Abstract
Single-molecule detection has long relied on fluorescent labeling with high quantum-yield fluorophores. Plasmon-enhanced detection circumvents the need for labeling by allowing direct optical detection of weakly emitting and completely nonfluorescent species. This review focuses on recent advances in single molecule detection using plasmonic metal nanostructures as a sensing platform, particularly using a single particle–single molecule approach. In the past decade two mechanisms for plasmon-enhanced single-molecule detection have been demonstrated: (1) by plasmonically enhancing the emission of weakly fluorescent biomolecules, or (2) by monitoring shifts of the plasmon resonance induced by single-molecule interactions. We begin with a motivation regarding the importance of single molecule detection, and advantages plasmonic detection offers. We describe both detection mechanisms and discuss challenges and potential solutions. We finalize by highlighting the exciting possibilities in analytical chemistry and medical diagnostics.
Keywords: plasmon sensing, single-molecule detection, fluorescence, label-free, nanoparticles, microscopy, functionalization, biosensing
Most single-molecule sensors exploit single-molecule fluorescence,1 which was first reported in the early 1990s.2,3 In the past decade several fluorescence-based technologies appeared on the market that exploit single-molecule sensitivity for, e.g., DNA sequencing4,5 and immunoassays based on single enzymes6,7 and single-molecule counting.8,9 The success of single-molecule fluorescence methods is due to the background-free nature of the signal resulting in a high signal-to-noise ratio. With modern optics the detection of single fluorophores has become routine in fields like materials science, biophysics, and sensing. Although widely employed, single-molecule fluorescence is fundamentally restricted to species that exhibit a sizable quantum yield (typically >0.1). Detection of weakly and nonemitting species is often done by fluorescently labeling the molecule of interest with a high quantum-yield fluorophore or quantum dot to generate the signal.10,11 However, such labeling is not feasible in applications like sensing where measurements are ideally conducted directly in a biological fluid with no washing steps.
This has sparked efforts to achieve so-called “label-free” sensors that do not require high-quantum-yield fluorescent labels to generate a signal. Solid-state and biological nanopores12 exploit changes in the current through a nanometer-sized pore when a molecule passes through it13,14 or binds to receptors near the pore.15 Carbon nanotubes have been employed as transducing elements that generate stepwise changes in conductance16,17 or exciton fluorescence18,19 upon interaction with single molecules. Whispering gallery modes in microresonators report local changes in refractive index or temperature induced by binding of analyte.20,21 Recently, plasmonic sensors based on individual gold nanoparticles have also been employed as sensors for single molecules, which is the focus of this review.
Biomolecular detection enhanced by plasmons was first achieved with a planar sensor, whereby prism coupling launches a propagating surface plasmon into a metallic film.22 On binding of biomolecules to the gold film, the local refractive index and thus the conditions for plasmon excitation are modified, causing changes in the intensity reflected from the film.23 Film based sensors are now commercially available and are used in many analytical laboratories to determine molecular interaction parameters and affinity. One of the drawbacks of planar plasmon sensors is their extended surface area that requires relatively large sample volumes and large numbers of molecular interactions to generate a detectable signal. A logical approach to overcome these limitations is the use of plasmonic nanoparticles, which exhibit a strongly reduced surface area compared to a metallic film.
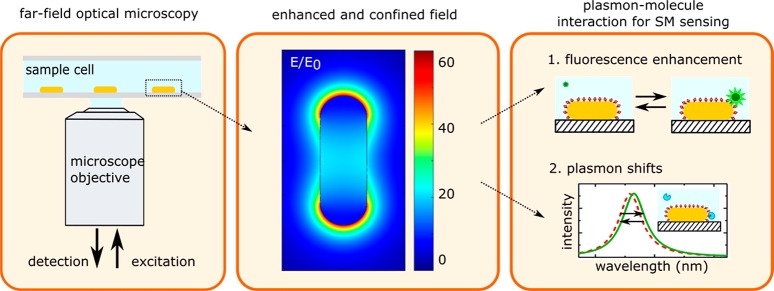
Plasmonic nanoparticles exhibit localized plasmon resonances due to the collective oscillation modes of the conduction electrons in the particle, which have to meet boundary conditions at the particle’s surface. The frequency of the plasmon resonance then depends on the shape, size, material, and environment of the particle. Localized plasmons in gold and silver particles occur at visible-NIR wavelengths (400–1000 nm) and are therefore compatible with standard optical microscopes (see Figure 1). Readout with far field optics enables sensing with low invasiveness, which is particularly advantageous in biological media, e.g., cells and tissue samples. Also, far-field optical microscopy systems are simple in design and can even be operated on a mobile-phone platform or an optical fiber.24
Figure 1.
Principle of plasmon-enhanced single-molecule sensing using nanoparticles. (left) The sensor is typically probed in a far-field optical microscope, where single particles are interrogated. (middle) The plasmon resonance induces a strongly enhanced and tightly confined local field around the particles. The field shown here is for a gold nanorod that is excited on resonance (calculated using the boundary element method). (right) This local field mediates plasmon–molecule interactions, enabling enhanced single-molecule detection by monitoring plasmon-induced changes of the molecule (resulting in, e.g., fluorescence enhancement) or by monitoring molecule-induced changes of the plasmon (resulting in frequency-shifts of the plasmon).
The ability to detect and study biomolecules using particle plasmons is facilitated by the electric field associated with the plasmon resonance (Figure 1b). This field penetrates the medium around the particle and enhances the interaction between a molecule and the particle’s plasmon. This plasmon–molecule interaction provides two pathways for sensing (Figure 1c): first, by exploiting the effect of the plasmon on the molecule leading to enhancement (or quenching) of the molecule’s fluorescence; or alternatively by exploiting the effect of the molecule on the plasmon leading to frequency shifts of the plasmon resonance.
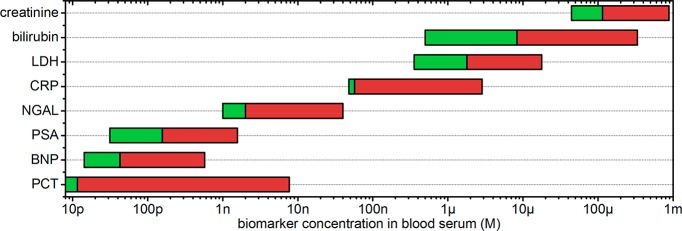
A major advantage of plasmon-mediated sensors is their compatibility with the highly concentrated molecular environment in cells and bodily fluids. Blood serum, for example, contains a total protein content of 60–80 g per liter, mainly consisting of albumin and globular proteins.25 Many clinically relevant biomarkers are present at concentrations ranging from several nanomolars to hundreds of micromolars (see Figure 2). An optical single-molecule sensor for these analytes thus needs a probe volume compatible with these high concentrations, ensuring that only a single protein is ever in the probe volume at once. The probe volume of a far-field microscope is typically 1 fL, restricting single-molecule detection to at most the low nanomolar range. In contrast, the probe volume of a plasmonic particle is determined by the extent of the locally enhanced field. This field typically extents over a few zeptoliters, 105 times smaller than the diffraction limit of light. This has already enabled single-molecule detection at concentrations as high as tens of micromolars.26,27
Figure 2.
Typical concentration range of clinically relevant biomarkers in blood serum.28 The green bars indicate reference values for healthy persons, whereas the red extension to the right indicates elevated values associated with disease. LDH: lactate dehydrogenase, CRP: c-reactive protein, NGAL: neutrophil gelatinase-associated lipocalin, PSA: prostate specific antigen, BNP: B-type natriuretic peptide, PCT: pro-calcitonin.
In this review we outline the recent progress in single-molecule sensing mediated by localized surface plasmons in metallic nanoparticles. We focus on approaches that are suitable to detect and characterize biomolecular interactions at the single-molecule level. We first summarize the available strategies to synthesize metal nanoparticles and their assemblies commonly used for sensing. Naturally, single-molecule sensing can only be achieved when individual plasmonic particles are resolved in the microscope, so we outline common strategies for optical single-particle detection. We then describe the detection of single weakly fluorescent molecules by plasmon-enhanced fluorescence, followed by the detection of completely nonfluorescent species by monitoring frequency shifts of the plasmon resonance. We conclude by outlining the current challenges in the field, and describe future applications.
Synthesis, Assembly, and Functionalization of Plasmon Sensors
Colloidal gold has been used since ancient Roman times to color glass with bright shades of yellow or red. The first scientific report on the synthesis of gold nanostructures dates from 1857 when Michael Faraday prepared gold colloids by reducing an aqueous solution of gold chloride with phosphorus.29 However, only in the past two decades have these methods been refined to generate high-quality colloidal samples with good control over the ensemble average size and shape. Moreover, the rapid development of nanofabrication facilities has led to new methods to fabricate nanosized metal structures using lithographic techniques. We will not describe all the available methods for preparation of metal particles, but rather highlight some of the methods and discuss their advantages in relation to sensing.
Wet-chemical synthesis of metal particles relies on the reduction of metal ions in solution. When a particular shape of metal particles is required, often chemicals are added that act as “shape directing” agents.30 The mechanisms underpinning these protocols have been developed empirically and there is no clear picture yet of how shape control works. The field has however developed rapidly in the past two decades and myriad shapes of metal particles can be synthesized with excellent yields.
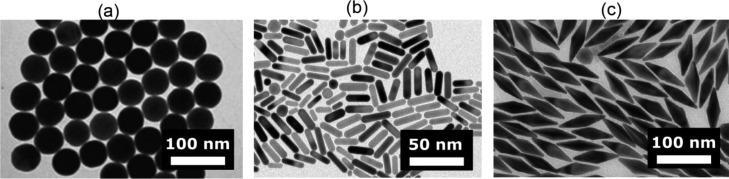
Nanospheres, nanorods, and nanobipyramids are the most commonly used geometries for plasmonic sensing because of their relatively narrow size distribution after synthesis, see Figure 3. This results in homogeneous plasmon line widths and near-field enhancements, which are crucial to reliable single-nanoparticle and single-molecule plasmon sensors. First synthesized by Turkevich in 195131 and later optimized by Brust in 197432 and others,33 gold nanospheres are straightforward to synthesize with near-unity yield and good size and shape homogeneity. Gold spheres, however, possess a plasmon resonance around 530 nm, close to the interband transitions in gold.34 This introduces significant losses that result in line broadening and a reduced local field enhancement, both detrimental if the particles are to be used as a sensor. For silver, the interband transitions are further away from a sphere’s plasmon resonance (around 400 nm), but silver is less commonly used due to its sensitivity to oxidation.35,36
Figure 3.
Particle shapes most commonly used for single-molecule plasmon-enhanced sensing. (a) Gold nanospheres. Figure adapted with permission from ref (33). Copyright 2013 American Chemical Society. (b) Gold nanorods synthesized by one of us following the protocol of El-Sayed.37 (c) Gold bipyramids. Figure adapted with permission from ref (38). Copyright 2015 John Wiley and Sons.
A red-shifted plasmon resonance away from the interband transitions in gold can be achieved by employing elongated particle shapes such as nanorods and nanobipyramids.39,40 Their longitudinal plasmon resonance lies conveniently in the visible to near-infrared wavelength range, making them ideal objects for biosensors.41−45 A purely wet-chemical method to synthesize nanorods was discovered by Murphy and co-workers,46 which was later modified by the group of El-Sayed.37 Gold nanorods with aspect ratios ranging from 2 to 5 can be routinely synthesized with nearly 100% yield, and the aspect ratio can be controlled by varying chemical conditions.
Liu and co-workers later showed47 that bipyramidal-shaped particles are formed with the same method when penta-twinned seeds stabilized by citrate are used instead of single-crystal seeds stabilized by amphiphilic surfactant. The yield of this protocol was later improved by Wang et al.,38 who employed shape-separation based on depletion forces to achieve near unity yield of bipyramids. These bipyramids are strikingly monodisperse in aspect ratio with a dispersion of only 5% (compared to 20% for nanorods), which results in a significantly narrower distribution of single-particle plasmon resonance wavelengths.45 Moreover, their sharp tips (typical tip radius of 3–5 nm) result in a higher field enhancement compared to their nanorod counterparts.40
Wet-chemical synthesis yields large quantities of well-dispersed particles with sizes down to several nanometers. Their crystalline nature47 ensures minimal losses of the plasmon due to scattering of electrons on surfaces and defects, resulting in narrow plasmon line widths and high field-enhancements.40 However, it is well-known that even higher field enhancements occur in nanometer-sized gaps between plasmonic particles or between a particle and a surface.50 This requires a post-synthesis, liquid-phase assembly protocol that usually employs molecular linkers such as DNA,51,52 zwitterions,53,54 or proteins.55,56
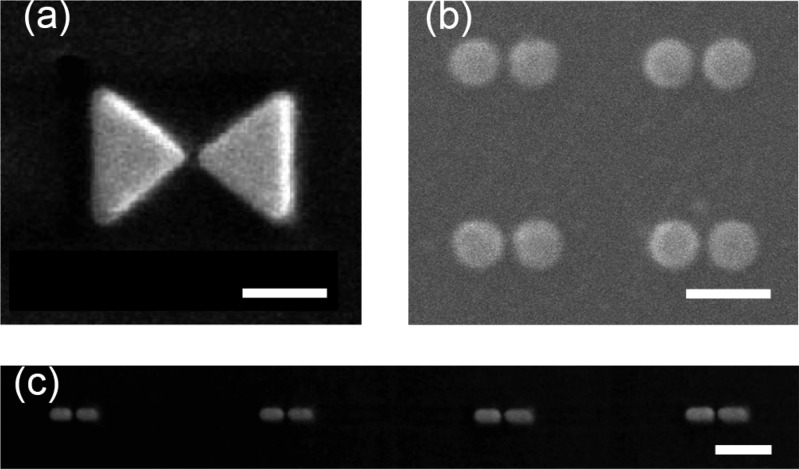
These assembly processes can create well-defined constructs of two or more metal particles. However, polydispersity is still present in the final colloidal solution; therefore the most used method to achieve well-controlled and reproducible assemblies of metallic particles is based on lithography. Lithographic methods employ a particle-beam (e.g., electrons or ions) to directly write nanostructures on a surface.57 Such methods have been employed to form nanostructures such as bow-tie48,58 and Yagi-Uda antennas,59 dimers of gold disks,49,60 and arrays of gold nanorods,48,61 with a few examples shown in Figure 4. Lithography gives control over the size, spacing and relative orientation of complex structures. The resolution of lithographic methods is determined by the quality of the particle-beam and the resist, but gaps <10 nm are usually difficult to realize.57 Importantly, lithographic structures typically exhibit broadened plasmon resonances and reduced near-field enhancements due to the amorphous crystal structure and the presence of adhesion layers.48
Figure 4.
Examples of nanoparticle pairs fabricated using lithographic techniques. (a) A bow-tie antenna. Figure adapted with permission from ref (48). Copyright 2010, Nature Publishing Group. (b) Gold-particle pairs. Figure adapted with permission from ref (49). Copyright 2009 American Chemical Society. (c) End-aligned gold nanorods. Figure adapted with permission from ref (48). Copyright 2010, Nature Publishing Group. All scale bars 200 nm.
