Abstract
Dynamic changes in chromatin structure play an important role in transcription regulation. Recent studies have revealed two mechanisms that alter chromatin structure. One involves ATP-dependent chromatin remodeling, and the other involves acetylation of the core histone tails. We have previously purified and characterized a multi-subunit protein complex, NuRD, which possesses both nucleosome remodeling and histone deacetylase activities. Despite extensive biochemical characterization of the complex, little is known about the functions of its individual components. In this study, we focused on Mi2, a component of the NuRD complex. We found that, similar to the native NuRD complex, recombinant Mi2 is a DNA-dependent, nucleosome-stimulated ATPase. Kinetic analysis of the ATP hydrolysis reaction indicated that the differential stimulation of the Mi2 ATPase by DNA and nucleosomes were primarily due to their differential effects on the turnover number of the reaction. Furthermore, we demonstrated that recombinant Mi2 is an efficient nucleosome remodeling factor when compared to that of the native NuRD complex. Our results define the biochemical function of Mi2 and set the stage for understanding the mechanism of nucleosome remodeling in a defined reconstituted system.
INTRODUCTION
Packaging of DNA into chromatin blocks access to DNA by proteins. Dynamic changes in chromatin structure play important roles in transcription, replication, recombination and repair (1,2). Studies in the past several years have identified at least two types of protein complexes that are capable of altering chromatin structure to allow protein factors access to nucleosomal DNA. One involves multiprotein complexes that use energy derived from ATP hydrolysis to ‘remodel’ nucleosomes (1); the other involves covalent modification, in particular acetylation, of core histone tails (3). A common feature of the nucleosome remodeling complexes is the presence of a subunit belonging to the SWI2/SNF2 family of ATPases (4). This subunit is postulated to function as a processive, ATP-driven motor to disrupt DNA–histone interactions (5). The purification and functional characterization of the nucleosome remodeling and histone deacetylase complex, NuRD, suggests that the two chromatin modifying enzymatic activities could be coupled (6–9).
NuRD is a multi-protein complex that possesses both nucleosome remodeling and histone deacetylase activities (9). In addition to the four subunit histone deacetylase core, HDAC1/2 and RbAp46/48, which is also present in the Sin3 histone deacetylase complex (10,11), NuRD contains at least three more subunits: MTA2, MBD3 and Mi2 (9,12). MTA2 is a novel protein that is highly similar (65% identical) to the candidate metastasis associated protein MTA1 (12,13). Biochemical characterization of MTA2 indicates that it plays an important role in modulating the histone deacetylase activity of the NuRD complex (12). Recently, MTA2 was also shown to negatively regulate p53-mediated cell growth arrest and apoptosis through affecting p53 acetylation (14). MBD3 is a methyl-CpG-binding domain-containing protein, similar to MBD2 (15). However, the function of MBD3 in the NuRD complex is not known (12). Mi2 is a SWI2/SNF2 type helicase/ATPase domain-containing protein that was first identified as a dermatomyositis-specific autoantigen (16), and has been postulated to be responsible for the chromatin remodeling activity of the NuRD complex (9). Supporting this prediction is the recent demonstration that recombinant SWI2/SNF2 family members, Drosophila ISWI, and human BRG1 and BRM, are capable of remodeling nucleosomes out of the context of their respective native complexes, the chromatin-accessibility complex (CHRAC) and the human SWI/SNF complex (17,18). However, the remodeling activity of recombinant Mi2 protein has not been demonstrated and it is not clear whether any other NuRD components play a role in chromatin remodeling.
To understand the biochemical function of Mi2 in the NuRD complex, we generated recombinant Mi2 protein and compared its activities with those of the native NuRD complex. We found that recombinant Mi2, like the NuRD complex, is a DNA-dependent, nucleosome-stimulated ATPase. Importantly, recombinant Mi2 efficiently disrupts histone–DNA contact in an ATP-dependent manner. Our results define the biochemical function of Mi2 and set the stage for understanding the mechanism of nucleosome remodeling in a defined reconstituted system.
