Abstract
The instability of microsatellite DNA repeats is responsible for at least forty neurodegenerative diseases. Recently, Mirkin and colleagues presented a novel mechanism for microsatellite expansions based on break-induced replication (BIR) at sites of microsatellite-induced replication stalling and fork collapse. The BIR model aims to explain single-step, large expansions of CAG/CTG trinucleotide repeats in dividing cells. BIR has been characterized extensively in S. cerevisiae as a mechanism to repair broken DNA replication forks (single-ended DSBs) and degraded telomeric DNA. However, the structural footprints of BIR-like DSB repair have been recognized in human genomic instability and tied to the etiology of diverse developmental diseases; thus, the implications of the paper by Kim et al. extend beyond trinucleotide repeat expansion in yeast and microsatellite instability in human neurological disorders. Significantly, insight into BIR-like repair can explain certain pathways of complex genome rearrangements (CGRs) initiated at non-B form microsatellite DNA in human cancers.
Keywords: break-induced replication, chromothripsis, DNA repair, DNA replication, FoSTeS, genome instability, microsatellite instability
Introduction – Microsatellite expansions lead to human disease
Microsatellite DNAs are runs of repetitive sequences in which short motifs, typically 1–6 base pairs in length, are repeated 5–50 times at numerous loci across the genome in wild type cells [1, 2]. These tandem repeats are zones of replication fork stalling and DNA breakage, and are frequently correlated with sites of mutations, copy number variation, replication template switches or structural variant breakpoints [3–8]. Although the repeat tracts responsible for microsatellite expansion-related disorders share the common tendency to form noncanonical DNA structures, they comprise diverse sequences. Thus, CAG/CTG microsatellites are found in the coding regions of polyglutamine (poly-Q) disease-related genes where trinucleotide repeat expansion causes Huntington disease (HTT; Online Mendelian Inheritance in Man (OMIM) 613004), dentatorubral–pallidoluysian atrophy (ATN1; OMIM 607462), spinal and bulbar muscular atrophy (AR; OMIM 313700) and several forms of spinocerebellar ataxia. In contrast, Fragile × mental retardation (OMIM 300624) is attributed to expansion of CGG/CCG repeats in the FMR1 gene, Freidrich ataxia (FXN, OMIM 229300) is caused by expansion of GAA/TTC repeats in the frataxin gene, and myotonic dystrophy type 2 (DM2, OMIM 602688) is due to expansions of CCTG/CAGG repeats in the zinc-finger protein-9 gene.
At the Huntington disease (HD) locus the range of CAG/CTG repeats in asymptomatic individuals is between nine and thirty-seven. Complete penetrance of HD was observed for tract sizes equal to or greater than forty-two CAG/CTG repeats, while incomplete penetrance was observed with repeat lengths of thirty-six to forty-one [9]. The phenomenon of ‘genetic anticipation’ is a hallmark of HD and other microsatellite expansion disorders, where earlier onset and increased severity of symptoms are correlated with intergenerational expansions of microsatellite tracts. Because expansions of diverse microsatellites are the causative factors in multiple neurodegenerative disorders, understanding the mechanisms of microsatellite instability is of significant clinical relevance [10–12].
The analysis of microsatellite instability and chromosome breakage has been complicated by the diversity of microsatellite repeats, differences between model systems, variable cellular responses to different microsatellite repeat lengths, and the contribution of chromosome context [13–23], as well as observations that microsatellite-induced replication fork stalling does not unavoidably cause instability [24]. Nevertheless, there is general agreement that microsatellite repeats as a group cause replication stress and chromosome fragility in a length-dependent manner [25, 26]. In human cell culture, expanded CAG/CTG tracts are sites of replicative polymerase stalling [27], hairpin structure formation on leading and lagging strand replication templates in vivo [28, 29], and chromosome DSBs [6, 30]. Neutralization of lagging strand hairpins by transfection of cells with oligonucleotides complementary to either CTG or CAG lagging strand templates simultaneously eliminated both leading and lagging strand hairpin formation and relieved polymerase stalling [27]. These observations suggest that replication is coordinated on leading and lagging templates such that barriers formed on the single stranded lagging template can impede leading strand polymerization and promote leading strand hairpin formation. Hence, CAG/CTG microsatellite hairpin structures are foci of replication fork stalling which generate substrates susceptible to fork collapse and DNA breaks [6, 14, 31, 32].
Multiple replication- and repair-based mechanisms have been proposed to contribute to microsatellite instability including slippage/stuttering of DNA polymerases [33–36], and hairpin formation in nascent strand DNA during replication [28, 37] (Figure 1). Aberrant DNA damage signaling [38], replisome destabilization [14, 39], collisions with transcription machinery [40], R-loop formation [41, 42], POLβ/δ DNA synthesis during base excision repair (BER) of oxidized nucleotides [43–45] and binding of mismatch repair (MMR) proteins [10, 46]) have additionally been proposed to exacerbate microsatellite instability.
Figure 1.
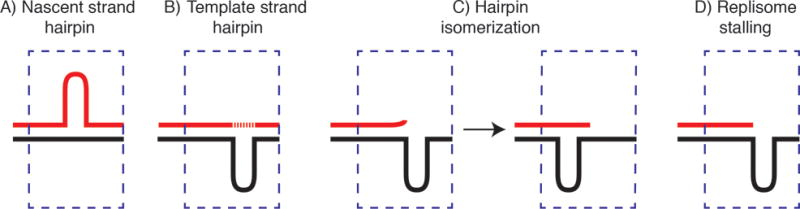
Hairpin formation during DNA replication or repair. A) Polymerase stuttering at microsatellite repeats leads to excess nascent strand microsatellite sequence (expansion). B) Terminal transferase-like nontemplated synthesis (dashed line) across a hairpin abasic gap [114–120] or C) Template hairpin isomerization following destabilization of a stalled polymerase [121] leads to contraction. D) Polymerase stalling leads to replication stress, fork collapse, single-ended DSB (seDSB).