An exciting development is the use of DNA origami structures to assemble colloidal particles on a scaffold of DNA.62−66 The scaffold can be designed to expose, e.g., thiolated strands of DNA that bind and immobilize gold particles at defined locations. This approach combines the unique optical properties of crystalline colloidal particles with the ability to assemble them with a controlled spacing and orientation. In such systems the average gap size is determined by the thickness of the DNA origami spacer, which can be controlled with nanometer accuracy. DNA origami has already been employed to assemble dimers of gold spheres (Figure 5a),63,65 chiral chains,63 and linear chains (Figure 5b),67 and to control the spacing and relative orientation of gold nanorods (Figure 5c).64,68
Figure 5.
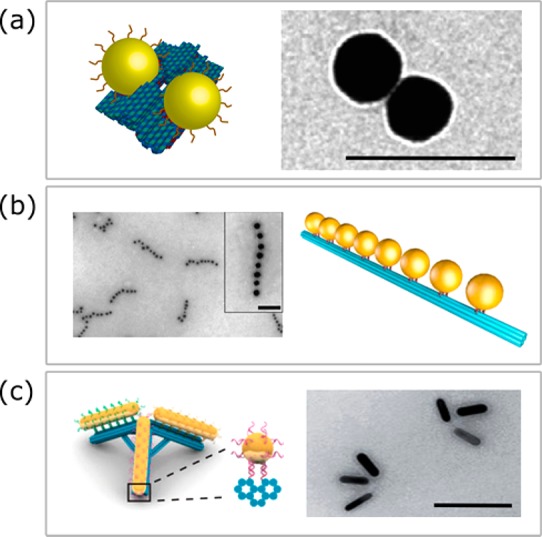
Examples of nanoparticle assemblies created on DNA-origami using thiolated docking strands. (a) Gold sphere dimers. Figure adapted with permission from ref (62). Copyright 2014 Nature Publishing Group. (b) Gold sphere chains. Figure adapted with permission from ref (67). Copyright 2016 American Chemical Society. (c) Gold nanorod trimers. Figure adapted with permission from ref (64). Copyright 2017 American Chemical Society. All scale bars 100 nm.
The detection of a specific biomolecule in a mixture typically requires functional groups on the surface of the sensor that exhibit a certain affinity for the analyte. Receptors can be attached to the surface of plasmonic particles using covalent thiol–chemistry. Commonly employed receptors include antibodies, oligonucleotides, aptamers, and peptides. Especially small receptor molecules such as aptamers and peptides are attractive because the sensitivity of a plasmon sensor scales with the local electric field intensity that rapidly decays away from the particle surface (see, e.g., Figure 14). Directing the analyte to bind close to the particle surface thus ensures maximum sensitivity. In addition, the local electric field intensity is inhomogeneously distributed on the particle surface, imparting regions with greater sensitivity compared to others. Often, sharp features and protrusions exhibit the highest field enhancements, such as the tips of a nanorod or the gap-region of a nanoparticle dimer. For that reason, functionalization protocols have been developed that specifically target the surface areas with high local field enhancement.
Figure 14.
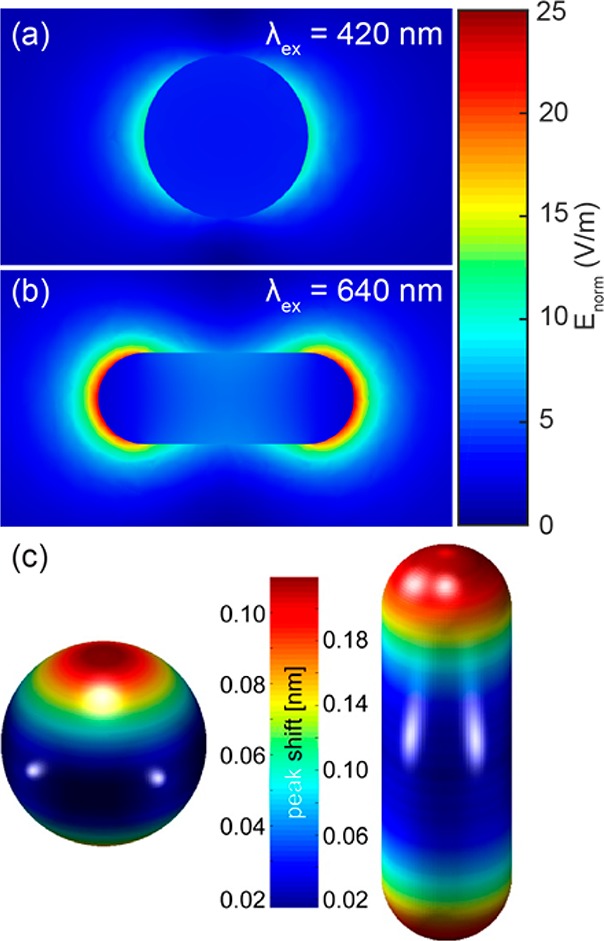
Local electric field intensity around a silver sphere (a) and a silver nanorod (b) immersed in water, simulated using the boundary element method. (c) Position dependent plasmon peak shifts for a dielectric nanosphere (radius 3 nm) binding to the surface of the particle in water. Figure adapted with permission from ref (78). Copyright 2016 American Chemical Society.
Several strategies have been reported to achieve such site-specific functionalization. Introducing polymeric blocking agents to inhibit the rate of receptor functionalization on “flat” facets results in selective functionalization of highly curved facets, such as the tips of a nanorod,69 the edges of a nanoplate,70 or the corners of a nanocube.71 Alternatively, the locally enhanced intensity can be employed directly for a photochemical reaction to achieve site-selective functionalization.72 If applicable, a clever combination of different substrate materials that offer orthogonal functionalization protocols has been employed to selectively functionalize the gap region in a gold particle dimer.73 Finally, the anistropic distribution of hot electrons in a nanoparticle can be employed to functionalize the gap region in a plasmonic dimer.74
Clearly many complementary strategies exist to synthesize and assemble plasmonic particles to be used as sensors. Most approaches result in a dispersion of particle sizes and assembly geometries, highlighting the need to correctly identify the geometry of the sensor to predict its sensing characteristics. This is often done by optical spectroscopy, which is the topic of the following section.
Optical Spectroscopy of Single Plasmonic Particles
The presence of plasmonic nanoparticles in the solution phase is easily identified by vivid solution colors, originating from the plasmonic extinction of a section of the visible spectrum. The extinction spectrum of these nanoparticles is inhomogeneously broadened due to geometric polydispersity inherent to any synthesis protocol.37 Naturally, single-molecule sensing requires the ability to resolve individual particles and their assemblies in a microscope, as probing a large ensemble of particles results in averaging of the asynchronous single-molecule events.75 The sensor output such as amplitude of the signal and binding kinetics strongly depend on the nanoscale geometry of the sensor.69,76−78 Therefore, a precise characterization of the optical and geometrical properties of the sensing nanoparticle is vital to the reliability of a plasmon sensor.
Here we describe the general optics required for single-particle detection and spectroscopy. Optical spectroscopy of single nanoparticles is usually done by immobilizing the particles on a substrate at a low density and isolating the particle’s signal from the orders of magnitude stronger fundamental excitation. Extensive reviews on the topic exist,41,81−83 so here we only summarize the aspects that are crucial to single-molecule plasmon sensing, i.e., how to unambiguously characterize the optical properties and geometry of the single-particle sensor.
Dark-field microscopy is the most commonly used method for single-particle detection, whereby scattering from immobilized nanoparticles is collected by an objective, while the fundamental excitation is prevented from entering the objective acceptance cone.84 Usually a dark-field condenser excites the sample with a cone of light (Figure 6a), while nanoparticle scattering is collected on the opposite side of the sample by a lower numerical aperture objective. If possible, background scatter and reflection from interfaces can be suppressed under index-matched conditions. Alternatively, total internal reflection excitation through an objective or an opposing prism (Figure 6b) can be used to set up a nonpropagating evanescent field that is scattered into the far field by a nanoparticle. The scattered photons are collected by the imaging objective, resulting in signals from single objects on a dark background (Figure 6c). By directing the signal from a single particle to a spectrometer, the scattering spectrum of individual particles can be recorded (Figure 6d).
Figure 6.
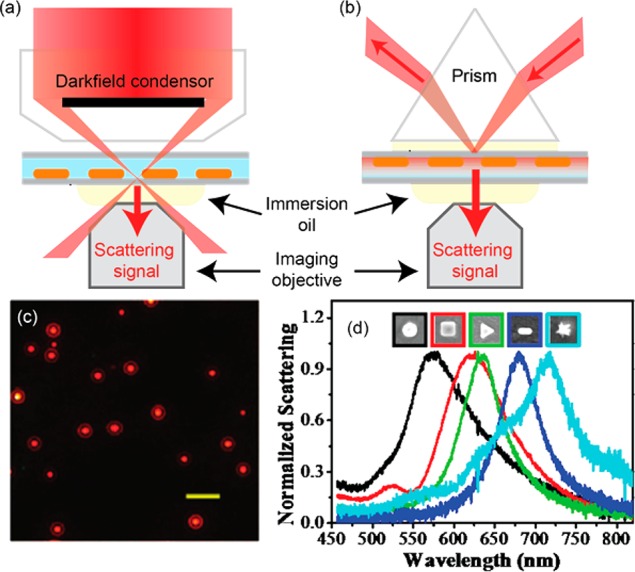
Dark-field excitation of nanoparticles (a) using a dark-field condenser and (b) by prism-style total internal reflection. (c) Dark-field scattering image of gold nanorods excited at 780 nm. Scale bar is 5 μm. Figure adapted with permission from ref (79). Copyright 2008 American Chemical Society. (d) Dark-field scattering spectra of differently shaped individual metallic nanoparticles. Figure adapted with permission from ref (80). Copyright 2014 American Chemical Society.
The scattering cross section scales with the square of the particle volume (for small enough particles), making scattering-based techniques less suitable for particles with an effective radius below 10 nm.81 The absorption cross section on the other hand scales as the volume of the particle, which has been exploited for detection of small particles that are difficult to image using scattering-based techniques.77,85 Photothermal detection exploits the fact that absorption of incident light results in photothermal heating of the nanoparticle, which induces a slight change of the refractive index around the particle that can be detected with a probe beam.85 Even though these refractive index changes are small, photothermal microscopy offers a high signal-to-noise ratio by employing an intense probe beam that is not resonant with the plasmon absorption. The ability of photothermal detection to resolve very small, individual metallic particles has been exploited for one of the first demonstrations of label-free single-molecule sensing, which we will describe later on.
Photoluminescence detection of single nanoparticles has been demonstrated using both one-86,87 and two-photon excitation.88,89 In contrast to scattering-based methods, here the fundamental excitation can be separated from the wavelength-shifted emission by optical filters. One-photon photoluminescence is induced by the absorption of a photon that promotes sub-Fermi-level electrons above the Fermi level.90 Two-photon excited photoluminescence (TPPL) is generated by two sequential single-photon absorption events exciting a single electron–hole pair.91 These excitations rapidly decay via electron–electron and electron–phonon scattering, with a very low probability of radiative decay in bulk gold (quantum yield 10–1090). In plasmonic nanoparticles, the radiative decay rate can be strongly enhanced by coupling to the plasmon resonance, boosting the quantum yield to 10–5–10–6.92,93 Despite the low quantum yield, the large absorption cross section of a single metallic particle results in photoluminescence signals that exceed those of single fluorophores.88
Other approaches to detect single metallic nanoparticles are by interferometric scattering microscopy,94 second and third harmonic generation,95 transient absorption spectroscopy,96 or four-wave mixing.97 In biosensing applications, however, dark-field scattering is the most commonly used method for single-particle detection due to its high signal-to-background ratio and straightforward implementation on commercial microscope platforms.41,81
In the described techniques, nanoparticles are imaged as diffraction limited spots distributed sparsely on a substrate. While the sparse arrangement suggests isolated nanoparticles or assemblies, some spots may correspond to multiparticle clusters with a physical dimension below the diffraction limit. These clusters can, for example, originate from the sample preparation that may involve deposition from solution and solvent evaporation. If clustered, the plasmons of the particles may hybridize,50 or if the interparticle spacing is larger than the diameter of the particles, the spectral response may be a superposition of multiple single-particle spectra.98,99 Uncontrolled clustering may also modulate near-field enhancements and field confinement.100 These clusters therefore lead to aberrant sensor responses and signal heterogeneity. It is therefore vital that the underlying particle geometry is identified somehow, which is often done using (1) spectroscopy or (2) correlative microscopy.
First, spectroscopy is often performed by sending the emitted light to a spectrometer.101 For example, in scattering spectroscopy the scattered signal is diffracted by a grating in the spectrometer and imaged on a camera. However, obtaining spectra is relatively slow because they are recorded particle-by-particle, complicating the acquisition of many spectra for statistical analysis. Acquisition rates can be improved by using an imaging spectrograph, which enables the acquisition of several spectra simultaneously.101 Highly parallelized spectroscopy has been achieved with several methods. Wide-field detection of the scattered light using a color camera enabled spectra of all nanoparticles in the field of view to be approximated algorithmically using RGB pixel values.102 Employing instead a tunable wavelength source, nanoparticle spectra have been reconstructed from multiple wide-field images recorded over a range of excitation wavelengths.103 Conversely in hyperspectral imaging, broadband excitation is used, but the scattering signal is passed through a tunable narrowband wavelength filter, and imaged on a camera.76,104 Capturing a series of images over a range of filter wavelengths enables the extraction of the spectra of all nanoparticles in the field of view in parallel. Tunable filtering can be implemented by either a continuously tunable acousto-optical device,104 or discretely by a filter wheel containing a range of narrow band-pass wavelength filters.76 Fitting extracted photon energy dependent scattering signals with a Lorentzian function then reliably extracts single particle line width and peak LSPR wavelength.76
By using spectroscopy the underlying geometry can be identifed to some degree by establishing, e.g., deviations in expected plasmon line width, peak wavelength, or signal intensity.81 For example, clustering of two or more nanospheres consistently leads to strongly red-shifted plasmon resonances compared to isolated spheres105 and integer distributions of signal intensity depending on the number of particles present.106 Identification of single gold nanorods is often done based on their well-defined plasmon line width,98 with greater line widths often indicating a cluster of nanoparticles. In some cases polarization dependent excitation or emission profiles can also reveal the presence of clusters.99
If optical spectroscopy alone proves insufficient to verify the underlying particle geometry, atomic force microscopy (AFM) and electron microscopy enable the nanoparticles to be imaged directly. This advantage comes at the cost of experimental complexity, given that electron microscopy images and optical properties of single nanoparticles must be correlated unambiguously.107 This correlation can be achieved using fiducial markers, such as large gold spheres, or by employing lithographic techniques based on templated evaporation,108 focused ion beam milling,109 or electron beam lithography.110 These techniques are used to fabricate alignment markers that can be used to compare the same sample-region in both modalities. An example is shown in Figure 7, where correlated optical and electron microscopy enabled silver nanospheres in single, dimer, or trimer configurations to be compared unambiguously.105 Such correlations have also been used to study dependencies of photothermal stability on nanoparticle geometry111 and to correlate the underlying geometry to two-photon luminescence signals.112
Figure 7.