MATERIALS AND METHODS
NuRD and rMi2 purification
The NuRD complex was purified as previously described (9). To generate recombinant Mi2 (rMi2), human Mi2 cDNA (accession no. X86691) with a 6× his C-terminal tag was cloned into the NotI and SmaI sites of the baculovirus expression vector pVL1392 (PharMingen), and was co-transfected with BaculoGoldTM AcNPV DNA. After isolation and plaque-purification, the recombinant baculovirus was amplified and used for infection of SF9 cells. Three days after infection, cells were collected and washed with phosphate-buffered saline before suspending in buffer C (20 mM HEPES pH 7.9, 1 mM DTT, 0.2 mM EDTA, 0.2 mM PMSF, 10% glycerol) containing 500 mM KCl and 0.1% NP-40. After homogenization, the supernatant was recovered by spinning at 25 000 r.p.m. (70 Ti, Beckman) for 20 min and dialyzed into buffer C containing 100 mM KCl (BC100) before loading onto a cellulose phosphate column. The bound proteins were eluted with a 10-column volume (10 cv) linear gradient from BC100 to BC1000. Fractions containing rMi2 were pooled and loaded onto a DEAE-52 column. The bound proteins were eluted with a 10 cv linear gradient from BC100 to BC400. Fractions containing rMi2 were pooled and bound to Ni-resin, the bound proteins were eluted and further separated on a Superose 6 gel filtration column. The purification process was monitored by western blot analysis. This purification procedure yielded rMi2 estimated to be >98% pure with ∼35% of recovery.
Nucleosome assembly, ATPase and mononucleosome disruption assays
Nucleosome assembly was performed with the salt dilution method (19). Successful assembly of nucleosomes was verified by their slower migration in agarose gels relative to naked DNA, and their characteristic DNase I protection pattern. The ATPase assay was performed as described (9). DNA or histone octamers (0.5 µg) or an equivalent amount of nucleosomes was used in each reaction. For mononucleosome disruption assays, mononucleosomes were assembled with HeLa core histones and an equal molar ratio of DNA, consisting of a 9:1 mass ratio of sonicated herring sperm DNA (Boehringer Mannheim) to labeled Xenopus 5S DNA. Assembled nucleosomes (5 µl) and various amounts of rMi2 and NuRD were mixed in 20 µl of reaction containing 10 mM HEPES pH 7.9, 100 mM KCl, 3 mM MgCl2, 2 mM ATP, 1 mM DTT, 0.5 mM EDTA and 10% glycerol. The reactions were incubated at 30°C for 1 h before addition of CaCl2 to a final concentration of 10 mM for DNase I digestion. After removing protein by phenol extraction, the digested DNA fragments were resolved in a 7% sequencing gel.
RESULTS
Mi2 is a DNA-dependent, nucleosome-stimulated ATPase
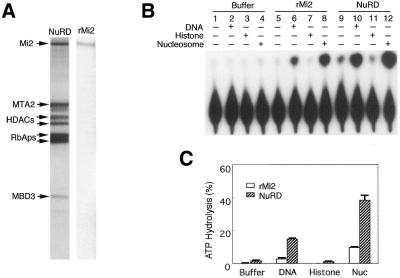
We and others have purified and characterized a nucleosome remodeling and histone deacetylase complex, NuRD (6–9). Analysis of this complex indicates that it contains seven major polypeptides (Fig. 1A). In addition to the two histone deacetylases HDAC1/2 and the two histone binding proteins RbAp46/48, which are also present in the Sin3 histone deacetylase complex (10,11), this complex contains three unique polypeptides including the SWI2/SNF2 ATPase domain-containing protein Mi2, the metastasis-associated protein MTA2 and the methyl-CpG-binding domain containing protein MBD3. Since Mi2 is likely to be the ATPase responsible for the remodeling activity of the NuRD complex, we decided to characterize its biochemical activity outside the context of the NuRD complex. To generate recombinant Mi2 protein, a baculovirus expressing human Mi2-β (also known as CHD4) was generated and used to infect SF9 cells. Recombinant Mi2 protein was purified to homogeneity from baculovirus infected SF9 cells (Fig. 1A).
Figure 1.
