limitation of the ferret model, because animal models do not always reflect human cases. Whereas the clinical outcome of two isolates was different, it may indicate that background and conditions of infected individuals are important for determining the virulence as well as molecular determinants. To understand the pathogenicity and transmissibility of viruses for preparation for future risks, it is necessary to observe and accumulate the information of potential factors as well as signature sequences to determine the phenotype of influenza viruses.
CONFLICT OF INTERES
None to declare.
Supplementary Material
Acknowledgments
We would like to thank Dr. Heinz Feldmann, Dr. Emmie de Wit, Dr. Vincent J Munster, Dr. Dawn Clifton, Mr. Mark Dewald and Mr. Trenton Bushmaker, Laboratory of Virology, DIR, NIAID, NIH for their technical support and intellectual contributions. We also thank Dr. Dana Scott and members of Rocky Mountain Veterinary Branch, DIR, NIAID, NIH for assistance with animal care. This research was supported by the Intramural Research Program of the NIH.
REFERENCES
- 1.Barnard D. L.2009. Animal models for the study of influenza pathogenesis and therapy. Antiviral Res. 82: A110–A122. doi: 10.1016/j.antiviral.2008.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chowell G., Bertozzi S. M., Colchero M. A., Lopez-Gatell H., Alpuche-Aranda C., Hernandez M., Miller M. A.2009. Severe respiratory disease concurrent with the circulation of H1N1 influenza. N. Engl. J. Med. 361: 674–679. doi: 10.1056/NEJMoa0904023 [DOI] [PubMed] [Google Scholar]
- 3.Dawood F. S., Jain S., Finelli L., Shaw M. W., Lindstrom S., Garten R. J., Gubareva L. V., Xu X., Bridges C. B., Uyeki T. M., Novel Swine-Origin Influenza A(H1N1) Virus Investigation Team2009. Emergence of a novel swine-origin influenza A(H1N1) virus in humans. N. Engl. J. Med. 360: 2605–2615. doi: 10.1056/NEJMoa0903810 [DOI] [PubMed] [Google Scholar]
- 4.Dawood F. S., Iuliano A. D., Reed C., Meltzer M. I., Shay D. K., Cheng P. Y., Bandaranayake D., Breiman R. F., Brooks W. A., Buchy P., Feikin D. R., Fowler K. B., Gordon A., Hien N. T., Horby P., Huang Q. S., Katz M. A., Krishnan A., Lal R., Montgomery J. M., Mølbak K., Pebody R., Presanis A. M., Razuri H., Steens A., Tinoco Y. O., Wallinga J., Yu H., Vong S., Bresee J., Widdowson M. A.2012. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect. Dis. 12: 687–695. doi: 10.1016/S1473-3099(12)70121-4 [DOI] [PubMed] [Google Scholar]
- 5.Fodor E., Crow M., Mingay L. J., Deng T., Sharps J., Fechter P., Brownlee G. G.2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76: 8989–9001. doi: 10.1128/JVI.76.18.8989-9001.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garten R. J., Davis C. T., Russell C. A., Shu B., Lindstrom S., Balish A., Sessions W. M., Xu X., Skepner E., Deyde V., Okomo-Adhiambo M., Gubareva L., Barnes J., Smith C. B., Emery S. L., Hillman M. J., Rivailler P., Smagala J., de Graaf M., Burke D. F., Fouchier R. A., Pappas C., Alpuche-Aranda C. M., López-Gatell H., Olivera H., López I., Myers C. A., Faix D., Blair P. J., Yu C., Keene K. M., Dotson P. D., Jr, Boxrud D., Sambol A. R., Abid S. H., St George K., Bannerman T., Moore A. L., Stringer D. J., Blevins P., Demmler-Harrison G. J., Ginsberg M., Kriner P., Waterman S., Smole S., Guevara H. F., Belongia E. A., Clark P. A., Beatrice S. T., Donis R., Katz J., Finelli L., Bridges C. B., Shaw M., Jernigan D. B., Uyeki T. M., Smith D. J., Klimov A. I., Cox N. J.2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325: 197–201. doi: 10.1126/science.1176225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatta M., Gao P., Halfmann P., Kawaoka Y.2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293: 1840–1842. doi: 10.1126/science.1062882 [DOI] [PubMed] [Google Scholar]