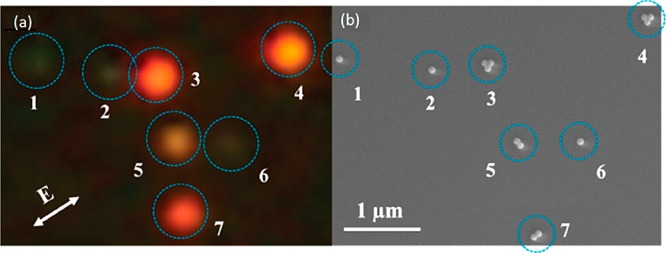
(a) Dark-field scattering image of single or clustered silver nanospheres. (b) A correlated scanning electron microscopy image revealing the underlying geometry and configuration of the nanospheres. Figure adapted with permission from ref (105). Copyright 2014 American Chemical Society.
In addition to electron microscopy, AFM has also been employed to establish particle geometries.87,113,114 The advantage of AFM is that it can be more easily integrated into an optical setup, although tip-convolution limits the accuracy with which nanoparticle dimensions can be extracted. AFM has also been used to manipulate individual particles to create particle clusters, e.g., dimers of nanospheres115 and orientation-controlled dimers of bipyramids.116 After manipulation with a high force, the resulting geometry can be imaged with a lower force.
Many methods are available to optically detect plasmonic nanoparticle sensors, alongside methods to reveal the underlying sensor geometry and stoichiometry. It will become clear in the following two sections that a precise knowledge of the underlying geometry is crucial in designing a sensor with the desired sensitivity and kinetic response.
Single-Molecule Sensing by Fluorescence Enhancement
Plasmon resonances in a metallic nanoparticle can be used to enhance and redirect the emission of biomolecules, which has enabled the detection of weakly emitting species and single-molecule Raman scattering signals.117 Without enhancement these processes are too inefficient to reach the single-molecule level. The investigations of plasmon-enhanced emission began in the 1960s when Drexhage found that a fluorescent molecule close to a metallic film exhibits modified decay times and angular distribution of fluorescence. Plasmon-enhanced fluorescence is widely employed in ensemble-averaged biosensors for the detection of RNA, DNA, and proteins,118,119 and has recently been extended to the single-molecule regime. Here we focus on single-molecule sensing applications only, and refer to recent reviews for ensemble-averaged biosensors.118−120 Since the early 2000s much effort has been spent to characterize the achievable enhancements in varying combinations of nanoparticles and emitters, down to the single-emitter and single-photon level. These efforts have been reviewed extensively, see, e.g., refs (117 and 121−123), so here we only summarize the mechanisms that determine the enhancement factors. We then redirect our focus to applications of plasmon-enhanced emission for single-molecule sensing.
The emission of a biomolecule is a function of the properties of the molecule and its interaction with the environment. Modification of the environment by, e.g., dielectric interfaces or metallic nanostructures can alter the local electric field, which directly affects the excitation rate and the emission properties. Overall, this can result in fluorescence quenching or enhancement, depending on the geometry and the orientation of the biomolecule with respect to the excitation polarization. Enhancement of the emission intensity of biomolecules in the near-field of a plasmonic particle is dominated by two factors, namely, (1) an increased excitation rate caused by the enhanced local electric field and (2) a modified quantum yield due to changes of the radiative and nonradiative decay-rate by coupling of the emitter to the particle. Basic theoretical treatment of these mechanisms has been published.124 Recently, charge transfer of hot carriers to nearby molecules has also shown to result in modifications of signal strength in select molecular compounds.125
The increased excitation rate of a fluorophore close to a metal structure is a direct consequence of the enhanced local electric field. Analytical and numerical calculations have shown that the resonant local field enhancement ranges from 5 (15) for a gold (silver) sphere to ∼50 for a gold nanorod and up to 150 for a gold bipyramid.40,126 The field enhancement in the gap of a dimer of gold spheres increases for smaller gap sizes and can reach values >100,127 whereas higher order aggregates of spheres128 and dimers of gold nanorods129 warrant even higher field enhancements. This field enhancement cannot be increased arbitrarily, however, with quantum mechanical effects and electron tunneling across small gaps posing intrinsic limits.127,130
The modified quantum yield is a consequence of coupling between the plasmon and the fluorophore that alters both the radiative and nonradiative rate of the complex. This can lead to enhancement of the quantum yield for intermediate distances from the particle, and quenching for distances smaller than a few nanometers due to coupling to so-called “dark modes” in the metal structure.131 The exact enhancement strongly depends on the spectral overlap between the plasmon and the spectrum of the fluorophore and the “unenhanced” quantum yield of the fluorophore.131 Direct measurement of the quantum yield enhancement is far from trivial and requires independent measurements of the excitation and emission rates.92,117,132
It is worthwhile to note that the excitation and emission enhancements are governed by the wavelength dependence of the plasmonic near-field, which is spectrally red-shifted from the far-field spectrum (i.e., the scattering and extinction spectrum observed in a spectrometer). This detuning can be understood by approximating the plasmon resonance as a driven damped harmonic oscillator. When damping is present the maximum displacement amplitudes occur at a lower energy than the resonance frequency, with a red shift that depends on the degree of damping.133 For plasmons this damping is caused by, e.g., the metal’s bulk absorption or radiative decay.133,134 In Figure 8 we show the calculated detuning between the near- and far-field spectra for a 150 nm gold sphere.133 The spectral shifts between near- and far-fields have later been experimentally confirmed by, e.g., surface-enhanced Raman scattering134 and near-field optical microscopy.135 This detuning should be considered when choosing the particles and laser wavelength for maximum fluorescence enhancement and detection sensitivity.
Figure 8.
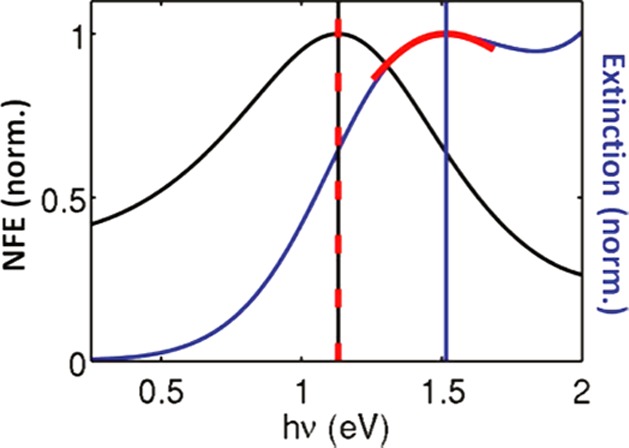
Calculated spectral profile of the near-field enhancement (NFE) and far-field extinction for a 150 nm gold sphere. The vertical black and blue lines denote the peak positions in the near-field and far-field spectra, respectively. Figure adapted with permission from ref (133). Copyright 2008 American Chemical Society.
The fluorescence enhancements are mediated by the near-field that is strongly confined compared to the diffraction limit of a conventional far-field microscope. Here the plasmon effectively converts freely propagating optical radiation into highly localized energy. The effective probe volume is on the order of a few zeptoliters, which is over 5 orders of magnitude smaller than that of a conventional microscope. This allows for single-molecule detection at concentrations of tens of micromolars and possibly higher. These concentrations approach the conditions inside a cell, where biomolecular concentrations of hundreds of micromolars are common. Plasmonic nanoparticles may therefore provide an opportunity to detect and study single biomolecules intracellularly, without requiring a high quantum yield.
Much effort has been spent characterizing the fluorescence enhancements and probe volume,117,121,123,132 particularly using fluorescence correlation spectroscopy (FCS) experiments. Herein a fluorophore or labeled biomolecule (DNA, protein) freely diffuses through the near-field around a plasmonic particle or particle dimer. The resulting fluorescence bursts are recorded on a camera or photon counter, which allows one to extract the overall enhancement factor by analyzing the amplitude of the bursts compared to the background emission from unenhanced molecules. The autocorrelation function contains information on the concentration of analyte and the diffusion time through the near-field. The latter can be used to estimate the probe volume using FCS theory, although accurate values are hard to obtain because the probe volume does not exhibit a Gaussian intensity profile anymore.
Orrit’s group found enhancements exceeding 1000-fold for single fluorophores with a quantum yield of 2% diffusing near single gold nanorods immobilized on a substrate.124,136 These studies showed that the overall enhancement factor strongly depends on the spectral overlap between the dye excitation and emission spectra, and the excitation-laser wavelength with respect to the plasmon resonance (see Figure 9). The autocorrelation functions displayed two characteristic times associated with free diffusion of the dye through the near-field of the particle, and a slower time constant due to dyes that stick to the particle or the glass and subsequently bleach. This highlights the need for coatings that suppress nonspecific interactions with the sensor, which we will discuss in more detail under Challenges.
Figure 9.
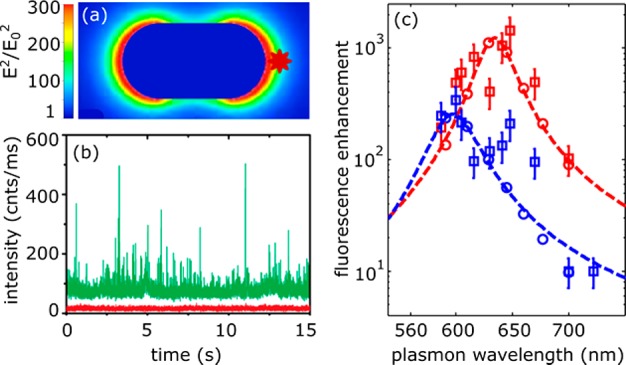
Single-molecule fluorescence enhancements of crystal violet (quantum-yield 2%) by a single gold nanorod. (a) Numerical calculation of the enhanced intensity around a single gold nanorod excited on resonance with its longitudinal plasmon. (b) Time trace of dye fluorescence with (green) and without (red) the presence of a gold nanorod. The nanorod is immobilized on a substrate while crystal violet diffuses freely in solution, causing spikes in intensity when it enters the particle's near-field. (c) Measured fluorescence enhancements for several individual gold nanorods, each with a different plasmon wavelength. The blue squares indicate an excitation wavelength of 594 nm, the red squares 633 nm. The open circles and dashed lines indicate the numerically calculated enhancement. Figure adapted with permission from ref (124). Copyright 2014 American Chemical Society.
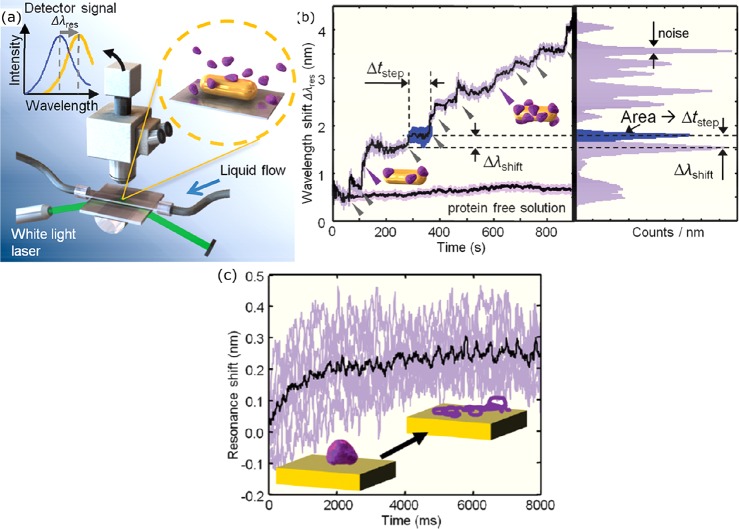
Moerner’s group employed bowtie nanoantennas fabricated by e-beam lithography that consist of two triangular particles spaced by a 10 nm gap. These antennas yielded fluorescence enhancements in excess of 1000 for a dye with a quantum yield of 2.5%, and their near-field volume of several tens of zeptoliters.58 Recently, Wenger’s group introduced an “antenna-in-box” platform that consists of a dimer of plasmonic spheres surrounded by a square aperture milled in a gold film137 (see Figure 10). This aperture further suppressed the background from surrounding molecules and yielded a fluorescence intensity enhancement of over 1000-fold for a dye with a quantum yield of 8%.137 Also, this platform exhibited a zeptoliter probe volume, which allowed for single-molecule detection at 30 μM concentrations.
Figure 10.
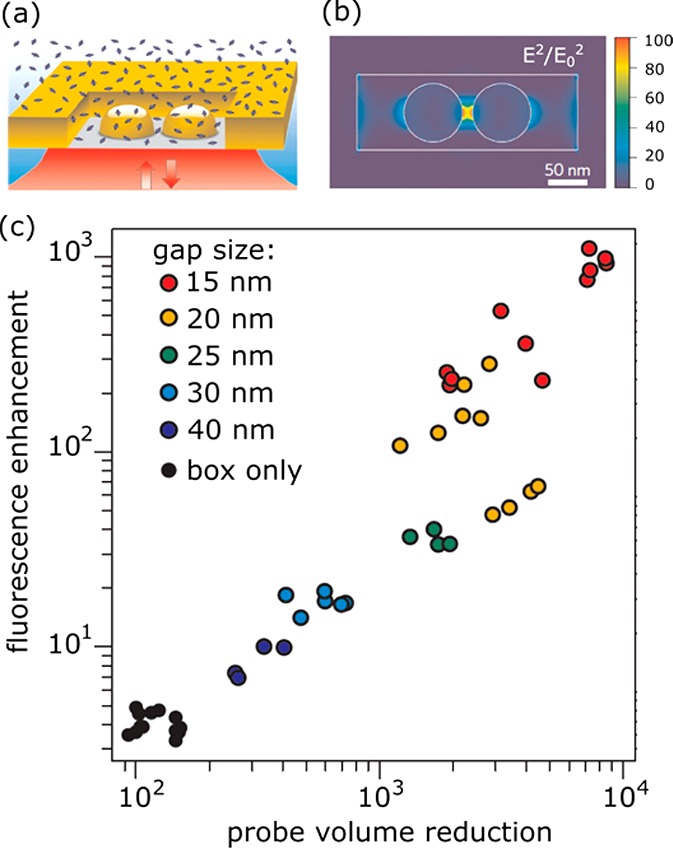
Single-molecule fluorescence enhancement of Alexa647 (quantum yield 8% in the presence of a quencher) by an antenna-in-box platform. (a) The experimental layout showing a particle dimer milled in a gold film. Alexa647 diffuses freely in solution. (b) Numerical calculation of the enhanced intensity around a nanostructure excited at 633 nm. (c) Measured fluorescence enhancements and probe-volume reduction (compared to a diffraction-limited microscope) for several structures with different gap sizes, compared to the structure in which the dimer is absent (box only). Figure adapted with permission from ref (137). Copyright 2013 Nature Publishing Group.