Mi2 is a DNA-dependent, nucleosome-stimulated ATPase. (A) Silver-stained SDS–PAGE containing the NuRD complex and recombinant Mi2. The identities of the proteins are indicated. (B) A comparison of the ATPase activity stimulated by DNA, histones or nucleosomes of equal amounts of Mi2 alone (rMi2) or in a complex (NuRD). (C) Quantification of the results shown in (B). Error bars represent the average deviation of two independent experiments.
ATPase assays were performed in the presence of DNA, core histones or nucleosomes to determine whether the purified recombinant Mi2 protein is functional (Fig. 1B). Buffer and the native NuRD complex were used as negative and positive controls, respectively. Like the NuRD complex, the ATPase activity of rMi2 was stimulated by both naked DNA and nucleosomes (Fig. 1B, compare lanes 6 and 8 to lanes 10 and 12). However, core histones had no effect on the ATPase activities under the same conditions (Fig. 1B, lanes 7 and 11). In addition, naked DNA was less efficient in stimulating the ATPase activity than was the same DNA that had been assembled into nucleosomes (Fig. 1B, compare lane 6 to 8 and lane 10 to 12). Quantification of the results from two independent experiments indicated that the ATPase activity of rMi2 was about one-third that of the native NuRD complex when equal numbers of Mi2 molecules were used (Fig. 1C). This result indicates that other NuRD components may have a role in assisting Mi2 to achieve optimal ATPase activity.
Kinetic analysis of rMi2 and NuRD ATPases
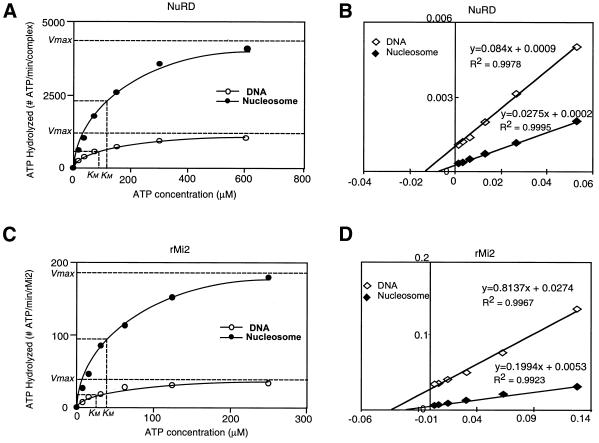
The above results indicate that DNA and nucleosomes have different stimulatory effects on the ATPase activity of the NuRD complex. In addition, the ATPase activity of rMi2 was only one-third that of the native NuRD complex. To understand how DNA and nucleosomes have different stimulatory effects on the ATPase of the NuRD complex and rMi2, and why rMi2 hydrolyzes ATP less efficiently than does the native NuRD complex, we performed a kinetic analysis of ATP hydrolysis by rMi2 and NuRD. A time course of ATP hydrolysis by the NuRD complex established that the reaction was linear for 1 h (data not shown), hence all ATPase reactions were allowed to progress for 30 min. By varying ATP concentration, we determined the velocities of ATP hydrolysis by the NuRD complex in the presence of DNA or nucleosomes (Fig. 2A). The maximal velocity (Vmax) and the Michaelis–Menten constant (Km) were then determined by curve fitting (Fig. 2A) and double-reciprocal plot (Fig. 2B) and were compared with those of other remodeling factors (Table 1). This analysis revealed that nucleosomes have a higher stimulatory effect on the NuRD ATPase activity than naked DNA because nucleosomes induced 4-fold higher Vmax and a 1.2-fold higher Km. Since Vmax measures the turnover number, our results indicate that nucleosomes stimulate the ATPase activity of the NuRD complex primarily by increasing its turnover number.
Figure 2.
Kinetic analysis of the ATP hydrolysis reaction catalyzed by NuRD and rMi2. (A and C) Plots of the ATP hydrolysis velocity, number of ATP molecules hydrolyzed by each molecule of rMi2 (C) or each NuRD (A) complex in 1 min, as a function of the ATP concentration. The plot for reactions in the presence of DNA or nucleosomes is indicated. (B and D) Double-reciprocal plots for NuRD (B) and rMi2 (D) kinetic analysis. The regression equations are shown in the Figures.