Replication- and repair-based mechanisms of microsatellite instability are not mutually exclusive in dividing cells, whereas DNA repair pathways presumably account for expansions in postmitotic tissues [17, 44, 46, 47]. Indeed, the MSH2-MSH3 (MutSβ) MMR proteins have been shown to contribute to the expansion of long CAG/CTG tracts, irrespective of replication polarity [24, 44, 48–52]. In HeLa cell extracts MutSβ promotes expansion of CAG/CTG repeats by interaction with hairpin structures and recruitment of DNA polymerases β and δ [44]. In humans, MMR deficiency leads to short expansions and contractions of microsatellites (microsatellite instability (MSI, MIN)) in the cancer predisposition disorders Lynch syndrome (OMIM 120435) and Muir-Torre syndrome (OMIM 158320) in dividing cells of the colon and elsewhere [53–55], presumably due to the inability of MMR to resolve nascent and template strand DNA hairpins arising during replication, repair, or transcription.
The break-induced replication fork is unstable and highly mutagenic
The process of break-induced replication (BIR) is a homologous recombination pathway conserved from phage to eukaryotes [56–58] which serves to repair single-ended double-stranded breaks (seDSBs) such as might arise at collapsed or broken replication forks (Figure 2A). BIR has also been implicated in the recombination-dependent alternative lengthening of telomeres (ALT) [59]. In theory, any process that leads to breakage at single stranded DNA (e.g. stalled replication forks or transcription complexes, base/nucleotide excision repair tracts, non‐B DNA secondary structures) could lead to BIR once a replisome collides with the end of the broken DNA template.
Figure 2.
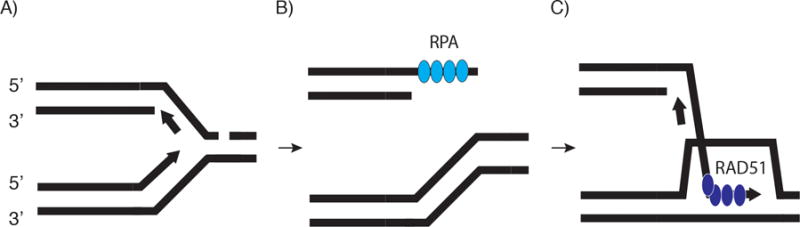
Model of break-induced replication. A) A single-ended DSB leads to BIR. A break in the lagging strand template is shown for simplicity, but other causes of fork collapse or nuclease cleavage (e.g. HO endonuclease [68] and camptothecin inhibition of topoisomerase I [85], have been used to produce seDSB. B) Displacement of the lagging strand template allows leading strand ligation to form an intact chromatid. The seDSB is subject to 5′ end resection and RPA binding. C) RPA is replaced by RAD51 to form and invading (acceptor) filament. The acceptor DNA released by branch migration of the unstable D-loop is a template for lagging strand conservative DNA synthesis.
BIR has been extensively characterized in molecular detail in budding yeast [57, 60–64], where 5′ DNA end resection at a seDSB is followed by RPA binding to the newly exposed 3′ single strand DNA (ssDNA) (Figure 2B) [62]. RPA is subsequently replaced by the RAD51 recombinase (Figure 2C), which directs strand invasion and D-loop formation at a complementary sequence. The complementary sequence is most often acquired from the proximal sister chromatid [62, 63], although annealing to non-allelic homologous or homeologous sequences is possible [65]. Establishment of a replisome containing the canonical CMG (CDC45-MCM-GINS) replicative helicase, PCNA and DNA polymerase at the 3′ ssDNA end [66] leads to extensive break-induced replication from the site of invasion, which can progress hundreds of kilobases [61, 63] but can be limited by resolvase (MUS81/YEN1) cleavage of BIR intermediates or fusion with an oncoming replication fork [67].
The initiation of BIR is slow following a seDSB, requiring several hours [68], and involving multiple template switches [69]. Although the components of the replicative helicase and the three major replicative DNA polymerases (α, δ, ε) are required for BIR, additional observations indicate that the BIR replication fork differs from a typical S-phase replication fork. Thus, BIR requires the nonessential POL32 (human POLD3) subunit of POLδ and POLζ, and the PIF1 helicase for processive DNA synthesis, and certain alleles of the PCNA clamp that can support semiconservative replication are dominant negative for BIR [66].
During BIR, POLδ is the primary polymerase for extension of the RAD51-coated invading strand, as the N-terminal polymerase catalytic domain of POLε is not essential in S. cerevisiae, and yeast mutants lacking this region of POLε are not hypersensitive to DSBs [70]. Additionally, yeast POLε is unable to perform displacement synthesis required to extend the invading strand D-loop in vitro, although a template unwinding function might be provided by the PIF1 helicase in vivo [71]. Nevertheless, based on the pattern of mutations during BIR in cells containing a proofreading-deficient allele, POLε has been implicated in second-strand synthesis [72].
Polymerization from the 3′ end of the invading strand is non-processive during the initial stages of break-induced replication [73], which include repeated cycles of invasion, synthesis, and D-loop dissociation, as well as multiple template switches [69]. Template switches are associated with modification of the PCNA processivity clamp by RAD18-dependent ubiquitination and SIZ1-dependent sumoylation [74, 75], as well as the recruitment of postreplication repair (PRR) translesion polymerases to the replication fork [76, 77]. Indeed, both RAD18 and SIZ1 are essential for efficient BIR [78].
The initial instability of the BIR D-loop suggested that translesion polymerases (Table 1) might contribute to the 1,000-fold greater mutagenicity of BIR vs. semiconservative replication [69, 79, 80]. Moreover, in vitro studies using yeast or human proteins to model homology-dependent DNA synthesis showed that POLη could extend RAD51-mediated D-loops [81–83] and efficiently utilize 3′ ends in the D-loop to synthesize several hundred base pairs of DNA. Unlike POLδ, D-loop extension by POLη did not depend on PCNA. In budding yeast, however, deletion of RAD30 (encoding DNA POLη) or REV3 (encoding the catalytic subunit of DNA POLζ) did not decrease the BIR-dependent repair of a segmented drug marker gene [66]. Nor did these TLS polymerase gene deletions have major effects on the frequency of frame shift mutations at A/T homopolymer tracts during BIR [84]. While these results leave open the possibility of other types of TLS mutations at alternative sequence motifs, the high mutation rate during BIR has been attributed instead to increased dNTP pools, an increased rate of POLδ errors due to decreased proofreading, and decreased efficiency of mismatch repair (MMR). Following leading strand synthesis, replication of the nascent strand displaced from the migrating D-loop is conservative, and serves to fix BIR mutations (Figure 2C) [61].
Table 1.