These characterization studies have revealed that fluorescence enhancements in excess of 1000 combined with probe volumes of several tens of zeptoliters are routinely achieved. Progress has been rapid in the past decade, and the field is now shifting from characterization of molecule-plasmon coupling to its applications. Here we focus on applications in single-molecule sensing of weakly fluorescent species.
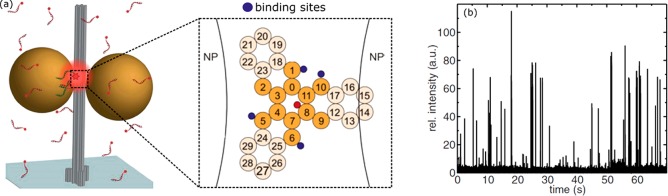
In 2012 Acuna et al. employed DNA origami structures66 to precisely space two colloidal gold spheres at a distance of approximately 20 nm (see Figure 11a). This relatively large gap provided a compromise between fluorescence enhancement and sufficient space for a biomolecular assay in the gap.
Figure 11.
Colloidal gold-nanosphere dimers assembled by DNA origami for single-molecule sensing of association and dissociation. (a) The gold dimers are assembled around a DNA origami scaffold with an interparticle spacing of 20 nm. In the gap region 4 binding sites for ssDNA are incorporated (dark blue spheres in inset). (b) Upon transient binding of complementary ssDNA fluorescence bursts are observed, originating from the emission of single att0655 labels enhanced by the dimer antenna. This allowed for single-molecule binding kinetics to be measured at concentrations of several tens of micromolars. Figure adapted with permission from ref (66). Copyright 2012 The American Association for the Advancement of Science.
They found a fluorescence enhancement of >100 for a high-quantum-yield dye (Atto647N) which was primarily attributed to excitation enhancement due to the high local intensity in the gap. They then attached ssDNA strands to the DNA origami in the gap of the dimer, which were used to bind complementary ssDNA (labeled with Atto655) in solution. They observed transient binding and unbinding in the gap, demonstrating that biomolecular assays can be performed in the dimer-gap (see Figure 11b). This assay was performed at analyte concentrations of 100 nM,66 and after optimizations at concentrations of 25 μM,26 well beyond the capabilities of conventional far-field optics.
In contrast to a single metallic particle, the inverted structures (holes in a metal film) have also been used for single-molecule sensing. Pioneered by the groups of Craighead and Webb in 2004, these so-called zero-mode waveguides (ZMW) act as a subwavelength pinhole that confines the incoming light in an evanescent mode.138 Later anisotropic holes139 and double nanohole apertures140 have also been explored. The evanescent nature of the near-field in holes with a diameter of 30–200 nm gives access to probe volumes of ∼10 zeptoliter (see Figure 10). Such ZMWs have subsequently been used to study single-molecule interactions at concentrations inaccessible to diffraction-limited optics.
In most studies single receptors are stochastically immobilized in the apertures at low concentration to obtain a large fraction of apertures with a single receptor at a random location. Recently, DNA origami141 and atomic force microscopy142 have been employed to deterministically place a single receptor at a well-defined location in the aperture. In 2008 Miyake et al. demonstrated the sensing of single-molecule protein interactions of GroEL with fluorescently labeled GroEs proteins.143 Transient association and dissociation events of fluorescently labeled GroES resulted in intensity bursts that gave access to single-molecule distributions of waiting times between subsequent association and dissociation events (see Figure 12b). These waiting-time distributions revealed that two rate constants dominate the interaction, something that was not yet observed at such high concentrations.143 Later ZMWs were used to study DNA–protein and protein–protein interactions with T4 replisomal proteins,144 and interactions between nucleotides and GTPases.145 To avoid the need to fluorescently label the analyte, the assay can also be reversed: the medium solution is mixed with fluorophores that are present in the apertures at a high concentration. Unlabeled analyte that diffuses into the aperture displaces the fluorophores resulting in a transient drop in fluorescence.146
Figure 12.
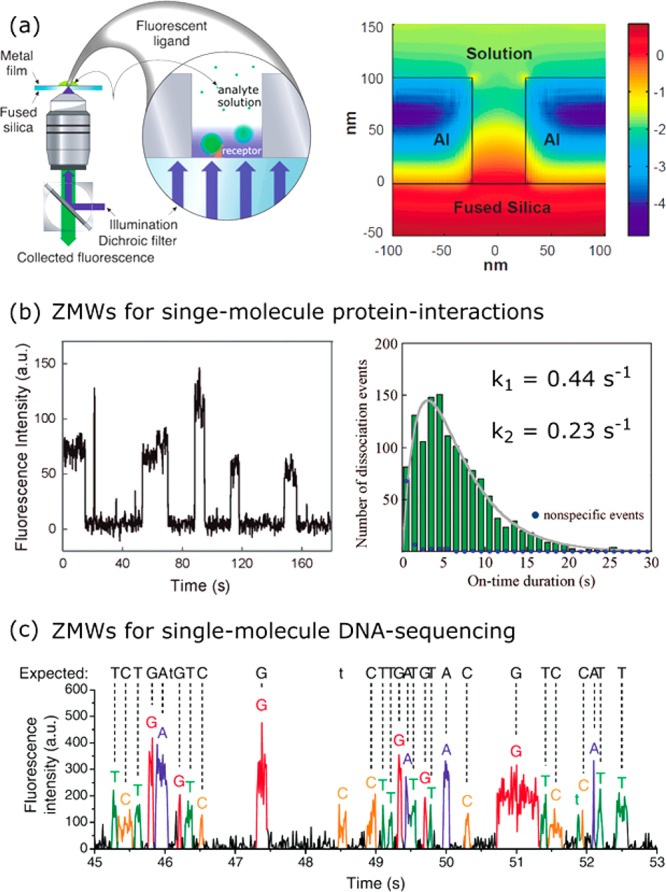
ZMW as single-molecule sensors. (a) Schematic of a far-field optical microscope with a zero-mode waveguide fabricated in an aluminum film. The right cartoon shows the local field around the nanohole (logarithmic scale). Figure adapted with permission from ref (138). Copyright 2013 The American Association for the Advancement of Science. (b) ZMWs used to detect single-molecule interactions between GroEL and GroES. The waiting-time distribution exhibits two populations with a different dissociation rate, indicating multiple dissociation pathways. Figure adapted with permission from ref (143). Copyright 2008 American Chemical Society. (c) ZMWs used for single-molecule DNA sequencing. Each nucleic acid is labeled with a distinct color fluorophore. The sequence of fluorescence bursts associated with incorporation of a nucleic acid by DNA polymerase reveals the sequence of the DNA template. Figure adapted with permission from ref (4). Copyright 2009 The American Association for the Advancement of Science.
In 2009, Eid et al. demonstrated the probing of enzyme–substrate interactions in ZMWs at micromolar substrate concentrations.4 They immobilized a DNA polymerase in a nanoaperture, and monitored fluorescence bursts due to incorporation of the four nucleotides each labeled with a different fluorophore (see Figure 12c). The order in which differently colored bursts are observed then corresponds to the sequence of the DNA template bound to the DNA polymerase. Each incorporation takes a few milliseconds, and several thousand nanopartures can be observed in real time allowing for parallelization and error-correction. The data also contained the kinetics of single DNA polymerases and revealed a temporally heterogeneous reaction rate that was attributed to DNA polymerase pausing due to the secondary structure of the DNA template.4 Single-molecule sequencing platforms based on ZMWs are a third-generation sequencing technology that is now commercially available. Recently, it has been shown that this commercial platform can also be applied to single-molecule sensing of DNA147,148 and biomolecular composition.149
Despite the enhancement of the emission of weakly emitting species, the above approaches cannot be applied to the majority of biomolecules because they do not emit at all at visible wavelengths. To tackle the challenge of detecting these nonfluorescent molecules, several label-free approaches based on plasmonics have been developed in the past decade, which are the topic of the next section.
Single-Molecule Sensing by Plasmon Shifts
The first reports on label-free plasmon sensing using single metallic particles stem from the early 2000s. Single silver and gold particles were used to detect the adsorption kinetics of thiols to bare particles150 and the interactions between avidin and a biotin-functionalized particle.151 Time-dependent measurements of the plasmon spectrum of the individual particles revealed the kinetics of binding (see Figure 13). The particles used were typically spherical with diameters of 40 nm or more to allow for detection in a dark-field microscope. It was later realized that other structures such as nanoshells,152 triangular particles,153 nanorods,42,154 nanocubes,155 and bipyramids43 exhibit higher sensitivities to molecular binding than spherical particles. Of these shapes, gold nanorods are now most employed as plasmon sensors42,44,79 due to their ease of synthesis, single-crystalline nature, and narrow plasmon resonance (limited by bulk absorption of gold).
Figure 13.
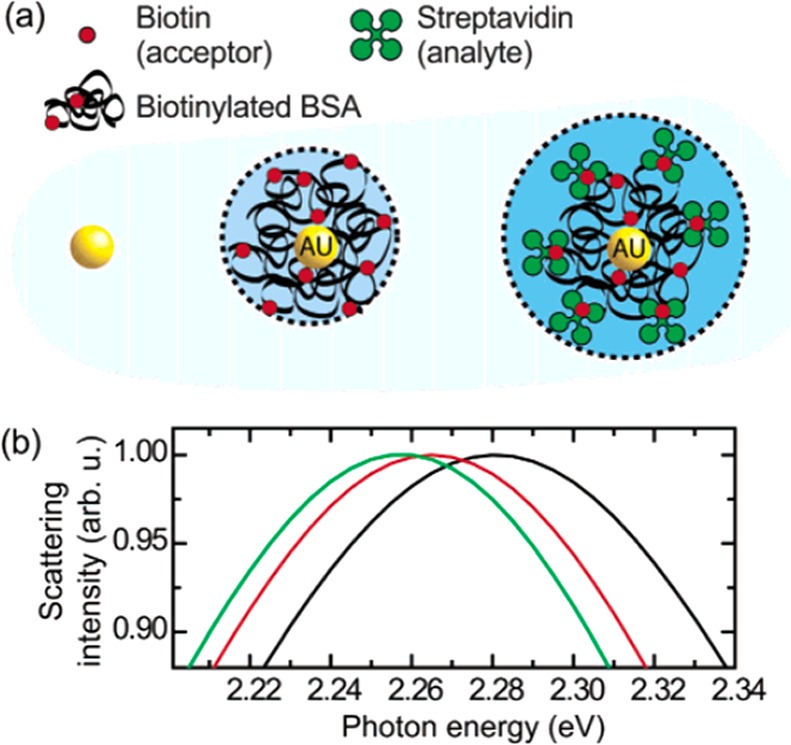
Plasmon sensing using individual gold nanospheres. (a) The nanospheres are functionalized with biotinylated bovine serum albumin (BSA) to allow for the detection of streptavidin. (b) Upon molecular binding the plasmon of individual particles was observed to shift to lower energy. Black line: before functionalization, red line: after functionalization with biotinylated BSA, green line: after incubation in streptavidin. Figure adapted with permission from ref (151). Copyright 2003 American Chemical Society.
Nusz et al. employed individual gold nanorods to monitor streptavidin binding to biotin functionalized particles.79 They detected plasmon shifts by recording single-particle scattering spectra using a dark-field microscope and found concentration-dependent shifts of several nanometers.79 Kinetics can also be monitored with subsecond time resolution by recording spectra over time. Multiplexed detection with single gold nanorods was demonstrated by Rosman et al.156 They employed aptamer functionalized particles that were flown into a flow-cell resulting in deposition in the sample chamber. By imaging the location of deposition in an optical microscope, they obtained a map of particle locations. This process was repeated for four different populations of nanorods, each functionalized with an aptamer against a different target. This allowed them to perform multiplexed detection of four different analytes: fibronectin, streptavidin, IgE, and thrombin.156 An alternative approach to multiplexed detection is to use different aspect ratios each with their own distinct plasmon resonance wavelength.157
Single particle sensing offers advantages in sensitivity over ensembles of particles because inhomogeneous broadening of the plasmon resonance is eliminated. However, it is essentially still an ensemble measurement, averaging the binding of many molecules to a single nanoparticle, and transducing it into an optical signal. As discussed before, molecular ensemble measurements cannot resolve events such as binding of rare molecular species or aberrant binding behaviors, which are averaged out in ensemble measurements.75 In order to resolve these heterogeneities among molecular populations, and to examine stochastic processes governing molecular interactions, single molecule sensing is essential.
Before describing experimental demonstrations of single-molecule plasmon sensing it is informative to estimate the plasmon shift induced by the binding of an individual molecule. Traditionally, the sensitivity of a plasmon sensor is expressed by dλ/dn that describes the shift of the resonance per refractive index unit.158 Although appropriate for bulk index sensing, the sensitivity to single-molecule events is far more complex. The plasmon shift induced by the binding of a single molecule is proportional to the local field intensity integrated over the volume of the molecule.159−161 The local field around a metallic particle is often strongly enhanced around tips and protrusions and quickly decays away from the particle surface (see Figure 14a). This means that the resulting plasmon shift is a function of the size of the particle and the molecule, the location of the molecule, and the distance of the molecule to the particle surface.
Theoretical estimates of the plasmon shift have been obtained using the electrostatic approximation,159 perturbation theory,160,161 and numerical simulations.159,162,163 Davis et al. modeled the shifts for gold nanorods using electrostatic coupling theory, modeling the molecule as a dipole contained within a dielectric sphere.161 In close proximity, the polarizable dielectric screens part of the charge on the nanoparticle leading to a red-shift of the plasmon.161 The largest plasmon shifts were found when the molecule rested directly on the nanorod surface in a regions with maximum electric field strength. Antosiewicz and Käll78 used numerical models to estimate the plasmon shift induced by the binding of a 3-nm-radius dielectric sphere to silver nanoparticles of different dimensions. They reported plasmon shifts that strongly varied with both nanoparticle geometry and binding position, as depicted in Figure 14. Typical plasmon shifts induced by single proteins are smaller than 0.5 nm (∼1% of the line width), and therefore require sensitive optical microscopy to be detectable.
Experimentally, in 2010 Mayer et al. demonstrated label-free detection of the unbinding of single IgG antibodies from gold bipyramids.164 However, measurement noise required averaging over tens of events to reveal a detectable binding signal. Discrete molecular binding and unbinding of single proteins without statistical averaging was demonstrated in 2012 simultaneously by Ament et al.165 and by Zijlstra et al.77
Ament et al. used dark-field spectroscopy of single gold nanorods to detect the nonspecific interaction of single proteins with the bare nanoparticle surface.165 As shown in Figure 15, discrete red-shifts of the peak wavelength were observed due to nonspecific attachment of single fibronectin proteins to the gold surface. Typical single-molecule plasmon shifts were found to be 0.3 nm (with measurement noise of 0.07 nm) with a waiting time between events of 50 s. The authors employed an intense white-light laser for dark-field illumination to achieve sufficient signal-to-noise ratio to resolve discrete shifts of the plasmon spectrum.165 This also allowed them to record the full spectrum of a single particle with millisecond time-resolution (see Figure 15c). Upon close inspection of the time-dependence of the plasmon shift it was found that the plasmon shifts on time scales of ∼2 s, which was attributed to a slow denaturing of the protein due to direct contact with the gold surface.