Table 1. Kinetic parameters of different nucleosome remodeling factors in ATP hydrolysis.
| Complex |
Cofactor |
Km (µM) |
Vmax (no.
ATP/min/complex) |
References |
| NuRD | DNA | 94 | 1111 | This work |
| Nucleosome | 112 | 4180 | This work | |
| rMi2 | DNA | 30 | 37 | This work |
| Nucleosome | 38 | 189 | This work | |
| xMi2 complex | Nucleosome | ND | <1 | 22 |
| ySWI/SNF | DNA | 100 ± 50 | 1000 ± 200 | 23 |
| Nucleosome | 100 ± 50 | 1000 ± 200 | 23 | |
| yRSC | DNA | 100 ± 50 | 1000 ± 200 | 23 |
| Nucleosome | 100 ± 50 | 1000 ± 200 | 23 |
rMi2, human recombinant Mi2; xMi2 complex, Xenopus Mi2 complex; no., number of ATP molecules hydrolysed in 1 min by each NuRD complex; ND, not determined.
To determine why rMi2 is less efficient than the NuRD complex in ATP hydrolysis, a similar kinetic analysis was performed using rMi2. Results shown in Figure 2C and D and Table 1 indicate that the Km values for rMi2 are comparable to those for NuRD (Table 1). However, the maximal velocities achieved by rMi2 are much lower than those attained by the NuRD complex regardless of the presence of naked DNA or nucleosomes (Table 1). Therefore, it is likely that other NuRD components modulate the ATPase activity of Mi2 by affecting its turnover number.
Mi2 remodels nucleosomes in an ATP-dependent manner
Having established that rMi2, like the NuRD complex, is a DNA-dependent, nucleosome-stimulated ATPase, we next asked whether rMi2, by itself, has nucleosome remodeling activity. To this end, rotationally phased mononucleosomes were assembled with HeLa core histone octamers and the 5S rDNA using the salt dilution method (19). The ability of the rMi2 to alter the DNase I digestion pattern of the nucleosomal DNA was tested. As shown in Figure 3, DNase I digestion of nucleosomal DNA produced a periodic pattern of enhanced cutting every 10 bp in the absence of remodeling (Fig. 3, lane 2), which contrasts the pattern of cutting for naked DNA (Fig. 3, lane 1) indicating successful assembly of nucleosomes. When increasing amounts of rMi2 were added to the remodeling reaction, the DNase I digestion pattern was altered (Fig. 3, compare lanes 3–6 to lane 2) indicating that the rMi2 alters the histone–DNA contact and therefore possesses nucleosome remodeling activity. The remodeling activity of rMi2 is ATP-dependent since the presence of rMi2 without ATP failed to alter the DNase I digestion pattern (Fig. 3, compare lanes 2 and 7). To compare the remodeling efficiency of rMi2 with that of the NuRD complex, parallel experiments with the NuRD complex were also performed (Fig. 3, lanes 8 and 9). Similar DNase I digestion was observed when equal concentrations of rMi2 and NuRD were compared (Fig. 3, lanes 4 and 9). Therefore, rMi2 itself has ATP-dependent nucleosome remodeling activity comparable to that of the NuRD complex.
Figure 3.
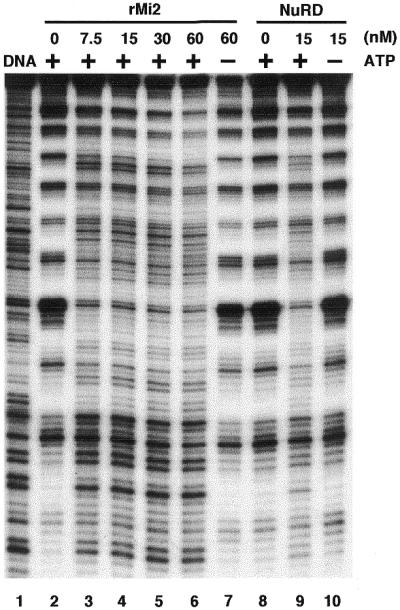
Mi2 is an ATP-dependent nucleosome remodeling factor. Mononucleosome disruption assay comparing the efficiency of rMi2 (lanes 2–7) with the NuRD complex (lanes 8–10). End-labeled mononucleosomes with a concentration of 20 nM were incubated with increasing concentrations of rMi2 as indicated, followed by DNase I digestion. The presence or absence of ATP in the reactions is indicated. The concentration of NuRD in the reactions is also indicated.