- 8.Herfst S., Chutinimitkul S., Ye J., de Wit E., Munster V. J., Schrauwen E. J., Bestebroer T. M., Jonges M., Meijer A., Koopmans M., Rimmelzwaan G. F., Osterhaus A. D., Perez D. R., Fouchier R. A.2010. Introduction of virulence markers in PB2 of pandemic swine-origin influenza virus does not result in enhanced virulence or transmission. J. Virol. 84: 3752–3758. doi: 10.1128/JVI.02634-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh Y., Shinya K., Kiso M., Watanabe T., Sakoda Y., Hatta M., Muramoto Y., Tamura D., Sakai-Tagawa Y., Noda T., Sakabe S., Imai M., Hatta Y., Watanabe S., Li C., Yamada S., Fujii K., Murakami S., Imai H., Kakugawa S., Ito M., Takano R., Iwatsuki-Horimoto K., Shimojima M., Horimoto T., Goto H., Takahashi K., Makino A., Ishigaki H., Nakayama M., Okamatsu M., Takahashi K., Warshauer D., Shult P. A., Saito R., Suzuki H., Furuta Y., Yamashita M., Mitamura K., Nakano K., Nakamura M., Brockman-Schneider R., Mitamura H., Yamazaki M., Sugaya N., Suresh M., Ozawa M., Neumann G., Gern J., Kida H., Ogasawara K., Kawaoka Y.2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460: 1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayaraman A., Pappas C., Raman R., Belser J. A., Viswanathan K., Shriver Z., Tumpey T. M., Sasisekharan R.2011. A single base-pair change in 2009 H1N1 hemagglutinin increases human receptor affinity and leads to efficient airborne viral transmission in ferrets. PLoS ONE 6: e17616. doi: 10.1371/journal.pone.0017616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobinger G. P., Meunier I., Patel A., Pillet S., Gren J., Stebner S., Leung A., Neufeld J. L., Kobasa D., von Messling V.2010. Assessment of the efficacy of commercially available and candidate vaccines against a pandemic H1N1 2009 virus. J. Infect. Dis. 201: 1000–1006. doi: 10.1086/651171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linderman S. L., Chambers B. S., Zost S. J., Parkhouse K., Li Y., Herrmann C., Ellebedy A. H., Carter D. M., Andrews S. F., Zheng N. Y., Huang M., Huang Y., Strauss D., Shaz B. H., Hodinka R. L., Reyes-Terán G., Ross T. M., Wilson P. C., Ahmed R., Bloom J. D., Hensley S. E.2014. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013-2014 influenza season. Proc. Natl. Acad. Sci. U.S.A. 111: 15798–15803. doi: 10.1073/pnas.1409171111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maines T. R., Jayaraman A., Belser J. A., Wadford D. A., Pappas C., Zeng H., Gustin K. M., Pearce M. B., Viswanathan K., Shriver Z. H., Raman R., Cox N. J., Sasisekharan R., Katz J. M., Tumpey T. M.2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325: 484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez-Briseño D., Torre-Bouscoulet L., Herrera-Zamora J. J., Díaz-Rico J., Sandoval-Macías G., Pérez-Padilla R., Hernández-Cárdenas C., Regalado-Pineda J., Salas-Hernández J., Santillán-Doherty P.2016. Clinical Characteristics and Mortality of Influenza A H1N1 and Influenza-Like Illness in Mexico City in the 2013-2014 Winter Season. Rev. Invest. Clin. 68: 147–153. [PubMed] [Google Scholar]
- 15.Meunier I., Embury-Hyatt C., Stebner S., Gray M., Bastien N., Li Y., Plummer F., Kobinger G. P., von Messling V.2012. Virulence differences of closely related pandemic 2009 H1N1 isolates correlate with increased inflammatory responses in ferrets. Virology 422: 125–131. doi: 10.1016/j.virol.2011.10.018 [DOI] [PubMed] [Google Scholar]
- 16.Munster V. J., de Wit E., van den Brand J. M., Herfst S., Schrauwen E. J., Bestebroer T. M., van de Vijver D., Boucher C. A., Koopmans M., Rimmelzwaan G. F., Kuiken T., Osterhaus A. D., Fouchier R. A.2009. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 325: 481–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muramoto Y., Noda T., Kawakami E., Akkina R., Kawaoka Y.2013. Identification of novel influenza A virus proteins translated from PA mRNA. J. Virol. 87: 2455–2462. doi: 10.1128/JVI.02656-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann G., Zobel A., Hobom G.1994. RNA polymerase I-mediated expression of influenza viral RNA molecules. Virology 202: 477–479. doi: 10.1006/viro.1994.1365 [DOI] [PubMed] [Google Scholar]
- 19.Okada T., Morozumi M., Matsubara K., Komiyama O., Ubukata K., Takahashi T., Iwata S.2011. Characteristic findings of pediatric inpatients with pandemic (H1N1) 2009 virus infection among severe and nonsevere illnesses. J. Infect. Chemother. 17: 238–245. doi: 10.1007/s10156-010-0115-z [DOI] [PubMed] [Google Scholar]