Figure 15.
(a) Experimental setup for the detection of plasmon shifts induced by single-molecule binding to individual gold nanorods. (b) Typical time-dependence of the plasmon wavelength. The triangles indicate discrete (stepwise) shifts of the plasmon attributed to single-molecule events. (c) Temporal evolution of a single binding event, illustrating a red-shift over time, attributed to the slow denaturing of the protein on the gold surface (inset). Figure adapted with permission from ref (165). Copyright 2012 American Chemical Society.
The absence of a particle coating resulted in large plasmon shifts due to the close proximity of the protein to the particle surface, but does not allow for the study of specific ligand–receptor interactions. Also in 2012, Zijlstra et al. reported the detection of single-molecule binding events to receptor-functionalized gold nanorods.77 They employed short thiolated receptors (linker length 1.5 nm) to ensure that the proteins interact in close proximity to the nanoparticle surface. To resolve plasmon shifts with high signal-to-noise ratio the authors employed photothermal microscopy instead of scattering spectroscopy.
In contrast to the scattered intensity, the photothermal signal scales with the volume of the particle enabling the detection of smaller particles with high signal-to-noise ratio.166 Upon protein binding, the plasmon peak wavelength red-shifts yielding discrete changes in the absorption cross section at the photothermal probe wavelength (see Figure 16). Knowing the nanorod’s plasmon line width, peak shifts could be inferred from detected discrete changes in nanorod absorption at the pump wavelength. Mean peak shifts of 0.5 nm for streptavidin (53 kDa), 1.2 nm for antibiotin (150 kDa), and 1.6 nm for streptavidin-RPE (300 kDa) were observed. The plasmon shifts did not scale linearly with molecular weight, which was attributed to a reduced overlap between the molecule and near-fields for larger molecules.
Figure 16.
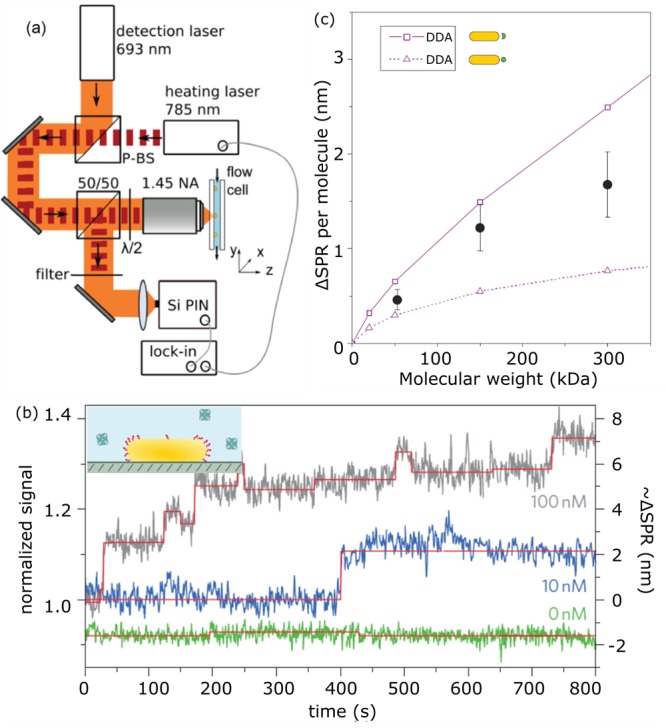
(a) Experimental setup for photothermal detection of plasmon shifts of single gold nanoparticles (nanorods sized 31 nm × 9 nm). (b) Time traces for different molecular concentrations showing discrete steps in the photothermal signal strength, indicating single-molecule interactions. (c) Measured shifts together with simulated shifts obtained from the discrete dipole approximation (DDA). Figure adapted with permission from ref (77). Copyright 2012 Nature Publishing Group.
In the work of both groups, single molecule interactions are monitored on one single nanorod, with wait times between binding events on the order of seconds to tens of seconds.77,165 While these techniques demonstrate single molecule detection, quantitative analysis of single molecule interactions is not possible because the acquisition of statistics is time-consuming. This can be overcome by measuring over long time scales or by making nanoparticle detection massively parallel.75 The latter approach was demonstrated by Beuwer et al., who imaged individual receptor-functionalized nanorods in parallel on a camera using a narrowband light-source. Upon addition of analyte discrete shifts of the plasmon resonance were observed as stepwise changes in the scattered intensity (see Figure 17). Positive and negative signals were observed, depending on the plasmon wavelength relative to the probe wavelength of 785 nm.
Figure 17.
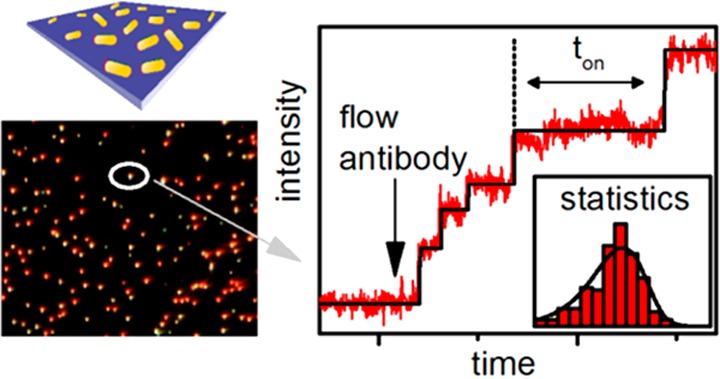
Wide-field monitoring of hundreds of single nanorods for single-molecule plasmon sensing. Each dot represents a single-molecule sensor. Multiplexing across many sensors in parallel allowed for the statistical analysis of single-molecule signals. Figure adapted with permission from ref (76). Copyright 2015 American Chemical Society.
By monitoring antibody interactions on hundreds of particles in parallel, the stochastic nature of the binding process could be examined. It was found that inverse wait times for molecule binding follow a first-order power law dependence on concentration. From these measurements, kon rates for the nanorod antibiotin system was calculated to be 8.3 × 105 M–1 s–1, in line with ensemble measurements for other antibody–antigen pairs. The low sensing volume of each nanorod, combined with the large number of sensors, theoretically allows concentration measurements with a 7 decade dynamic range. In this case, the high affinity between biotin and antibiotin precludes measurement of unbinding events, and thus measurement of equilibrium kinetics.
Challenges
Despite rapid progress in the past decade, several challenges remain before plasmon-enhanced sensing will be mature enough to be incorporated into real-life applications. Here we describe three of those challenges:
Low Signals from Small Molecules
Although sensing by plasmon-enhanced fluorescence allows for the detection of small molecules, labeling is not always practical. A drawback of the label-free approaches discussed above is that they rely on small changes in the local refractive index induced by molecular binding. These refractive index changes result in a plasmon shift which can currently be detected with a sensitivity of ∼0.5 nm (equivalent to ∼1% of the line width) on a single nanoparticle. This detection limit is the result of a trade-off: on one hand larger particles result in a higher signal-to-noise ratio for single-particle detection, but simultaneously in a reduced plasmon shift per molecule due to the larger extent of the locally enhanced field. An exception is the single molecule detection achieved with gold bipyramids.45 Although larger than nanorods, bipyramids exhibit high field-enhancements due to their sharp tips, and may be of interest in future single molecule detection.
This trade-off between noise and sensitivity currently results in a shot-noise limited detection limit for single molecules of ∼50 kDa,77 thus excluding many relevant biological targets such as brain natriuretic peptide (MW 4 kDa) and procalcitonin (MW 15 kDa). The signal-to-noise ratio in the shot-noise limited regime scales as √N, where N is the number of detected photons. This suggests that this detection limit is easily improved by increasing the excitation power. However, increasing power leads to additional plasmonic heating of the nanoparticle and its immediate surroundings.77 The 50 kDa detection limit was achieved while limiting heating to <2 °C, far below the temperature at which a protein denatures (ranging from 40 to 80 °C for globular proteins167). A second trade-off thus exists, where the shot-noise detection limit must be balanced against localized heating, which may perturb the interaction being studied.
A promising strategy to amplify the plasmon shift per molecule is to further increase the field-confinement by employing dimers of particles or particles coupled to a gold film. Theoretical studies confirmed that dimers of gold spheres49 or gold nanorods160 exhibit substantially larger plasmon shifts for molecular binding in the gap region compared to their monomer counterpart. A numerical study by Yang et al. showed that the sensitivity is nearly doubled for a dimer of gold nanorods spaced by 15 nm when a protein binds in the gap region.160 The difficulty in the implementation lies in the realization of controllable gaps that should be only slightly larger than the analyte to be detected. In addition, after assembly the gap should be accessible for the analyte and not be blocked by the linkers used for assembly. DNA origami scaffolds as outlined before may provide an attractive solution because they offer control over the spacing and orientation of the particles while leaving the gap region free for an immunoassay.68,129
Further field confinement on subnanometer length scales was demonstrated by Taylor et al., who employed cucurbiturils as linkers to immmoblize a gold sphere on top of a metallic film.168 Cucurbiturils are barrel-shaped macrocyclic molecules that form a well-defined 0.9 nm spacer between the gold particle and the film. Interestingly, the barrel shape of the cucurbituril allows for the binding of specific “guest” molecules that slightly deform the cucurbituril and modulate its SERS signal, thus providing a read-out mechanism for molecular binding.169 Recently, Benz et al. found that individual atomic features inside the gap of such a plasmonic assembly can localize light to volumes well below 1 cubic nanometer (therefore dubbed “picocavities”).170 These picocavities are formed by a single gold atom protruding from a much larger nanoparticle toward the underlying metallic film, and may in the future enable optical sensing on the atomic scale.
An alternative approach for the label-free detection of low-molecular-weight analyte may lie in the use of conformational changes. Lee et al. recently demonstrated the use of gold nanosphere dimers linked by an aptamer to monitor single-molecule aptamer–analyte interactions.171 Aptamers are single-stranded DNA oligonucleotides that fold into a well-defined tertiary structure. Their capability to bind a specific analyte is derived from key molecular interactions such as shape complementarity, hydrogen bonding, and electrostatic interactions.172 Aptamers undergo a large conformational change upon analyte binding, resulting in detectable plasmonic shifts dictated by the proximity of the two nanospheres. Approaches to assemble well-defined dimers linked with a single aptamer are available as described in the section on synthesis and assembly, making this a promising strategy to enable label-free detection of single small molecules.
Nonspecific Interactions in Biological Media
The application of single-molecule sensing in a biological environment requires robust resistivity of the particles against nonspecific interactions. The total macromolecular concentration in, for example, blood exceeds 100 μM, whereas biomarkers such as listed in Figure 2 are present at concentrations that are several orders of magnitude lower. This large concentration difference implies that even weak nonspecific interactions can dominate the response of a sensor, as they effectively compete with specific interactions. In biological environments this can even lead to the formation of a so-called protein-corona around a nanoparticle,173 which may shield its surface-bound receptors and inhibit its sensitivity.
Antifouling coatings are often employed to minimize nonspecific interactions with the exposed sensor surface. Polyethylene glycols (PEGs) are widely used because they are easily synthesized and commercially available. They are usually coupled to metallic particles using reactive thiol groups because the covalent bond ensures a stable conjugation.175 Although the thermodynamic and molecular mechanisms that determine the antifouling properties of PEGs are not completely understood, it is believed that chain length, surface density, conformation, and hydrophilicity of the polymer play important roles in resisting protein adhesion. An alternative class of antifouling materials are zwitterionic polymers that maintain electric neutrality with an equivalent number of positively and negatively charged groups. It is proposed that the superior antifouling properties of zwitterions is related to their stronger hydration induced by electrostatic interactions instead of hydrogen bonding.176 Zwitterionic polymers however still have limited commercial availability, which makes their use less widespread than PEGs.
In ensemble-averaged biosensors the detrimental effects of nonspecific interactions are partially countered by employing capture probes (antibodies, aptamers, peptides) with a high affinity and specificity for the analyte. This ensures that a sizable fraction of receptors is occupied by the analyte and dominates the sensor response. Sensing at the single-molecule level has the advantage that the kinetic parameters (i.e., time between binding events and residence time) are also available for each event. This extra information can be exploited to further enhance the specificity of the sensor by analyzing distributions of residence times.75,174 Nonspecific interactions are ideally weak, whereas specific interactions exhibit a well-defined and longer residence time due to their molecularly determined interaction potential. This principle is referred to as kinetic fingerprinting and was already exploited by Johnson-Buck et al. to distinguish on-target DNA-microRNA interactions from single base-pair mismatches by statistical analysis of residence times (see Figure 18).174
Figure 18.
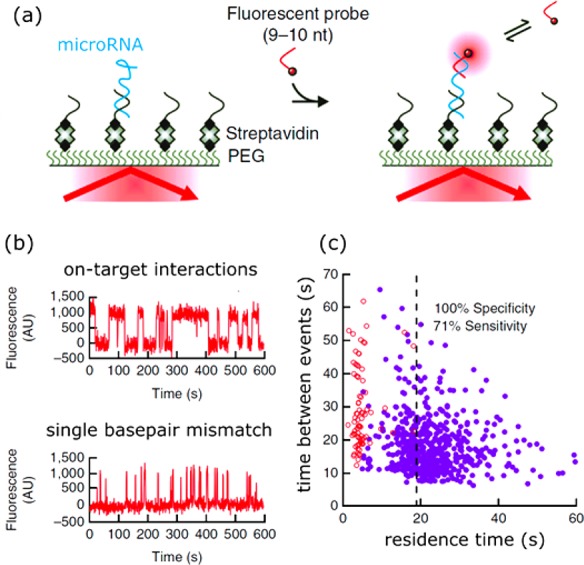
Distinguishing specific from nonspecific interactions by single-molecule analysis of kinetic parameters. (a) A surface coated with biotin-labeled LNA capture molecules is functionalized with microRNA. Incubation with fluorescent probes induces kinetic binding and unbinding with the microRNA. (b) This kinetic binding and unbinding is observed as a digital signal in a single-molecule fluorescence assay. Clear differences exist in the residence times for complementary interactions and single base-pair mismatches. (c) Correlation between residence time and time between events. Two populations are observed of which the one with short residence time corresponds to nonspecific interactions. Figure adapted with permission from ref (174). Copyright 2015 Nature Publishing Group.
The ability of kinetic fingerprinting has the interesting consequence that the optimum capture probe is not necessarily the one with the highest affinity. Instead it is the probe that exhibits a distribution of residence times that (a) can be measured with good statistics in a reasonable time and (b) can be easily distinguished from the nonspecific background. Choosing the right capture probe therefore requires detailed understanding of nonspecific interactions with functionalized nanoparticles at the single-molecule level. There are no studies yet that address nonspecific interactions with nanoparticles at the single-molecule level, but it is clear that these will be crucial to optimize the sensor for operation in biological media.
Statistics and Multiplexing
The main motivation to expend the effort to develop single-molecule sensors lies in the ability to reveal populations of molecular interaction parameters and identify rare species in a sample volume. The reliability with which these properties can be measured depends on the amount of statistics that can be gathered in a reasonable time. Single-molecule waiting-time distributions exhibit Poisson statistics, and the error in determining the mean of a distribution thus scales as 1/√N. This implies that the ability to distinguish different populations depends on the amount of statistics gathered.