TLS DNA polymerases potentially involved in BIR
| DNA polymerase | Human gene (S. cerevisiae gene) | Biological activity | References |
|---|---|---|---|
| POL ζ | POLZ (REV3) hREV7 (REV7) POLD3 (POL31) POLD4 (POL32) |
Lower fidelity than other B family polymerases; extends mismatches; interacts with REV1, PCNA, POL δ (POL31, POL32); replication from nascent DNA template during MMBIR. | [70, 113–116] |
| POL η | POLH (RAD30) | Low fidelity polymerase; bypass of pyrimidine dimers; extensive D-loop synthesis; knockdown decreases HR in DT40 cells; interacts with nonubiquitinated and ubiquitinated PCNA | [74, 76, 115, 117–119] |
| POL κ | POLK | Highly error-prone polymerase (T → G transversions); strong mismatch-extending ability; extends RAD51-dependent D-loops | [76, 117] |
| REV1 | REV1L (REV1) | Deoxycytidyl transferase; inserts CdR across from abasic sites; interacts with multiple TLS polymerases and monoubiquitinated PCNA; promotes MMBIR | [70, 116, 120–122] |
| POL μ | POLM (N/A) | Template-independent terminal deoxynucleotidyl transferase (TdT) activity, able to accept template distortions | [110, 111] |
| POL θ | POLQ (N/A) | Template-independent terminal transferase (TdT) activity, implicated in alt-EJ/MMEJ | [106–109] |
Break-induced replication can lead to large expansions of CAG/CTG microsatellites
Now, the work by Kim et al. [85] adds the novel observation that break-induced replication is among the mechanisms leading to expansion of CAG/CTG microsatellites in budding yeast. The authors have engineered a system in which large CAG expansions lead to the transcriptional inactivation of a neighboring CAN1 arginine permease gene that can be scored by the conversion to canavanine resistance. In contrast to other potential causes of replication fork stalling and breakage such as R-loop formation or protein binding, the combination of two characteristics of microsatellites promote expansion by BIR; first, the tendency of repetitive sequences to form noncanonical structures that stall replication and cause chromosome fragility [5, 19, 25, 32, 38], and second, the likelihood of stochastic, homology-mediated repeat misalignment between sister chromatid microsatellites during strand invasion.
In the most parsimonius model of Kim et al., nuclease cleavage generates a seDSB at a replication fork stalled by stable hairpin formation (Figure 3A). 5′ excision is followed by RPA binding to the resected seDSB (Figure 3B). RPA is then replaced by RAD51 to form the invading 3′ end nucleoprotein filament and D-loop (Figure 3C). Base pairing of a 3′ CAG repeat to a template 3′ CTG repeat results in microsatellite expansion. Additional expansions may occur through hairpin formation during synthesis of the nascent leading or lagging strands (Figure 3D).
Figure 3.
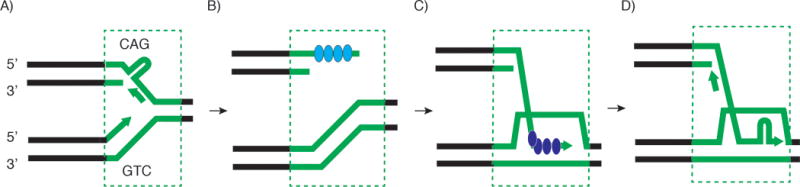
Model for (CAG/CTG) microsatellite expansion by break-induced replication. A) Replication stalling at a (CAG/CTG) repeat tract (green). B) DNA cleavage (MUS81-EME2) at the site of stalling leads to a single-ended double strand break, fork collapse (replisome dissociation), resection of the 5′ end of the seDSB and binding of RPA to the extended 3′ ssDNA. C) Replacement of RPA by RAD51 and homology-dependent invasion of the sister chromatid repeat forms a displacement loop (D-loop). Misalignment of the acceptor (CAG) and donor (CTG) repeats at the start or middle of the template repeat tract leads to large expansions. D) Repeat expansions larger than the initial repeat tract length arise after continued template misalignment, mutation-prone replication fork slippage and hairpin formation across the repeat. The acceptor DNA released from the unstable D-loop is a template for conservative lagging strand replication. Break-induced replication is subsequently terminated by fusion with a leftward moving replication fork and/or resolvase (MUS81, YEN1) cleavage.
Kim et al. have shown that genetic knockout of proteins involved in homologous recombination (RAD51, RAD52, MUS81/YEN1, MRE11) and break-induced-replication (POL32, PIF1) markedly decreased large CAG/CTG expansions, while knockout of proteins that positively (MSH2, MSH3, MSH6) or negatively (SRS2) affect small scale expansions did not affect large expansions. In the context of the blocking effects of rad51Δ and rad52Δ, the observation that deletion of the SRS2 helicase did not increase homology-dependent BIR [66, 85, 86] suggests that the SRS2 anti-recombinase activity is targeted to other forms of homologous recombination.
Treatment with HU to slow replication, or with camptothecin to introduce single strand DNA breaks at topoisomerase I cleavage sites, led to an approximate four-fold increase in large CAG expansions, indicating that replication stalling contributes to the BIR process, which can then continue into G2 or M phases [61, 87]. The authors also observed that transcription of the CAG/CTG repeat enhanced BIR, suggesting that RNA synthesis or an altered chromatin structure of the repeat tract may promote breakage. These observations raise the question of whether replisome components present in highly transcribed regions of postmitotic nuclei (e.g. in neurons) could promote BIR [88–90].
Under nonselective growth, large (BIR-related) expansions were observed at ~1 × 10−5 per replication, while small expansions (~1 × 10−2 per replication) and small contractions (~5–17 × 10−2 per replication) were approximately 1000-fold more frequent. In the experiments of Kim et al. [85], the expanded CAG tracts that were sequenced were found not to be mutated aside from changes in length, possibly because of the small number of tracts that were sequenced or the short length of the sequenced DNAs. Significantly, the authors showed that yeast containing an expanded (CAG/CTG)140 microsatellite could add large (>20 repeats) expansions, some of which exceeded the initial length of the (CAG/CTG)140 tract. Since It is well known that the tendency towards repeat instability increases with tract length, and repeated rounds of template switching occur during the early stages of BIR, it is possible that the largest expansions resulted from iterative rounds of strand invasion [69], as has been observed in synthesis-dependent strand annealing (SDSA) [91] and single-strand annealing (SSA) [92]. Additionally, alternative expansion mechanisms including polymerase stuttering, fork slippage and hairpin formation may also be at work during BIR of the leading or lagging nascent strands [93].