- 20.Otte A., Marriott A. C., Dreier C., Dove B., Mooren K., Klingen T. R., Sauter M., Thompson K. A., Bennett A., Klingel K., van Riel D., McHardy A. C., Carroll M. W., Gabriel G.2016. Evolution of 2009 H1N1 influenza viruses during the pandemic correlates with increased viral pathogenicity and transmissibility in the ferret model. Sci. Rep. 6: 28583. doi: 10.1038/srep28583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Padilla R., de la Rosa-Zamboni D., Ponce de Leon S., Hernandez M., Quiñones-Falconi F., Bautista E., Ramirez-Venegas A., Rojas-Serrano J., Ormsby C. E., Corrales A., Higuera A., Mondragon E., Cordova-Villalobos J. A., INER Working Group on Influenza2009. Pneumonia and respiratory failure from swine-origin influenza A(H1N1) in Mexico. N. Engl. J. Med. 361: 680–689. doi: 10.1056/NEJMoa0904252 [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez A., Pérez-González A., Nieto A.2007. Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II. J. Virol. 81: 5315–5324. doi: 10.1128/JVI.02129-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Safronetz D., Rockx B., Feldmann F., Belisle S. E., Palermo R. E., Brining D., Gardner D., Proll S. C., Marzi A., Tsuda Y., Lacasse R. A., Kercher L., York A., Korth M. J., Long D., Rosenke R., Shupert W. L., Aranda C. A., Mattoon J. S., Kobasa D., Kobinger G., Li Y., Taubenberger J. K., Richt J. A., Parnell M., Ebihara H., Kawaoka Y., Katze M. G., Feldmann H.2011. Pandemic swine-origin H1N1 influenza A virus isolates show heterogeneous virulence in macaques. J. Virol. 85: 1214–1223. doi: 10.1128/JVI.01848-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanz-Ezquerro J. J., de la Luna S., Ortín J., Nieto A.1995. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J. Virol. 69: 2420–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi M., Jagger B. W., Wise H. M., Digard P., Holmes E. C., Taubenberger J. K.2012. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J. Virol. 86: 12411–12413. doi: 10.1128/JVI.01677-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith H., Sweet C.1988. Lessons for human influenza from pathogenicity studies with ferrets. Rev. Infect. Dis. 10: 56–75. doi: 10.1093/clinids/10.1.56 [DOI] [PubMed] [Google Scholar]
- 27.WHO 2010. H1N1 in post-pandemic period. http://www.who.int/mediacentre/news/statements/2010/h1n1_vpc_20100810/en/ [accessed December 29, 2016].
- 28.WHO 2015. Surveillance and monitoring Influenza update. http://www.who.int/influenza/surveillance_monitoring/updates/latest_update_GIP_surveillance/en/ [accessed December 29, 2016].
- 29.Wright P. F., Nerumanm G., Kawaoka Y.2007. Orthomyxoviruses. pp. 1691–1740. In: Fields Virology, 5th ed. (Knipe, D. M. and Howley, P. M. eds.), Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- 30.Yewdell J. W., Ince W. L.2012. Virology. Frameshifting to PA-X influenza. Science 337: 164–165. doi: 10.1126/science.1225539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Zhang Q., Gao Y., He X., Kong H., Jiang Y., Guan Y., Xia X., Shu Y., Kawaoka Y., Bu Z., Chen H.2012. Key molecular factors in hemagglutinin and PB2 contribute to efficient transmission of the 2009 H1N1 pandemic influenza virus. J. Virol. 86: 9666–9674. doi: 10.1128/JVI.00958-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Z., Yi C., Zhao L., Wang S., Zhou L., Hu Y., Zou W., Chen H., Jin M.2014. PB2-588I enhances 2009 H1N1 pandemic influenza virus virulence by increasing viral replication and exacerbating PB2 inhibition of beta interferon expression. J. Virol. 88: 2260–2267. doi: 10.1128/JVI.03024-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou B., Li Y., Halpin R., Hine E., Spiro D. J., Wentworth D. E.2011. PB2 residue 158 is a pathogenic determinant of pandemic H1N1 and H5 influenza a viruses in mice. J. Virol. 85: 357–365. doi: 10.1128/JVI.01694-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou B., Pearce M. B., Li Y., Wang J., Mason R. J., Tumpey T. M., Wentworth D. E.2013. Asparagine substitution at PB2 residue 701 enhances the replication, pathogenicity, and transmission of the 2009 pandemic H1N1 influenza A virus. PLoS ONE 8: e67616. doi: 10.1371/journal.pone.0067616 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.