For the single and very small sensors described here, the number of molecular interactions detected per unit time depends on the sensor size and the diffusion coefficient of the analyte.78,177−179 For analyte present at high concentrations (>nM), the frequency of detection is typically more than several Hz for a hemispherical sensor with a radius of 50 nm.177 This implies that a distribution can typically be clearly resolved within minutes. However, at 1 fM it takes nearly a day for the first molecule to diffuse to the single-particle sensor.177 In a sensing device these mass-transport limitations result in unrealistically long waiting times between events and thus a slow response and limited accuracy in determining concentrations and populations.
Several solutions have been proposed in the literature to tackle mass-transport limitations and improve statistics. The first solution is to enhance the concentration of analyte in proximity to the sensor. De Angelis et al. exploited superhydrophobic surfaces made of silicon micropillar arrays for the evaporation and local enrichment of analyte (see Figure 19a and ref (180)). Barik et al. on the other hand employed dielectrophoretic forces on charged analyte to enhance its concentration near a gold nanohole film.181 Both approaches result in an increased analyte concentration near the sensor surface, which is then detected by fluorescence, transmission, or surface-enhanced Raman spectroscopy on embedded plasmonic structures. These approaches allow for the molecular detection of a range of analytes at atto- to picomolar concentrations, equivalent to a few molecules per drop only.
Figure 19.
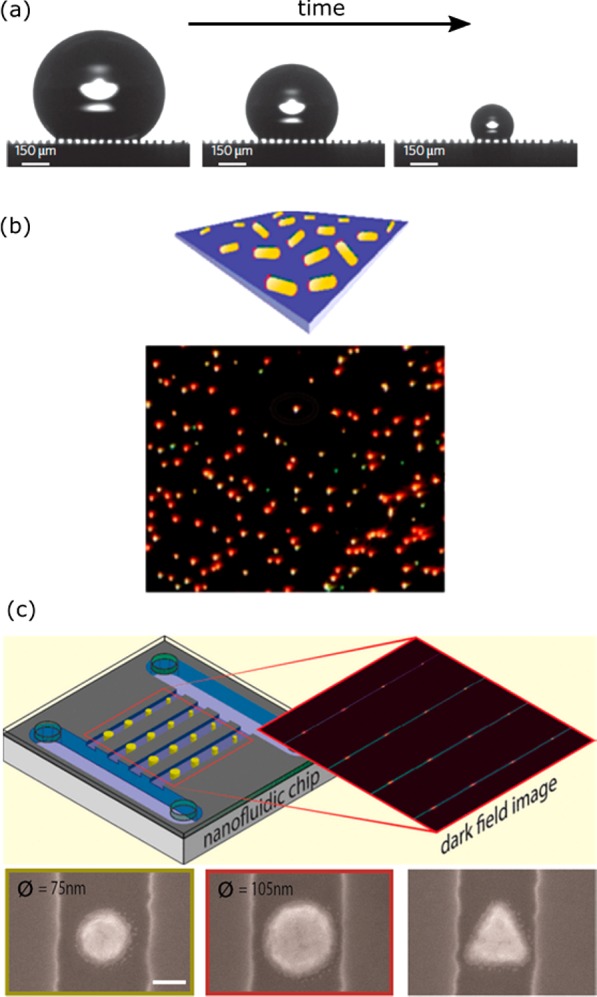
Approaches to increase the event frequency to allow for acquisition of relevant statistics. (a) Evaporation of a drop of analyte on a superhydrophobic surface results in local enrichment of biomarkers. Figure adapted with permission from ref (180). Copyright 2011 Nature Publishing Group. (b) Parallel detection of many single-molecule sensors in a wide-field microscope. Figure adapted with permission from ref (76). Copyright 2015 American Chemical Society. (c) Array of nanoparticles in nanometer-sized fluidic channels. Figure adapted with permission from ref (182). Copyright 2016 American Chemical Society.
An alternative solution that would enable the detection at low analyte concentrations was proposed by Beuwer et al. (see Figure 19b and ref (76)). They probed hundreds of single-molecule sensors simultaneously in a wide-field optical microscope to increase the number of detected events. Highly parallel detection is enabled here by modern high-resolution cameras used in combination with a low-magnification microscope objective. The lowest concentrations that can then be probed are limited by the number of sensors that can be imaged simultaneously. Assuming that approximately 100 events are required to establish a concentration, they estimated that concentrations <1 pM would be measurable within 10 min by simultaneous probing of 105 single-molecule sensors.
Instead of waiting for diffusion to transport the analyte to a nanoscale sensor, Fritzsche et al. employed nanofluidic sensors with dimensions similar to the plasmonic sensors (see Figure 19c and ref (182)). The nanochannels were fabricated with a combination of electron beam lithography and reactive ion etching, resulting in channels with a width of 130 to 280 nm. They incorporated reference channels to correct for baseline drift and showed the detection of analyte binding to the gold structures. Herein, analyte was actively pumped past the nanosensors by a large pressure difference between the inlet and the outlet. The proximity of the analyte to the sensor increases the probability of detection, reducing the wait time between detection events, and lowering the concentration limits that be probed.
The challenges defined here, namely, sensitivity, specificity, and statistics, are all highly active fields of research, and we hypothesize that many of these challenges will be soon overcome with novel developments.
Outlook
Quantification of biomolecular interactions at the single-molecule level has greatly expanded the scope of biosensors and analytical technologies. In contrast to ensemble-averaged approaches, single-molecule sensitivity gives access to the underlying heterogeneity of molecular properties. This heterogeneity could originate from, e.g., the presence of different species within a sample, or the presence of different conformations of the same species. Recent experiments demonstrated the power of single-molecule sensing by identifying multiple protein-homologues within a mixture,15,183 distinguishing specific interactions from nonspecific background signals,174 and characterizing a small population of partially unfolded proteins in a population of properly folded molecules.184 Single-molecule sensitivity also gives access to equilibrium dynamics that are averaged out in ensemble measurements because they are usually not synchronized in time. This has allowed for the study of, e.g., the conformational dynamics of a protein185 and measurement of molecular interactions at equilibrium.174
These single-molecule studies have so far largely been conducted using fluorescence microscopy. With the advent of label-free techniques the range of biomolecules that can be studied is greatly expanded because fluorescent labeling is not needed. Furthermore, the biomolecule can be studied in its native state without chemical modifications, potentially enabling real-time sensing in biological fluids. Plasmonic approaches have an additional advantage in their small probe volume (determined by the extent of the near-field) that allows for the detection of single molecules in highly concentrated solutions. This may extend single-molecule biophysical experiments to concentrated matrices that mimic intracellular conditions. It is known that molecular interactions and function are affected by high concentrations of macromolecules that induce excluded volume effects,186 but the effects at the single-molecule level remain elusive.
A method for measuring heterogeneity by single-molecule sensing may not only benefit academic research into the fundamentals of protein-interactions and function, but may also find application in pharmaceutical companies. A major challenge for the next generation of therapeutics is to, e.g., improve the homogeneity of therapeutic antibodies to yield more efficient and potent drugs.187 Moreover, characterization of small populations of partially unfolded proteins involved in diseases188 may be possible because these partially unfolded proteins exhibit a distinct kinetic fingerprint that is revealed uniquely at the single-molecule level.183 This may yield novel and more detailed insight into interactions of drugs with their target.
Beyond applications in analytical laboratories we also foresee exciting developments in biosensors worn in or on the body that continuously report on biomarker concentrations.189,190 A well-known example is the continuous monitoring of glucose that allows for accurate dosing of insulin.191 A drawback of current ensemble-averaged sensors is that they require regular calibration due to baseline drift induced by, e.g., nonspecific interactions.192 In contrast, single-molecule sensors report association and dissociation events at equilibrium, and may allow one to distinguish between nonspecific (short-lived) interactions and specific (long-lived) interactions on a molecule-by-molecule basis.174 Such kinetic fingerprinting alleviates the requirements of an antifouling coating because a certain level of nonspecific interactions can be tolerated in the sensor. In addition, different homologues of analyte can be discriminated based on their kinetic fingerprint.
These applications will depend on the ability to miniaturize the device. Currently, single-molecule plasmon sensors employ an optical microscope such as found in many laboratories, consisting of a laser as the excitation source, a high-quality objective lens, interference filters, and a sensitive detector, either an avalanche photodiode (APD) or a CCD camera for parallel imaging of many objects in a wide field of view. Recently, several developments have allowed for the miniaturization of optical microscopes to platforms as small as a smartphone.24,193,194 Laser sources can be replaced by light-emitting diodes, which are no bigger than a few millimeters and generate multiple watts of output power. Complementary metal-oxide-semiconductor (CMOS) cameras have rapidly evolved over the past years and allow for sensitive detection down to single fluorophores without the need for amplification as is done in an electron-multiplying charge coupled device (EMCCD). CMOS cameras in mobile phones already have sufficient sensitivity to detect single metal particles and small fluorescent beads.24,193,194 Improving sensitivity and resolution of the device would give access to fluorescence or scattering images of even smaller individual objects, perhaps down to single fluorophores.
The required research to achieve practical applications is highly multidisciplinary and spans the fields of physics, chemistry, biology, mathematics, and electrical engineering. Therefore, not only is fundamental research needed, but also multidisciplinary and strategic collaborations are crucial to achieve this goal. Ultimately, the prospect of fully integrated biosensors that continuously monitors biomarker levels in vivo is a tempting one that may revolutionize personalized healthcare.
Acknowledgments
The authors thank Menno Prins for critical reading of the manuscript. P.Z. acknowledges financial support from The Netherlands Organisation for Scientific Research (NWO VIDI). This work is part of the research programme of the Foundation for Fundamental Research on Matter (FOM), which is financially supported by The Netherlands Organisation for Scientific Research (NWO).
Glossary
Vocabulary
- surface plasmon resonance
the resonant oscillation of conduction electrons stimulated by incident light
- plasmonic nanoparticles
nanosized particles whose conduction electrons couple with electromagnetic radiation (light)
- near-field
the electromagnetic field around the nanoparticle at distances shorter than the wavelength
- radiative decay
the loss of energy due to photon emission
- nonradiative decay
any other loss mechanism wherein no photons are emitted
- biosensing
the use of transducers incorporating biologically active capture molecules to detect the presence of particular biomolecules
The authors declare no competing financial interest.
This paper was originally published on August 8, 2017, with errors in the citations of the Figure 3b and Figure 5b captions. The corrected version was reposted on August 15, 2017.
References
- Ma F.; Li Y.; Tang B.; Zhang C. Y. Fluorescent Biosensors Based on Single-Molecule Counting. Acc. Chem. Res. 2016, 49 (9), 1722–1730. 10.1021/acs.accounts.6b00237. [DOI] [PubMed] [Google Scholar]
- Orrit M.; Bernard J. Single Pentacene Molecules Detected by Fluorescence Excitation in a P-Terphenyl Crystal. Phys. Rev. Lett. 1990, 65 (21), 2716–2719. 10.1103/PhysRevLett.65.2716. [DOI] [PubMed] [Google Scholar]
- Moerner W. E.; Kador L. Optical Detection and Spectroscopy of Single Molecules in a Solid. Phys. Rev. Lett. 1989, 62 (21), 2535–2538. 10.1103/PhysRevLett.62.2535. [DOI] [PubMed] [Google Scholar]
- Eid J.; Fehr A.; Gray J.; Luong K.; Lyle J.; Otto G.; Peluso P.; Rank D.; Baybayan P.; Bettman B.; Bibillo A.; Bjornson K.; Chaudhuri B.; Christians F.; Cicero R.; Clark S.; Dalal R.; deWinter A.; Dixon J.; Foquet M.; Gaertner A.; Hardenbol P.; Heiner C.; Hester K.; Holden D.; Kearns G.; Kong X.; Kuse R.; Lacroix Y.; Lin S.; Lundquist P.; Ma C.; Marks P.; Maxham M.; Murphy D.; Park I.; Pham T.; Phillips M.; Roy J.; Sebra R.; Shen G.; Sorenson J.; Tomaney A.; Travers K.; Trulson M.; Vieceli J.; Wegener J.; Wu D.; Yang A.; Zaccarin D.; Zhao P.; Zhong F.; Korlach J.; Turner S. Real-Time DNA Sequencing from Single Polymerase Molecules. Science 2009, 323 (5910), 133–138. 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Pacific Biosciences; www.pacb.com, (accessed 19-5-2017).
- Rissin D. M.; Kan C. W.; Campbell T. G.; Howes S. C.; Fournier D. R.; Song L.; Piech T.; Patel P. P.; Chang L.; Rivnak A. J.; Ferrell E. P.; Randall J. D.; Provuncher G. K.; Walt D. R.; Duffy D. C. Single-Molecule Enzyme-Linked Immunosorbent Assay Detects Serum Proteins at Subfemtomolar Concentrations. Nat. Biotechnol. 2010, 28 (6), 595–599. 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quanterix; www.quanterix.com, accessed 19-5-2017.
- Todd J.; Freese B.; Lu A.; Held D.; Morey J.; Livingston R.; Goix P. Ultrasensitive Flow-Based Immunoassays Using Single-Molecule Counting. Clin. Chem. 2007, 53 (11), 1990–1995. 10.1373/clinchem.2007.091181. [DOI] [PubMed] [Google Scholar]
- Singulex; www.singulex.com, accessed 19-5-2017.