DISCUSSION
Both nucleosome remodeling factors and histone deacetylases have been isolated as multi-subunit protein complexes. The identification and characterization of the NuRD–Mi2 complex suggests that the two enzymatic activities involved in nucleosome modification may be functionally coupled (6–9). To understand the relationship between the two enzymatic activities, both need to be reconstituted. Here we demonstrated that Mi2, out of the context of the NuRD complex, is an efficient nucleosome remodeling factor.
The presence of an ATPase domain in the Mi2 protein suggests that Mi2 is likely responsible for the ATPase activity of the NuRD complex. This supposition was confirmed by the demonstration that highly purified recombinant Mi2, similar to the NuRD complex, was a DNA-dependent, nucleosome-stimulated ATPase (Fig. 1). Consistent with our finding, the ATPase activity of the Xenopus Mi2 complex was also found to be stimulated by both DNA and nucleosomes (20). While this manuscript was being prepared, Brehm and coworkers reported that recombinant Drosophila Mi2 (dMi2) is an ATPase (21). Interestingly, the ATPase activity of dMi2 was found to be stimulated exclusively by nucleosomes (21). Whether the discrepancy between human/Xenopus Mi2 and Drosophila Mi2 reflects a species-specific difference remains to be determined. Comparison of the abilities of DNA and nucleosomes in stimulating the ATPase activity of Mi2 and the NuRD complex indicated that nucleosomes are more efficient than naked DNA. The Km and Vmax values increased ∼1.2- and 4-fold, respectively, when the ATP hydrolysis reactions were performed in the presence of nucleosomes compared to when the reactions were performed in the presence of naked DNA (Table 1). Since Vmax measures the turnover number of an enzyme, our kinetic data indicate that nucleosomes increase the efficiency of ATP hydrolysis by rMi2 and NuRD primarily by increasing the turnover number. Therefore, Mi2 and NuRD are likely to recognize a specific feature of nucleosomes that is not found either on free histones or free DNA. Similarly, the Km and Vmax values increased 3- and 26-fold, respectively, when reactions performed with NuRD were compared to those with equal molecules of rMi2 (Table 1). Therefore, other components of the NuRD complex are likely to facilitate ATP hydrolysis by increasing the turnover number of Mi2. Comparison of the kinetic data of the NuRD complex with other nucleosome remodeling factors (Table 1) indicated that NuRD is one of the most active ATPases. With one exception (22), all the tested remodeling complexes hydrolyze ATP with a maximum velocity between 1000 and 4000 molecules/minute/complex in the presence of nucleosomes (Table 1).
Mononucleosome disruption assays indicated that the ability of recombinant Mi2 to disrupt rotationally phased mononucleosomes is comparable to that of the native NuRD complex (Fig. 3) although recombinant Mi2 is relatively less efficient in ATP hydrolysis compared to that of the NuRD complex (Fig. 1). It has been established that ATP hydrolysis is required for nucleosome disruption. However, whether the extent of nucleosome disruption is directly proportional to the ATPase activity is not clear. Nevertheless, our current studies, coupled with an earlier demonstration that other SWI2/SNF2 family members (the Drosophila ISWI, and human BRG1 and BRM) are also capable of remodeling nucleosomes out of the context of the CHRAC and hSWI/SNF complexes (17,18), indicate that the SWI2/SNF2 family members, found in all the three families of nucleosome remodeling factors, are capable of remodeling nucleosomes out the context of their respective protein complexes. Thus, the SWI2/SNF2 family members have an essential role in nucleosome remodeling. Other components of the different remodeling factors are likely to be responsible for regulating and targeting the different SWI2/SNF2 family proteins to achieve tissue- or gene-specific regulation. A major challenge is to understand how the different subunits coordinate to regulate chromatin function.