The demonstration by Kim et al. that BIR can lead to expansion of microsatellite DNAs takes on added significance because repeated sequences are prone to forming noncanonical DNA secondary structures (hairpins, slipped strands, triplexes, G-quadruplexes) that are hotspots of replication fork stress and DNA breakage [4, 94–97]. Thus, the same structures that interfere with semiconservative replication and promote BIR may also lead to stalling or collapse of a BIR replication fork, causing a transition to microhomology-mediated break-induced replication (MMBIR) [77]. Significantly, in contrast to the relatively long homology tracts necessary for RAD51-dependent BIR (>100–200 nucleotides [98]), MMBIR involves multiple template switching events to non-allelic sequences of 0–6 nucleotide homology or homeology and utilizes the error-prone POLζ and REV1 translesion polymerases [77].
Does BIR/MMBIR occur in human cells?
As discussed above, a break in the replication template can initiate BIR [57, 61]. Particularly relevant to breaks at non-B DNA structures, once homology-dependent invasion of a donor chromosome has occurred, microsatellite-induced replisome stress in the donor template, template breaks or replication fork collapse [77, 79] could trigger the transition to MMBIR, especially if levels of RAD51 are limiting [73] (Figure 4A, B). Further rounds of template switching can occur to nonallelic templates containing 0–6 bases of homology or homeology, including snapping back to use the nascent strand as template (Figure 4C, D). Template switching may be promoted by DNA damage or noncanonical structures in the MMBIR template, imperfect primer synthesis by TLS polymerases, the lack of TLS polymerase processivity, D-loop instability, or collapse of the MMBIR fork.
Figure 4.
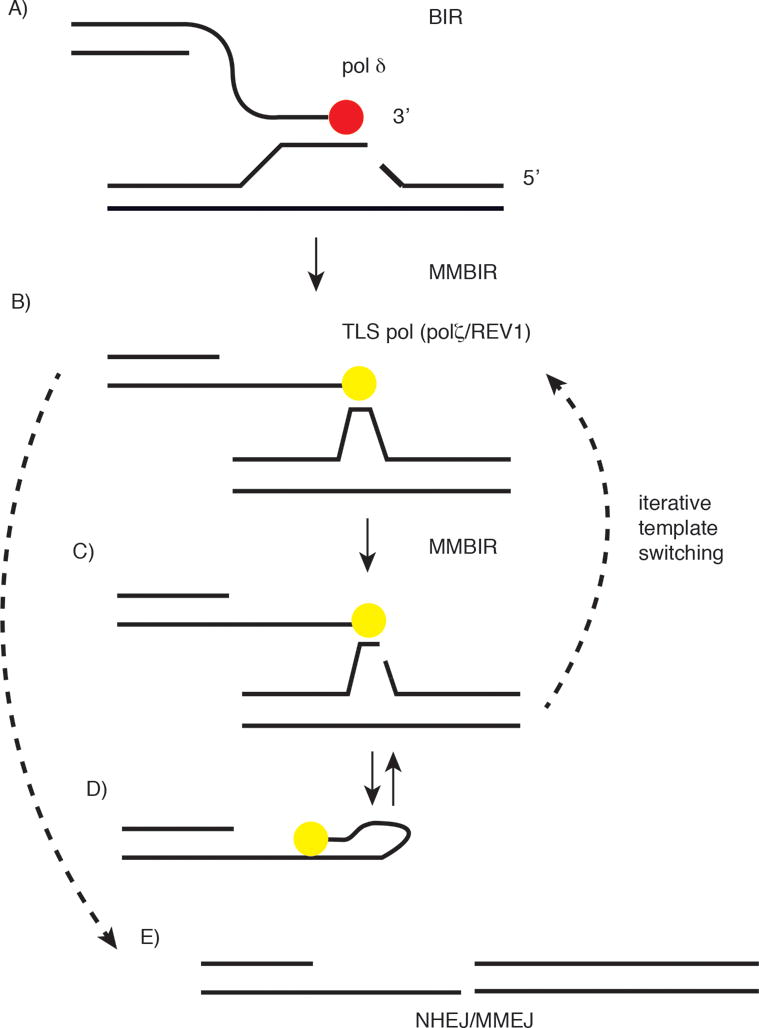
Template switching and the transition from BIR to MMBIR. A) Fork stalling within or beyond microsatellite sequences causes fork collapse/breakage. B) TLS polymerases (Polζ/Rev1) enable microhomology-mediated BIR (MMBIR). TLS polymerase synthesis is not processive and fork collapse leads to template switching and microhomology-mediated BIR at a new site. C) Successive cycles of fork stalling and template switching (FoSTeS) lead to complex genomic rearrangements (CGRs). D) Self-annealing and DNA synthesis at microhomologies in nascent DNA. E) DSBs at simultaneously broken microsatellites may recombine by nonhomologous end joining (NHEJ) or microhomology-mediated end joining (MMEJ).
MMBIR has been proposed to be an important mechanism responsible for non-recurrent chromosomal rearrangements (i.e. rearrangements at the same genetic locus that differ in size and sequence between individuals) associated with developmental disorders and cancer [73, 99–101]. In contrast to models in which cancer results from the gradual accumulation of driver gene mutations that successively enhance tumorigenesis [102, 103], a crucial finding was the discovery that tens to hundreds of complex genomic rearrangements comprising at least two breakpoint junctions can be formed during a single catastrophic DNA repair event termed chromothripsis [99, 104]. Thus, in contrast to the fusion of distal DNA sequences after “chromosome shattering”, BIR/MMBIR models posit that breakpoint junctions result from fork stalling and template switching (FoSTeS) [100]. The complex genomic rearrangements characteristic of chromothripsis are observed in 2%–3% of all cancers [104]. The MMBIR mechanism is proposed to be common to chromothripsis, chromoanasynthesis (inherited, constitutive CGRs) [105], and kataegis (localized clusters of single nucleotide somatic hypermutation) [106] in that these chromosome catastrophes arise from local and long range template switching and ordinarily are localized to only one or a few chromosomes.