- Giepmans B. N. G.; Adams S. R.; Ellisman M. H.; Tsien R. Y. The Fluorescent Toolbox for Assessing Protein Location and Function. Science 2006, 312 (5771), 217–224. 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Medintz I. L.; Uyeda H. T.; Goldman E. R.; Mattoussi H. Quantum Dot Bioconjugates for Imaging, Labelling and Sensing. Nat. Mater. 2005, 4, 435–446. 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- Miles B. N.; Ivanov A. P.; Wilson K. A.; Doğan F.; Japrung D.; Edel J. B. Single Molecule Sensing with Solid-State Nanopores: Novel Materials, Methods, and Applications. Chem. Soc. Rev. 2013, 42 (1), 15–28. 10.1039/C2CS35286A. [DOI] [PubMed] [Google Scholar]
- Cherf G. M.; Lieberman K. R.; Rashid H.; Lam C. E.; Karplus K.; Akeson M. Automated Forward and Reverse Ratcheting of DNA in a Nanopore at 5-Å Precision. Nat. Biotechnol. 2012, 30 (4), 344–348. 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrao E. A.; Derrington I. M.; Laszlo A. H.; Langford K. W.; Hopper M. K.; Gillgren N.; Pavlenok M.; Niederweis M.; Gundlach J. H. Reading DNA at Single-Nucleotide Resolution with a Mutant MspA Nanopore and phi29 DNA Polymerase. Nat. Biotechnol. 2012, 30 (4), 349–353. 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.; Gatterdam V.; Wieneke R.; Tampé R.; Rant U. Stochastic Sensing of Proteins with Receptor-Modified Solid-State Nanopores. Nat. Nanotechnol. 2012, 7 (4), 257–263. 10.1038/nnano.2012.24. [DOI] [PubMed] [Google Scholar]
- Guo X. Single-Molecule Electrical Biosensors Based on Single-Walled Carbon Nanotubes. Adv. Mater. 2013, 25 (25), 3397–3408. 10.1002/adma.201301219. [DOI] [PubMed] [Google Scholar]
- Goldsmith B. R.; Coroneus J. G.; Khalap V. R.; Kane A. A.; Weiss G. A.; Collins P. G. Conductance-Controlled Point Functionalization of Single-Walled Carbon Nanotubes. Science 2007, 315 (5808), 77–81. 10.1126/science.1135303. [DOI] [PubMed] [Google Scholar]
- Cognet L.; Tsyboulski D. A.; Rocha J.-D. R.; Doyle C. D.; Tour J. M.; Weisman R. B. Stepwise Quenching of Exciton Fluorescence in Carbon Nanotubes by Single-Molecule Reactions. Science 2007, 316 (5830), 1465–1468. 10.1126/science.1141316. [DOI] [PubMed] [Google Scholar]
- Jin H.; Heller D. A.; Kim J. H.; Strano M. S. Stochastic Analysis of Stepwise Fluorescence Quenching Reactions on Single-Walled Carbon Nanotubes: Single Molecule Sensors. Nano Lett. 2008, 8 (12), 4299–4304. 10.1021/nl802010z. [DOI] [PubMed] [Google Scholar]
- Baaske M. D.; Foreman M. R.; Vollmer F. Single-Molecule Nucleic Acid Interactions Monitored on a Label-Free Microcavity Biosensor Platform. Nat. Nanotechnol. 2014, 9 (11), 933–939. 10.1038/nnano.2014.180. [DOI] [PubMed] [Google Scholar]
- Dantham V. R.; Holler S.; Barbre C.; Keng D.; Kolchenko V.; Arnold S. Label-Free Detection of Single Protein Using a Nanoplasmonic-Photonic Hybrid Microcavity. Nano Lett. 2013, 13 (7), 3347–3351. 10.1021/nl401633y. [DOI] [PubMed] [Google Scholar]
- Pockrand I.; Swalen J. D.; Gordon J. G.; Philpott M. R. Surface Plasmon Spectroscopy of Organic Monolayer Assemblies. Surf. Sci. 1978, 74 (1), 237–244. 10.1016/0039-6028(78)90283-2. [DOI] [Google Scholar]
- Kretschmann E.; Raether H. Radiative Decay of Non Radiative Surface Plasmons Excited by Light. Z. Naturforsch., A: Phys. Sci. 1968, 23 (12), 2135–2136. 10.1515/zna-1968-1247. [DOI] [Google Scholar]
- Wei Q.; Qi H.; Luo W.; Tseng D.; Ki S. J.; Wan Z.; Göröcs Z.; Bentolila L. A.; Wu T. T.; Sun R.; Ozcan A. Fluorescent Imaging of Single Nanoparticles and Viruses on a Smart Phone. ACS Nano 2013, 7 (10), 9147–9155. 10.1021/nn4037706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson N. L.; Anderson N. G. The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Mol. Cell. Proteomics 2002, 1 (11), 845–867. 10.1074/mcp.R200007-MCP200. [DOI] [PubMed] [Google Scholar]
- Puchkova A.; Vietz C.; Pibiri E.; Wünsch B.; Sanz Paz M.; Acuna G. P.; Tinnefeld P. DNA Origami Nanoantennas with over 5000-Fold Fluorescence Enhancement and Single-Molecule Detection at 25 μM. Nano Lett. 2015, 15 (12), 8354–8359. 10.1021/acs.nanolett.5b04045. [DOI] [PubMed] [Google Scholar]
- Khatua S.; Yuan H.; Orrit M. Enhanced-Fluorescence Correlation Spectroscopy at Micro-Molar Dye Concentration around a Single Gold Nanorod. Phys. Chem. Chem. Phys. 2015, 17 (33), 21127–21132. 10.1039/C4CP03057E. [DOI] [PubMed] [Google Scholar]
- Mayo Medical Laboratories; www.mayomedicallaboratories.com, accessed 19-5-2017.
- Faraday M. The Bakerian Lecture: Experimental Relations of Gold (and Other Metals) to Light. Philos. Trans. R. Soc. London 1857, 147, 145–181. 10.1098/rstl.1857.0011. [DOI] [Google Scholar]
- Grzelczak M.; Pérez-Juste J.; Mulvaney P.; Liz-Marzán L. M. Shape Control in Gold Nanoparticle Synthesis. Chem. Soc. Rev. 2008, 37 (9), 1783–1791. 10.1039/b711490g. [DOI] [PubMed] [Google Scholar]
- Turkevich J.; Cooper Stevenson P.; Hillier J. A Study of the Nucleation and Growth Processes In the Synthesis of Colloidal Gold. Discuss. Faraday Soc. 1951, 11, 55–57. 10.1039/df9511100055. [DOI] [Google Scholar]
- Brust M.; Walker M.; Bethell D.; Schiffrin D. J.; Whyman R. Synthesis of Thiol-Derivatised Gold Nanoparticles. J. Chem. Soc., Chem. Commun. 1994, 801–802. 10.1039/C39940000801. [DOI] [Google Scholar]
- Lee Y. J.; Schade N. B.; Sun L.; Fan J. A.; Bae D. R.; Mariscal M. M.; Lee G.; Capasso F.; Sacanna S.; Manoharan V. N.; Yi G. R. Ultrasmooth, Highly Spherical Monocrystalline Gold Particles for Precision Plasmonics. ACS Nano 2013, 7 (12), 11064–11070. 10.1021/nn404765w. [DOI] [PubMed] [Google Scholar]
- Johnson P. B.; Christy R. W. Optical Constants of the Noble Metals. Phys. Rev. B 1972, 6, 4370–4379. 10.1103/PhysRevB.6.4370. [DOI] [Google Scholar]
- Yu K.; Sader J. E.; Zijlstra P.; Hong M.; Xu Q. H.; Orrit M. Probing Silver Deposition on Single Gold Nanorods by Their Acoustic Vibrations. Nano Lett. 2014, 14 (2), 915–922. 10.1021/nl404304h. [DOI] [PubMed] [Google Scholar]
- Gallardo O. A. D.; Moiraghi R.; Macchione M. A.; Godoy J. A.; Pérez M. A.; Coronado E. A.; Macagno V. A. Silver Oxide Particles/silver Nanoparticles Interconversion: Susceptibility of Forward/backward Reactions to the Chemical Environment at Room Temperature. RSC Adv. 2012, 2 (7), 2923–2929. 10.1039/c2ra01044e. [DOI] [Google Scholar]
- Nikoobakht B.; El-Sayed M. A. Preparation and Growth Mechanism of Gold Nanorods (NRs) Using Seed - Mediated Growth Method. Chem. Mater. 2003, 15 (16), 1957–1962. 10.1021/cm020732l. [DOI] [Google Scholar]
- Li Q.; Zhuo X.; Li S.; Ruan Q.; Xu Q. H.; Wang J. F. Production of Monodisperse Gold Nanobipyramids with Number Percentages Approaching 100% and Evaluation of Their Plasmonic Properties. Adv. Opt. Mater. 2015, 3 (6), 801–812. 10.1002/adom.201400505. [DOI] [Google Scholar]
- Sönnichsen C.; Franzl T.; Wilk T.; von Plessen G.; Feldmann J.; Wilson O.; Mulvaney P. Drastic Reduction of Plasmon Damping in Gold Nanorods. Phys. Rev. Lett. 2002, 88 (7), 077402. 10.1103/PhysRevLett.88.077402. [DOI] [PubMed] [Google Scholar]
- Liu M.; Guyot-Sionnest P.; Lee T. W.; Gray S. K. Optical Properties of Rodlike and Bipyramidal Gold Nanoparticles from Three-Dimensional Computations. Phys. Rev. B: Condens. Matter Mater. Phys. 2007, 76 (23), 235428. 10.1103/PhysRevB.76.235428. [DOI] [Google Scholar]
- Sriram M.; Zong K.; Vivekchand S. R. C.; Gooding J. Single Nanoparticle Plasmonic Sensors. Sensors 2015, 15 (10), 25774–25792. 10.3390/s151025774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusz G. J.; Curry A. C.; Marinakos S. M.; Wax A.; Chilkoti A. Rational Selection of Gold Nanorod Geometry for Label-Free Plasmonic Biosensors. ACS Nano 2009, 3 (4), 795–806. 10.1021/nn8006465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.; Mayer K. M.; Hafner J. H. Improved Localized Surface Plasmon Resonance Immunoassay with Gold Bipyramid Substrates. Anal. Chem. 2009, 81 (11), 4450–4455. 10.1021/ac900276n. [DOI] [PubMed] [Google Scholar]
- Becker J.; Trügler A.; Jakab A.; Hohenester U.; Sönnichsen C. The Optimal Aspect Ratio of Gold Nanorods for Plasmonic Bio-Sensing. Plasmonics 2010, 5 (2), 161–167. 10.1007/s11468-010-9130-2. [DOI] [Google Scholar]
- Peters S. M. E.; Prins M. W. J.; Zijlstra P. Strong Reduction of Spectral Heterogeneity of Gold Bipyramids for Single-Particle and Single-Molecule Plasmon Sensing. Nanotechnology 2016, 27, 024001. 10.1088/0957-4484/27/2/024001. [DOI] [PubMed] [Google Scholar]
- Jana N. R.; Gearheart L.; Murphy C. J. Wet Chemical Synthesis of High Aspect Ratio Cylindrical Gold Nanorods. J. Phys. Chem. B 2001, 105 (19), 4065–4067. 10.1021/jp0107964. [DOI] [Google Scholar]
- Liu M.; Guyot-Sionnest P. Mechanism of silver(I)-Assisted Growth of Gold Nanorods and Bipyramids. J. Phys. Chem. B 2005, 109 (47), 22192–22200. 10.1021/jp054808n. [DOI] [PubMed] [Google Scholar]
- Huang J.-S.; Callegari V.; Geisler P.; Brüning C.; Kern J.; Prangsma J. C.; Wu X.; Feichtner T.; Ziegler J.; Weinmann P.; Kamp M.; Forchel A.; Biagioni P.; Sennhauser U.; Hecht B. Atomically Flat Single-Crystalline Gold Nanostructures for Plasmonic Nanocircuitry. Nat. Commun. 2010, 1 (9), 150. 10.1038/ncomms1143. [DOI] [PubMed] [Google Scholar]
- Aćimović S. S.; Kreuzer M. P.; González M. U.; Quidant R. Plasmon near-Field Coupling in Metal Dimers as a Step toward Single-Molecule Sensing. ACS Nano 2009, 3 (5), 1231–1237. 10.1021/nn900102j. [DOI] [PubMed] [Google Scholar]
- Halas N. J.; Lal S.; Chang W. S.; Link S.; Nordlander P. Plasmons in Strongly Coupled Metallic Nanostructures. Chem. Rev. 2011, 111 (6), 3913–3961. 10.1021/cr200061k. [DOI] [PubMed] [Google Scholar]
- Mirkin C. A.; Letsinger R. L.; Mucic R. C.; Storhoff J. J. A DNA-Based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature 1996, 382, 607–609. 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- Alivisatos A. P.; Johnsson K. P.; Peng X.; Wilson T. E.; Loweth C. J.; Bruchez M. P.; Schultz P. G. Organization of “Nanocrystal Molecules” Using DNA. Nature 1996, 382, 609–611. 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- Zhang S. Z.; Kou X. S.; Yang Z.; Shi Q. H.; Stucky G. D.; Sun L. D.; Wang J. F.; Yan C. H. Nanonecklaces Assembled from Gold Rods, Spheres, and Bipyramids. Chem. Commun. 2007, 1816–1818. 10.1039/b618818d. [DOI] [PubMed] [Google Scholar]
- Sun Z.; Ni W.; Yang Z.; Kou X.; Li L.; Wang J. pH-Controlled Reversible Assembly and Disassembly of Gold Nanorods. Small 2008, 4 (9), 1287–1292. 10.1002/smll.200800099. [DOI] [PubMed] [Google Scholar]
- Nikitin M. P.; Zdobnova T. A.; Lukash S. V.; Stremovskiy O. A.; Deyev S. M. Protein-Assisted Self-Assembly of Multifunctional Nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (13), 5827–5832. 10.1073/pnas.1001142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen S. J.; Huang C. Z.; Wang J.; Li Y. F. End-to-End Assembly of Gold Nanorods on the Basis of Aptamer-Protein Recognition. J. Phys. Chem. C 2009, 113 (52), 21543–21547. 10.1021/jp905084e. [DOI] [Google Scholar]
- Gates B. D.; Xu Q.; Stewart M.; Ryan D.; Willson C. G.; Whitesides G. M. New Approaches to Nanofabrication: Molding, Printing, and Other Techniques. Chem. Rev. 2005, 105 (4), 1171–1196. 10.1021/cr030076o. [DOI] [PubMed] [Google Scholar]
- Kinkhabwala A.; Yu Z.; Fan S.; Avlasevich Y.; Müllen K.; Moerner W. E. Large Single-Molecule Fluorescence Enhancements Produced by a Bowtie Nanoantenna. Nat. Photonics 2009, 3 (11), 654–657. 10.1038/nphoton.2009.187. [DOI] [Google Scholar]
- Curto A. G.; Volpe G.; Taminiau T. H.; Kreuzer M. P.; Quidant R.; van Hulst N. F. Unidirectional Emission of a Quantum Dot Coupled to a Nanoantenna. Science 2010, 329 (5994), 930–933. 10.1126/science.1191922. [DOI] [PubMed] [Google Scholar]
- Rechberger W.; Hohenau A.; Leitner A.; Krenn J. R.; Lamprecht B.; Aussenegg F. R. Optical Properties of Two Interacting Gold Nanoparticles. Opt. Commun. 2003, 220, 137–141. 10.1016/S0030-4018(03)01357-9. [DOI] [Google Scholar]
- Ueno K.; Mizeikis V.; Juodkazis S.; Sasaki K.; Misawa H. Optical Properties of Nanoengineered Gold Blocks. Opt. Lett. 2005, 30 (16), 2158–2160. 10.1364/OL.30.002158. [DOI] [PubMed] [Google Scholar]