Acknowledgments
ACKNOWLEDGEMENTS
Y.Z. is a V-foundation scholar and is supported by the NIH (GM63067-01).
References
- 1.Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- 2.Schlissel M.S. (2000) Perspectives: transcription. A tail of histone acetylation and DNA recombination. Science, 287, 438–440. [DOI] [PubMed] [Google Scholar]
- 3.Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- 4.Eisen J.A., Sweder,K.S. and Hanawalt,P.C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res., 23, 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pazin M.J. and Kadonaga,J.T. (1997) SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein–DNA interactions? Cell, 88, 737–740. [DOI] [PubMed] [Google Scholar]
- 6.Tong J.K., Hassig,C.A., Schnitzler,G.R., Kingston,R.E. and Schreiber,S.L. (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature, 395, 917–921. [DOI] [PubMed] [Google Scholar]
- 7.Wade P.A., Jones,P.L., Vermaak,D. and Wolffe,A.P. (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol., 8, 843–846. [DOI] [PubMed] [Google Scholar]
- 8.Xue Y., Wong,J., Moreno,G.T., Young,M.K., Cote,J. and Wang,W. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell, 2, 851–861. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., LeRoy,G., Seelig,H.P., Lane,W.S. and Reinberg,D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95, 279–289. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y., Iratni,R., Erdjument-Bromage,H., Tempst,P. and Reinberg,D. (1997) Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell, 89, 357–364. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., Sun,Z.W., Iratni,R., Erdjument-Bromage,H., Tempst,P., Hampsey,M. and Reinberg,D. (1998) SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell, 1, 1021–1031. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y., Ng,H.H., Erdjument-Bromage,H., Tempst,P., Bird,A. and Reinberg,D. (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev., 13, 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toh Y., Pencil,S.D. and Nicolson,G.L. (1994) A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression and protein analyses. J. Biol. Chem., 269, 22958–22963. [PubMed] [Google Scholar]
- 14.Luo J., Su,F., Chen,D., Shiloh,A. and Gu,W. (2000) Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature, 408, 377–381. [DOI] [PubMed] [Google Scholar]
- 15.Hendrich B. and Bird,A. (1998) Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol., 18, 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seelig H.P., Moosbrugger,I., Ehrfeld,H., Fink,T., Renz,M. and Genth,E. (1995) The major dermatomyositis-specific Mi-2 autoantigen is a presumed helicase involved in transcriptional activation. Arthritis Rheum., 38, 1389–1399. [DOI] [PubMed] [Google Scholar]
- 17.Corona D.F., Langst,G., Clapier,C.R., Bonte,E.J., Ferrari,S., Tamkun,J.W. and Becker,P.B. (1999) ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell, 3, 239–245. [DOI] [PubMed] [Google Scholar]
- 18.Phelan M.L., Sif,S., Narlikar,G.J. and Kingston,R.E. (1999) Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell, 3, 247–253. [DOI] [PubMed] [Google Scholar]
- 19.Steger D.J., Eberharter,A., John,S., Grant,P.A. and Workman,J.L. (1998) Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc. Natl Acad. Sci. USA, 95, 12924–12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyer L.A., Logie,C., Bonte,E., Becker,P.B., Wade,P.A., Wolffe,A.P., Wu,C., Imbalzano,A.N. and Peterson,C.L. (2000) Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem., 275, 18864–18870. [DOI] [PubMed] [Google Scholar]
- 21.Brehm A., Langst,G., Kehle,J., Clapier,C.R., Imhof,A., Eberharter,A., Muller,J. and Becker,P.B. (2000) dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J., 19, 4332–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guschin D., Wade,P.A., Kikyo,N. and Wolffe,A.P. (2000) ATP-dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry, 39, 5238–5245. [DOI] [PubMed] [Google Scholar]
- 23.Logie C., Tse,C., Hansen,J.C. and Peterson,C.L. (1999) The core histone N-terminal domains are required for multiple rounds of catalytic chromatin remodeling by the SWI/SNF and RSC complexes. Biochemistry, 38, 2514–2522. [DOI] [PubMed] [Google Scholar]