Epidemiological analyses of diverse tumors across human populations estimate that well over one-third of oncogenic driver mutations arise from replication-based error mechanisms, as opposed to inherited or environmentally induced mutations [103]. DNA sequencing of structural variants from individuals exhibiting similar disease phenotypes has revealed signature features of MMBIR. In contrast to NHEJ, NAHR (nonallelic homologous recombination) or BIR, MMBIR is characterized by junction microhomologies, complex breakpoint junctions (duplications, triplications, inversions) attributed to short-range template switching in cis within the same replication fork, and juxtaposition of DNA sequences ordinarily separated by large genomic distances (>10–100 kb) due to FoSTeS in trans between distinct replication forks [101, 107]. DNA synthesis during CGR repair also exhibits increased single-nucleotide variation and enhanced indel (50–100 bp) mutation rates near breakpoint junctions (~2.1 × 10−4 mutations/bp, ~1.7 × 10−3 events/bp, respectively) [107], consistent with highly error-prone DNA synthesis following template switching.
Recently, Hickson and colleagues have reported BIR-like mitotic DNA synthesis (MiDAS) to occur at common fragile sites (CFS) [87]. Like MMBIR, MiDAS is not RAD51 dependent. Similar to BIR, MiDAS involves generation of a single-ended DSB by the MUS81-EME1 nuclease, RAD52 loading of the noncatalytic POLδ subunit POLD3, and conservative DNA synthesis. Costantino et al. have also reported that RAD51-dependent BIR occurs in replication-stressed (cyclin E overexpressing) human cells [60]. This form of BIR is nominally sensitive to siRNA knockdown of POLD3 (the human ortholog of POL32), or RAD52, or to genetic deletion of RAD52 [108], but not to knockdown of PIF1 [108].
Gu et al. have proposed that Alu-mediated recombination is the result of BIR-like recombination-coupled DNA replication [109]. Human Alu repeats contain fork-stalling homonucleotide (A/T) tracts of up to 100 base pairs, and are present at approximately one million copies per genome [110, 111], making them prime candidates for replication fork stalling, homology-dependent annealing and FoSTeS. While NAHR and NHEJ mechanisms of DNA repair have been associated Alu-mediated recombination leading to more than a dozen human diseases, including Fanconi anemia (OMIM 300514), Alzheimer’s disease (OMIM 157140) and Gaucher disease (OMIM 231000), Alu-mediated MMBIR/FoSTeS is specifically implicated in hereditary spastic paraplegia (OMIM 604277) and Waardenberg syndrome type 4 (OMIM 613266) [111].
Microsatellite repeats comprise approximately 3% of the human genome [112]. Taken with the results of Kim et al. [85], examples of microsatellite-dependent fork stalling and DSBs during replication stress enlarge the significance of BIR/MMBIR to include not only neurological and developmental diseases but the formation and progression of tumors. Many new questions are raised by these studies, viz., Does replication stress lead to microsatellite seDSBs at structure-prone repeats other than CAG/CTG tracts [30]? Do concurrent breaks at multiple microsatellites add to the complexity of FoSTeS and “chromosome catastrophes” [113]? What PCNA modifications occur during BIR/MMBIR to allow TLS polymerase recruitment and template switching? How does the distance from a replication origin affect the frequency of BIR, and D-loop stability? What are the effects of ataxia telangiectasia and RAD3-related (ATR) protein binding and downstream kinase signaling on BIR/MMBIR? Can BIR occur in postmitotic cells such as neurons? What is the role of BIR/MMBIR in the response to chemotherapy-induced ssDNA breaks? Thus, the questions raised by BIR in yeast and human cells have far reaching implications for genome stability in normal and pathological conditions.
Outlook
Several themes ought to be considered in the context of the conclusion by Kim et al. [85] that broken replication forks can lead to microsatellite instability by chromosome template switching, namely that microsatellites are prone to replication fork stalling and breakage; that microsatellites in human cells are hotspots of recombination; and that CGRs show evidence of BIR/MMBIR-driven recombination. One next step will be to construct human model systems that exhibit microsatellite breakage under replication stress to test the effects of non-B structures on fork collapse, quantitate the rate of BIR-derived mutagenesis, and characterize the genomic consequences of BIR/MMBIR initiated at specific loci.
Acknowledgments
This work was supported by a grant from the NIGMS (GM099874) to ML.
Abbreviations
- (MM)BIR
(microhomology-mediated) break-induced replication break-induced replication
- (se)DSB
(single-ended) double strand break
Footnotes
The author has no conflict of interest to declare.
References
- 1.Schlotterer C. Evolutionary dynamics of microsatellite DNA. Chromosoma. 2000;109:365–71. doi: 10.1007/s004120000089. [DOI] [PubMed] [Google Scholar]
- 2.Toth G, Gaspari Z, Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000;10:967–81. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacolla A, Jaworski A, Larson JE, Jakupciak JP, et al. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc Natl Acad Sci USA. 2004;101:14162–7. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bose P, Hermetz KE, Conneely KN, Rudd MK. Tandem repeats and G-rich sequences are enriched at human CNV breakpoints. PLoS One. 2014;9:e101607. doi: 10.1371/journal.pone.0101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr AM, Lambert S. Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J Mol Biol. 2013;425:4733–44. doi: 10.1016/j.jmb.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 6.Gadgil R, Barthelemy J, Lewis T, Leffak M. Replication stalling and DNA microsatellite instability. Biophys Chem. 2016 doi: 10.1016/j.bpc.2016.11.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen JM, Chuzhanova N, Stenson PD, Ferec C, et al. Complex gene rearrangements caused by serial replication slippage. Hum Mutat. 2005;26:125–34. doi: 10.1002/humu.20202. [DOI] [PubMed] [Google Scholar]
- 8.Leclercq S, Rivals E, Jarne P. DNA slippage occurs at microsatellite loci without minimal threshold length in humans: a comparative genomic approach. Genome Biol Evol. 2010;2:325–35. doi: 10.1093/gbe/evq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinkman RR, Mezei MM, Theilmann J, Almqvist E, et al. The likelihood of being affected with Huntington disease by a particular age, for a specific CAG size. Am J Hum Genet. 1997;60:1202–10. [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt MH, Pearson CE. Disease-associated repeat instability and mismatch repair. DNA Repair (Amst) 2016;38:117–26. doi: 10.1016/j.dnarep.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 11.Seriola A, Spits C, Simard JP, Hilven P, et al. Huntington’s and myotonic dystrophy hESCs: down-regulated trinucleotide repeat instability and mismatch repair machinery expression upon differentiation. Hum Mol Genet. 2011;20:176–85. doi: 10.1093/hmg/ddq456. [DOI] [PubMed] [Google Scholar]
- 12.Wells RD, Ashizawa T. Genetic instabilities and neurological diseases. 2nd. Amsterdam; Boston: Elsevier; 2006. [Google Scholar]
- 13.Freudenreich CH, Stavenhagen JB, Zakian VA. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol Cell Biol. 1997;17:2090–8. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freudenreich CH, Lahiri M. Structure-forming CAG/CTG repeat sequences are sensitive to breakage in the absence of Mrc1 checkpoint function and S-phase checkpoint signaling: implications for trinucleotide repeat expansion diseases. Cell Cycle. 2004;3:1370–4. doi: 10.4161/cc.3.11.1246. [DOI] [PubMed] [Google Scholar]
- 15.Farrell BT, Lahue RS. CAG*CTG repeat instability in cultured human astrocytes. Nucleic Acids Res. 2006;34:4495–505. doi: 10.1093/nar/gkl614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Razidlo DF, Lahue RS. Mrc1, Tof1 and Csm3 inhibit CAG.CTG repeat instability by at least two mechanisms. DNA Repair (Amst) 2008;7:633–40. doi: 10.1016/j.dnarep.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panigrahi GB, Lau R, Montgomery SE, Leonard MR, et al. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat Struct Mol Biol. 2005;12:654–62. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- 18.Panigrahi GB, Slean MM, Simard JP, Gileadi O, et al. Isolated short CTG/CAG DNA slip-outs are repaired efficiently by hMutSbeta, but clustered slip-outs are poorly repaired. Proc Natl Acad Sci USA. 2010;107:12593–8. doi: 10.1073/pnas.0909087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelletier R, Krasilnikova MM, Samadashwily GM, Lahue R, et al. Replication and expansion of trinucleotide repeats in yeast. Mol Cell Biol. 2003;23:1349–57. doi: 10.1128/MCB.23.4.1349-1357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mankodi A, Logigian E, Callahan L, McClain C, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–73. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 21.van den Broek WJ, Nelen MR, Wansink DG, Coerwinkel MM, et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet. 2002;11:191–8. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 22.Houseley JM, Wang Z, Brock GJ, Soloway J, et al. Myotonic dystrophy associated expanded CUG repeat muscleblind positive ribonuclear foci are not toxic to Drosophila. Hum Mol Genet. 2005;14:873–83. doi: 10.1093/hmg/ddi080. [DOI] [PubMed] [Google Scholar]
- 23.de Haro M, Al-Ramahi I, De Gouyon B, Ukani L, et al. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum Mol Genet. 2006;15:2138–45. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- 24.Viterbo D, Michoud G, Mosbach V, Dujon B, et al. Replication stalling and heteroduplex formation within CAG/CTG trinucleotide repeats by mismatch repair. DNA Repair (Amst) 2016;42:94–106. doi: 10.1016/j.dnarep.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Voineagu I, Freudenreich CH, Mirkin SM. Checkpoint responses to unusual structures formed by DNA repeats. Mol Carcinog. 2009;48:309–18. doi: 10.1002/mc.20512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voineagu I, Surka CF, Shishkin AA, Krasilnikova MM, et al. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat Struct Mol Biol. 2009;16:226–8. doi: 10.1038/nsmb.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu G, Chen X, Leffak M. Oligodeoxynucleotide Binding to (CTG)•(CAG) Microsatellite Repeats Inhibits Replication Fork Stalling, Hairpin Formation, and Genome Instability. Mol Cell Biol. 2013;33:571–81. doi: 10.1128/MCB.01265-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G, Chen X, Bissler JJ, Sinden RR, et al. Replication-dependent instability at (CTG) × (CAG) repeat hairpins in human cells. Nat Chem Biol. 2010;6:652–9. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Axford MM, Wang YH, Nakamori M, Zannis-Hadjopoulos M, et al. Detection of slipped-DNAs at the trinucleotide repeats of the myotonic dystrophy type I disease locus in patient tissues. PLoS Genet. 2013;9:e1003866. doi: 10.1371/journal.pgen.1003866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barthelemy J, Hanenberg H, Leffak M. FANCJ is essential to maintain microsatellite structure genome-wide during replication stress. Nucleic Acids Res. 2016;44:6803–16. doi: 10.1093/nar/gkw433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sundararajan R, Gellon L, Zunder RM, Freudenreich CH. Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics. 2010;184:65–77. doi: 10.1534/genetics.109.111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–6. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 33.Hartenstine MJ, Goodman MF, Petruska J. Weak strand displacement activity enables human DNA polymerase beta to expand CAG/CTG triplet repeats at strand breaks. J Biol Chem. 2002;277:41379–89. doi: 10.1074/jbc.M207013200. [DOI] [PubMed] [Google Scholar]
- 34.Viguera E, Canceill D, Ehrlich SD. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001;20:2587–95. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canceill D, Viguera E, Ehrlich SD. Replication slippage of different DNA polymerases is inversely related to their strand displacement efficiency. J Biol Chem. 1999;274:27481–90. doi: 10.1074/jbc.274.39.27481. [DOI] [PubMed] [Google Scholar]
- 36.Ohshima K, Wells RD. Hairpin formation during DNA synthesis primer realignment in vitro in triplet repeat sequences from human hereditary disease genes. J Biol Chem. 1997;272:16798–806. doi: 10.1074/jbc.272.27.16798. [DOI] [PubMed] [Google Scholar]
- 37.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–40. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 38.Lahiri M, Gustafson TL, Majors ER, Freudenreich CH. Expanded CAG repeats activate the DNA damage checkpoint pathway. Mol Cell. 2004;15:287–93. doi: 10.1016/j.molcel.2004.06.034. [DOI] [PubMed] [Google Scholar]
- 39.Liu G, Chen X, Gao Y, Lewis T, et al. Altered replication in human cells promotes DMPK (CTG)(n). (CAG)(n) repeat instability. Mol Cell Biol. 2012;32:1618–32. doi: 10.1128/MCB.06727-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Y, Hubert L, Jr, Wilson JH. Transcription destabilizes triplet repeats. Mol Carcinog. 2009;48:350–61. doi: 10.1002/mc.20488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y, Dent SY, Wilson JH, Wells RD, et al. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci USA. 2010;107:692–7. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan X, Jiang N, Chen X, Zhou X, et al. R-loop structure: the formation and the effects on genomic stability. Yi Chuan. 2014;36:1185–94. doi: 10.3724/SP.J.1005.2014.1185. [DOI] [PubMed] [Google Scholar]
- 43.Trushina E, McMurray CT. Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience. 2007;145:1233–48. doi: 10.1016/j.neuroscience.2006.10.056. [DOI] [PubMed] [Google Scholar]
- 44.Guo J, Gu L, Leffak M, Li GM. MutSbeta promotes trinucleotide repeat expansion by recruiting DNA polymerase beta to nascent (CAG)n or (CTG)n hairpins for error-prone DNA synthesis. Cell Res. 2016;26:775–86. doi: 10.1038/cr.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Wilson SH. DNA base excision repair: a mechanism of trinucleotide repeat expansion. Trends Biochem Sci. 2012;37:162–72. doi: 10.1016/j.tibs.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morales F, Vasquez M, Santamaria C, Cuenca P, et al. A polymorphism in the MSH3 mismatch repair gene is associated with the levels of somatic instability of the expanded CTG repeat in the blood DNA of myotonic dystrophy type 1 patients. DNA Repair (Amst) 2016;40:57–66. doi: 10.1016/j.dnarep.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Gomes-Pereira M, Hilley JD, Morales F, Adam B, et al. Disease-associated CAG.CTG triplet repeats expand rapidly in non-dividing mouse cells, but cell cycle arrest is insufficient to drive expansion. Nucleic Acids Res. 2014;42:7047–56. doi: 10.1093/nar/gku285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams GM, Surtees JA. MSH3 Promotes Dynamic Behavior of Trinucleotide Repeat Tracts In Vivo. Genetics. 2015;200:737–54. doi: 10.1534/genetics.115.177303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakatani R, Nakamori M, Fujimura H, Mochizuki H, et al. Large expansion of CTG*CAG repeats is exacerbated by MutSbeta in human cells. Sci Rep. 2015;5:11020. doi: 10.1038/srep11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tome S, Holt I, Edelmann W, Morris GE, et al. MSH2 ATPase domain mutation affects CTG*CAG repeat instability in transgenic mice. PLoS Genet. 2009;5:e1000482. doi: 10.1371/journal.pgen.1000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foiry L, Dong L, Savouret C, Hubert L, et al. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum Genet. 2006;119:520–6. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- 52.Tian L, Hou C, Tian K, Holcomb NC, et al. Mismatch recognition protein MutSbeta does not hijack (CAG)n hairpin repair in vitro. J Biol Chem. 2009;284:20452–6. doi: 10.1074/jbc.C109.014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinen CD. Mismatch repair defects and Lynch syndrome: The role of the basic scientist in the battle against cancer. DNA Repair (Amst) 2016;38:127–34. doi: 10.1016/j.dnarep.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz RA, Torre DP. The Muir-Torre syndrome: a 25-year retrospect. J Am Acad Dermatol. 1995;33:90–104. doi: 10.1016/0190-9622(95)90017-9. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto H, Imai K. Microsatellite instability: an update. Arch Toxicol. 2015;89:899–921. doi: 10.1007/s00204-015-1474-0. [DOI] [PubMed] [Google Scholar]
- 56.Shcherbakov VP, Plugina L, Shcherbakova T, Kudryashova E, et al. Double-strand break repair and recombination-dependent replication of DNA in bacteriophage T4 in the absence of UvsX recombinase: replicative resolution pathway. DNA Repair (Amst) 2012;11:470–9. doi: 10.1016/j.dnarep.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 57.Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annu Rev Genet. 2012;46:455–73. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- 58.Saveson CJ, Lovett ST. Tandem repeat recombination induced by replication fork defects in Escherichia coli requires a novel factor, RadC. Genetics. 1999;152:5–13. doi: 10.1093/genetics/152.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roumelioti FM, Sotiriou SK, Katsini V, Chiourea M, et al. Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep. 2016;17:1731–7. doi: 10.15252/embr.201643169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costantino L, Sotiriou SK, Rantala JK, Magin S, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saini N, Ramakrishnan S, Elango R, Ayyar S, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–92. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruff P, Donnianni RA, Glancy E, Oh J, et al. RPA Stabilization of Single-Stranded DNA Is Critical for Break-Induced Replication. Cell Rep. 2016;17:3359–68. doi: 10.1016/j.celrep.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donnianni RA, Symington LS. Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci USA. 2013;110:13475–80. doi: 10.1073/pnas.1309800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Llorente B, Smith CE, Symington LS. Break-induced replication: what is it and what is it for? Cell Cycle. 2008;7:859–64. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- 65.Jain S, Sugawara N, Haber JE. Role of Double-Strand Break End-Tethering during Gene Conversion in Saccharomyces cerevisiae. PLoS Genet. 2016;12:e1005976. doi: 10.1371/journal.pgen.1005976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lydeard JR, Lipkin-Moore Z, Sheu YJ, Stillman B, et al. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010;24:1133–44. doi: 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mayle R, Campbell IM, Beck CR, Yu Y, et al. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349:742–7. doi: 10.1126/science.aaa8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Malkova A, Naylor ML, Yamaguchi M, Ira G, et al. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol. 2005;25:933–44. doi: 10.1128/MCB.25.3.933-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–5. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 70.Kesti T, Flick K, Keranen S, Syvaoja JE, et al. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–85. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 71.Ganai RA, Zhang XP, Heyer WD, Johansson E. Strand displacement synthesis by yeast DNA polymerase epsilon. Nucleic Acids Res. 2016;44:8229–40. doi: 10.1093/nar/gkw556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–5. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–79. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 75.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–20. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 76.Stafa A, Donnianni RA, Timashev LA, Lam AF, et al. Template switching during break-induced replication is promoted by the Mph1 helicase in Saccharomyces cerevisiae. Genetics. 2014;196:1017–28. doi: 10.1534/genetics.114.162297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sakofsky CJ, Ayyar S, Deem AK, Chung WH, et al. Translesion Polymerases Drive Microhomology-Mediated Break-Induced Replication Leading to Complex Chromosomal Rearrangements. Mol Cell. 2015;60:860–72. doi: 10.1016/j.molcel.2015.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–3. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 79.Malkova A, Ira G. Break-induced replication: functions and molecular mechanism. Curr Opin Genet Dev. 2013;23:271–9. doi: 10.1016/j.gde.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kane DP, Shusterman M, Rong Y, McVey M. Competition between replicative and translesion polymerases during homologous recombination repair in Drosophila. PLoS Genet. 2012;8:e1002659. doi: 10.1371/journal.pgen.1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, et al. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20:783–92. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 82.Sneeden JL, Grossi SM, Tappin I, Hurwitz J, et al. Reconstitution of recombination-associated DNA synthesis with human proteins. Nucleic Acids Res. 2013;41:4913–25. doi: 10.1093/nar/gkt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sebesta M, Burkovics P, Juhasz S, Zhang S, et al. Role of PCNA and TLS polymerases in D-loop extension during homologous recombination in humans. DNA Repair (Amst) 2013;12:691–8. doi: 10.1016/j.dnarep.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A. Break-induced replication is highly inaccurate. PLoS biology. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim JC, Harris ST, Dinter T, Shah KA, et al. The role of break-induced replication in large-scale expansions of (CAG)n/(CTG)n repeats. Nat Struct Mol Biol. 2017;24:55–60. doi: 10.1038/nsmb.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu J, Renault L, Veaute X, Fabre F, et al. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature. 2011;479:245–8. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bhowmick R, Minocherhomji S, Hickson ID. RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol Cell. 2016;64:1117–26. doi: 10.1016/j.molcel.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 88.Widrow RJ, Hansen RS, Kawame H, Gartler SM, et al. Very late DNA replication in the human cell cycle. Proc Natl Acad Sci USA. 1998;95:11246–50. doi: 10.1073/pnas.95.19.11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci. 2007;8:368–78. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- 90.Raji NS, Krishna TH, Rao KS. DNA-polymerase alpha, beta, delta and epsilon activities in isolated neuronal and astroglial cell fractions from developing and aging rat cerebral cortex. Int J Dev Neurosci. 2002;20:491–6. doi: 10.1016/s0736-5748(02)00079-5. [DOI] [PubMed] [Google Scholar]
- 91.McVey M, Adams M, Staeva-Vieira E, Sekelsky JJ. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics. 2004;167:699–705. doi: 10.1534/genetics.103.025411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mott C, Symington LS. RAD51-independent inverted-repeat recombination by a strand-annealing mechanism. DNA Repair (Amst) 2011;10:408–15. doi: 10.1016/j.dnarep.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Iraqui I, Chekkal Y, Jmari N, Pietrobon V, et al. Recovery of arrested replication forks by homologous recombination is error-prone. PLoS Genet. 2012;8:e1002976. doi: 10.1371/journal.pgen.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang H, Freudenreich CH. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol Cell. 2007;27:367–79. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang G, Carbajal S, Vijg J, DiGiovanni J, et al. DNA structure-induced genomic instability in vivo. J Natl Cancer Inst. 2008;100:1815–7. doi: 10.1093/jnci/djn385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu S, Wang G, Bacolla A, Zhao J, et al. Short Inverted Repeats Are Hotspots for Genetic Instability: Relevance to Cancer Genomes. Cell Rep. 2015;10:1674–80. doi: 10.1016/j.celrep.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Balakumaran BS, Freudenreich CH, Zakian VA. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum Mol Genet. 2000;9:93–100. doi: 10.1093/hmg/9.1.93. [DOI] [PubMed] [Google Scholar]
- 98.Waldman AS, Liskay RM. Dependence of intrachromosomal recombination in mammalian cells on uninterrupted homology. Mol Cell Biol. 1988;8:5350–7. doi: 10.1128/mcb.8.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–47. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 100.Zhang F, Khajavi M, Connolly AM, Towne CF, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–53. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carvalho CM, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet. 2016;17:224–38. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, et al. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tomasetti C, Li L, Vogelstein B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science. 2017;355:1330–4. doi: 10.1126/science.aaf9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stephens PJ, Greenman CD, Fu B, Yang F, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kloosterman WP, Tavakoli-Yaraki M, van Roosmalen MJ, van Binsbergen E, et al. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 2012;1:648–55. doi: 10.1016/j.celrep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 106.Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, et al. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–8. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carvalho CM, Pehlivan D, Ramocki MB, Fang P, et al. Replicative mechanisms for CNV formation are error prone. Nat Genet. 2013;45:1319–26. doi: 10.1038/ng.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sotiriou SK, Kamileri I, Lugli N, Evangelou K, et al. Mammalian RAD52 Functions in Break-Induced Replication Repair of Collapsed DNA Replication Forks. Mol Cell. 2016;64:1127–34. doi: 10.1016/j.molcel.2016.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gu S, Yuan B, Campbell IM, Beck CR, et al. Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3. Hum Mol Genet. 2015;24:4061–77. doi: 10.1093/hmg/ddv146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–9. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 111.Kim S, Cho CS, Han K, Lee J. Structural Variation of Alu Element and Human Disease. Genomics Inform. 2016;14:70–7. doi: 10.5808/GI.2016.14.3.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu P, Erez A, Nagamani SC, Dhar SU, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Masset H, Hestand MS, Van Esch H, Kleinfinger P, et al. A Distinct Class of Chromoanagenesis Events Characterized by Focal Copy Number Gains. Hum Mutat. 2016;37:661–8. doi: 10.1002/humu.22984. [DOI] [PubMed] [Google Scholar]
- 115.Kent T, Mateos-Gomez PA, Sfeir A, Pomerantz RT. Polymerase theta is a robust terminal transferase that oscillates between three different mechanisms during end-joining. Elife. 2016;5:e13740. doi: 10.7554/eLife.13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Black SJ, Kashkina E, Kent T, Pomerantz RT. DNA Polymerase theta: A Unique Multifunctional End-Joining Machine. Genes (Basel) 2016;7:E67. doi: 10.3390/genes7090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, et al. A gradient of template dependence defines distinct biological roles for family × polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–66. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 118.Juarez R, Ruiz JF, Nick McElhinny SA, Ramsden D, et al. A specific loop in human DNA polymerase mu allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res. 2006;34:4572–82. doi: 10.1093/nar/gkl457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moon AF, Gosavi RA, Kunkel TA, Pedersen LC, et al. Creative template-dependent synthesis by human polymerase mu. Proc Natl Acad Sci USA. 2015;112:E4530–4536. doi: 10.1073/pnas.1505798112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Northam MR, Moore EA, Mertz TM, Binz SK, et al. DNA polymerases zeta and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Res. 2014;42:290–306. doi: 10.1093/nar/gkt830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hedglin M, Pandey B, Benkovic SJ. Stability of the human polymerase delta holoenzyme and its implications in lagging strand DNA synthesis. Proc Natl Acad Sci USA. 2016;113:E1777–1786. doi: 10.1073/pnas.1523653113. [DOI] [PMC free article] [PubMed] [Google Scholar]