- Thacker V. V.; Herrmann L. O.; Sigle D. O.; Zhang T.; Liedl T.; Baumberg J. J.; Keyser U. F. DNA Origami Based Assembly of Gold Nanoparticle Dimers for Surface-Enhanced Raman Scattering. Nat. Commun. 2014, 5, 3448. 10.1038/ncomms4448. [DOI] [PubMed] [Google Scholar]
- Schreiber R.; Luong N.; Fan Z.; Kuzyk A.; Nickels P. C.; Zhang T.; Smith D. M.; Yurke B.; Kuang W.; Govorov A. O.; Liedl T. Chiral Plasmonic DNA Nanostructures with Switchable Circular Dichroism. Nat. Commun. 2013, 4, 2948. 10.1038/ncomms3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan P.; Dutta P. K.; Wang P.; Song G.; Dai M.; Zhao S.; Wang Z.-G.; Yin P.; Zhang W.; Ding B.; Ke Y. Reconfigurable Three-Dimensional Gold Nanorod Plasmonic Nanostructures Organized on DNA Origami Tripod. ACS Nano 2017, 11, 1172–1179. 10.1021/acsnano.6b06861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung A. M.; Micheel C. M.; Bozano L. D.; Osterbur L. W.; Wallraff G. M.; Cha J. N. Large-Area Spatially Ordered Arrays of Gold Nanoparticles Directed by Lithographically Confined DNA Origami. Nat. Nanotechnol. 2010, 5 (2), 121–126. 10.1038/nnano.2009.450. [DOI] [PubMed] [Google Scholar]
- Acuna G. P.; Möller F. M.; Holzmeister P.; Beater S.; Lalkens B.; Tinnefeld P. Fluorescence Enhancement at Docking Sites of DNA-Directed Self-Assembled Nanoantennas. Science 2012, 338 (6106), 506–510. 10.1126/science.1228638. [DOI] [PubMed] [Google Scholar]
- Gür F. N.; Schwarz F. W.; Ye J.; Diez S.; Schmidt T. L. Toward Self-Assembled Plasmonic Devices: High-Yield Arrangement of Gold Nanoparticles on DNA Origami Templates. ACS Nano 2016, 10 (5), 5374–5382. 10.1021/acsnano.6b01537. [DOI] [PubMed] [Google Scholar]
- Dai G.; Lu X.; Chen Z.; Meng C.; Ni W.; Wang Q. DNA Origami-Directed, Discrete Three-Dimensional Plasmonic Tetrahedron Nanoarchitectures with Tailored Optical Chirality. ACS Appl. Mater. Interfaces 2014, 6 (8), 5388–5392. 10.1021/am501599f. [DOI] [PubMed] [Google Scholar]
- Zijlstra P.; Paulo P. M. R.; Yu K.; Xu Q. H.; Orrit M. Chemical Interface Damping in Single Gold Nanorods and Its near Elimination by Tip-Specific Functionalization. Angew. Chem., Int. Ed. 2012, 51 (33), 8352–8355. 10.1002/anie.201202318. [DOI] [PubMed] [Google Scholar]
- Beeram S. R.; Zamborini F. P. Selective Attachment of Antibodies to the Edges of Gold Nanostructures for Enhanced Localized Surface Plasmon Resonance Biosensing. J. Am. Chem. Soc. 2009, 131, 11689–11691. 10.1021/ja904387j. [DOI] [PubMed] [Google Scholar]
- Rycenga M.; Langille M. R.; Personick M. L.; Ozel T.; Mirkin C. A. Chemically Isolating Hot Spots on Concave Nanocubes. Nano Lett. 2012, 12 (12), 6218–6222. 10.1021/nl3032235. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Soppera O.; Plain J.; Jradi S.; Wei Sun X.; Volkan Demir H.; Yang X.; Deeb C.; Gray S. K.; Wiederrecht G. P.; Bachelot R. Plasmon-Based Photopolymerization: Near-Field Probing, Advanced Photonic Nanostructures and Nanophotochemistry. J. Opt. 2014, 16 (11), 114002. 10.1088/2040-8978/16/11/114002. [DOI] [Google Scholar]
- Feuz L.; Jonsson M. P.; Höök F. Material-Selective Surface Chemistry for Nanoplasmonic Sensors: Optimizing Sensitivity and Controlling Binding to Local Hot Spots. Nano Lett. 2012, 12, 873–879. 10.1021/nl203917e. [DOI] [PubMed] [Google Scholar]
- Cortés E.; Xie W.; Cambiasso J.; Jermyn A. S.; Sundararaman R.; Narang P.; Schlücker S.; Maier S. A. Plasmonic Hot Electron Transport Drives Nano-Localized Chemistry. Nat. Commun. 2017, 8, 14880. 10.1038/ncomms14880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding J. J.; Gaus K. Single-Molecule Sensors: Challenges and Opportunities for Quantitative Analysis. Angew. Chem., Int. Ed. 2016, 55, 11354–11366. 10.1002/anie.201600495. [DOI] [PubMed] [Google Scholar]
- Beuwer M. A.; Prins M. W. J.; Zijlstra P. Stochastic Protein Interactions Monitored by Hundreds of Single-Molecule Plasmonic Biosensors. Nano Lett. 2015, 15 (5), 3507–3511. 10.1021/acs.nanolett.5b00872. [DOI] [PubMed] [Google Scholar]
- Zijlstra P.; Paulo P. M. R.; Orrit M. Optical Detection of Single Non-Absorbing Molecules Using the Surface Plasmon Resonance of a Gold Nanorod. Nat. Nanotechnol. 2012, 7 (6), 379–382. 10.1038/nnano.2012.51. [DOI] [PubMed] [Google Scholar]
- Antosiewicz T. J.; Käll M. A Multiscale Approach to Modeling Plasmonic Nanorod Biosensors. J. Phys. Chem. C 2016, 120, 20692–20701. 10.1021/acs.jpcc.6b01897. [DOI] [Google Scholar]
- Nusz G. J.; Marinakos S. M.; Curry A. C.; Dahlin A.; Hook F.; Wax A.; Chilkoti A. Label-Free Plasmonic Detection of Biomolecular Binding by a Single Gold Nanorod. Anal. Chem. 2008, 80 (4), 984–989. 10.1021/ac7017348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N.; Chen Y.; Li L.; Guan Z.; Zhao T.; Zhou N.; Yuan P.; Yao S. Q.; Xu Q. H. Shape-Dependent Two-Photon Photoluminescence of Single Gold Nanoparticles. J. Phys. Chem. C 2014, 118 (25), 13904–13911. 10.1021/jp502038v. [DOI] [Google Scholar]
- Zijlstra P.; Orrit M. Single Metal Nanoparticles: Optical Detection, Spectroscopy and Applications. Rep. Prog. Phys. 2011, 74 (10), 106401. 10.1088/0034-4885/74/10/106401. [DOI] [Google Scholar]
- Olson J.; Dominguez-Medina S.; Hoggard A.; Wang L.-Y.; Chang W.-S.; Link S. Optical Characterization of Single Plasmonic Nanoparticles. Chem. Soc. Rev. 2015, 44 (1), 40–57. 10.1039/C4CS00131A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crut A.; Maioli P.; Del Fatti N.; Vallée F. Optical Absorption and Scattering Spectroscopies of Single Nano-Objects. Chem. Soc. Rev. 2014, 43 (11), 3921–3956. 10.1039/c3cs60367a. [DOI] [PubMed] [Google Scholar]
- Hu M.; Novo C.; Funston A.; Wang H.; Staleva H.; Zou S.; Mulvaney P.; Xia Y.; Hartland G. V. Dark-Field Microscopy Studies of Single Metal Nanoparticles: Understanding the Factors That Influence the Linewidth of the Localized Surface Plasmon Resonance. J. Mater. Chem. 2008, 18 (17), 1949–1960. 10.1039/b714759g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer D.; Tamarat P.; Maali A.; Lounis B.; Orrit M. Photothermal Imaging of Nanometer-Sized Metal Particles Among Scatterers. Science 2002, 297 (5584), 1160–1163. 10.1126/science.1073765. [DOI] [PubMed] [Google Scholar]
- Tcherniak A.; Dominguez-Medina S.; Chang W. S.; Swanglap P.; Slaughter L. S.; Landes C. F.; Link S. One-Photon Plasmon Luminescence and Its Application to Correlation Spectroscopy as a Probe for Rotational and Translational Dynamics of Gold Nanorods. J. Phys. Chem. C 2011, 115 (32), 15938–15949. 10.1021/jp206203s. [DOI] [Google Scholar]
- Gaiduk A.; Yorulmaz M.; Orrit M. Correlated Absorption and Photoluminescence of Single Gold Nanoparticles. ChemPhysChem 2011, 12 (8), 1536–1541. 10.1002/cphc.201100167. [DOI] [PubMed] [Google Scholar]
- Wang H.; Huff T. B.; Zweifel D. A.; He W.; Low P. S.; Wei A.; Cheng J. X. In Vitro and in Vivo Two-Photon Luminescence Imaging of Single Gold Nanorods. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 15752–15756. 10.1073/pnas.0504892102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imura K.; Nagahara T.; Okamoto H. Near-Field Two-Photon-Induced Photoluminescence from Single Gold Nanorods and Imaging of Plasmon Modes. J. Phys. Chem. B 2005, 109 (27), 13214–13220. 10.1021/jp051631o. [DOI] [PubMed] [Google Scholar]
- Mooradian A. Photoluminescence of Metals. Phys. Rev. Lett. 1969, 22 (5), 185–187. 10.1103/PhysRevLett.22.185. [DOI] [Google Scholar]
- Jiang X.; Pan Y.; Jiang C.; Zhao T.; Yuan P.; Venkatesan T.; Xu Q. Excitation Nature of Two-Photon Photoluminescence of Gold Nanorods and Coupled Gold Nanoparticles Studied by Two-Pulse Emission Modulation Spectroscopy. J. Phys. Chem. Lett. 2013, 4, 1634–1638. 10.1021/jz400582h. [DOI] [PubMed] [Google Scholar]
- Yorulmaz M.; Khatua S.; Zijlstra P.; Gaiduk A.; Orrit M. Luminescence Quantum Yield of Single Gold Nanorods. Nano Lett. 2012, 12 (8), 4385–4391. 10.1021/nl302196a. [DOI] [PubMed] [Google Scholar]
- Dulkeith E.; Niedereichholz T.; Klar T. A.; Feldmann J.; Von Plessen G.; Gittins D. I.; Mayya K. S.; Caruso F. Plasmon Emission in Photoexcited Gold Nanoparticles. Phys. Rev. B: Condens. Matter Mater. Phys. 2004, 70 (20), 205424. 10.1103/PhysRevB.70.205424. [DOI] [Google Scholar]
- Ortega-Arroyo J.; Kukura P. Interferometric Scattering Microscopy (iSCAT): New Frontiers in Ultrafast and Ultrasensitive Optical Microscopy. Phys. Chem. Chem. Phys. 2012, 14 (45), 15625–15636. 10.1039/c2cp41013c. [DOI] [PubMed] [Google Scholar]
- Lippitz M.; van Dijk M. A.; Orrit M. Third-Harmonic Generation from Single Gold Nanoparticles. Nano Lett. 2005, 5 (4), 799–802. 10.1021/nl0502571. [DOI] [PubMed] [Google Scholar]
- Hartland G. V. Ultrafast Studies of Single Semiconductor and Metal Nanostructures through Transient Absorption Microscopy. Chem. Sci. 2010, 1 (3), 303–309. 10.1039/c0sc00243g. [DOI] [Google Scholar]
- Jung Y.; Chen H.; Tong L.; Cheng J. Imaging Gold Nanorods by Plasmon-Resonance-Enhanced Four Wave Mixing Imaging Gold Nanorods by Plasmon-Resonance-Enhanced Four Wave Mixing. J. Phys. Chem. C 2009, 113, 2657–2663. 10.1021/jp810852c. [DOI] [Google Scholar]
- Ruijgrok P. V.; Verhart N. R.; Zijlstra P.; Tchebotareva A. L.; Orrit M. Brownian Fluctuations and Heating of an Optically Aligned Gold Nanorod. Phys. Rev. Lett. 2011, 107 (3), 037401. 10.1103/PhysRevLett.107.037401. [DOI] [PubMed] [Google Scholar]
- Kumar J.; Wei X.; Barrow S.; Funston A. M.; Thomas K. G.; Mulvaney P. Surface Plasmon Coupling in End-to-End Linked Gold Nanorod Dimers and Trimers. Phys. Chem. Chem. Phys. 2013, 15 (12), 4258–4264. 10.1039/c3cp44657c. [DOI] [PubMed] [Google Scholar]
- Willets K. A.; Wilson A. J.; Sundaresan V.; Joshi P. B. Super-Resolution Imaging and Plasmonics. Chem. Rev. 2017, 117, 7538. 10.1021/acs.chemrev.6b00547. [DOI] [PubMed] [Google Scholar]
- Sönnichsen C.; Geier S.; Hecker N. E.; von Plessen G.; Feldmann J.; Ditlbacher H.; Lamprecht B.; Krenn J. R.; Aussenegg F. R.; Chan V. Z.-H.; Spatz J. P.; Möller M. Spectroscopy of Single Metallic Nanoparticles Using Total Internal Reflection Microscopy. Appl. Phys. Lett. 2000, 77 (19), 2949–2951. 10.1063/1.1323553. [DOI] [Google Scholar]
- Gu Z.; Jing C.; Ying Y. L.; He P.; Long Y. T. In Situ High Throughput Scattering Light Analysis of Single Plasmonic Nanoparticles in Living Cells. Theranostics 2015, 5 (2), 188–195. 10.7150/thno.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J. Michaelis-Menten Equation and Detailed Balance in Enzymatic Networks. J. Phys. Chem. B 2011, 115 (18), 5493–5498. 10.1021/jp110924w. [DOI] [PubMed] [Google Scholar]
- Bingham J. M.; Willets K. A.; Shah N. C.; Andrews D. Q.; Van Duyne R. P. LSPR Imaging: Simultaneous Single Nanoparticle Spectroscopy and Diffusional Dynamics. J. Phys. Chem. C 2009, 113 (39), 16839–16842. 10.1021/jp907377h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X.; Zhou Y.; Thota S.; Zou S.; Zhao J. Plasmonic Coupling in Single Silver Nanosphere Assemblies by Polarization-Dependent Dark-Field Scattering Spectroscopy. J. Phys. Chem. C 2014, 118 (25), 13801–13808. 10.1021/jp503505x. [DOI] [Google Scholar]
- Schopf C.; Noonan E.; Quinn A.; Iacopino D. Self-Assembly of Gold Nanocrystals into Discrete Coupled Plasmonic Structures. Crystals 2016, 6 (9), 117. 10.3390/cryst6090117. [DOI] [Google Scholar]
- Olson J.; Dominguez-Medina S.; Hoggard A.; Wang L.-Y.; Chang W.-S.; Link S. Optical Characterization of Single Plasmonic Nanoparticles. Chem. Soc. Rev. 2015, 44 (1), 40–57. 10.1039/C4CS00131A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehl C. L.; Grady N. K.; Goodrich G. P.; Tam F.; Halas N. J.; Hafner J. H. Scattering Spectra of Single Gold Nanoshells. Nano Lett. 2004, 4, 2355–2359. 10.1021/nl048610a. [DOI] [Google Scholar]
- Novo C.; Funston A. M.; Pastoriza-Santos I.; Liz-Marzán L. M.; Mulvaney P. Spectroscopy and High-Resolution Microscopy of Single Nanocrystals by a Focused Ion Beam Registration Method. Angew. Chem., Int. Ed. 2007, 46 (19), 3517–3520. 10.1002/anie.200700033. [DOI] [PubMed] [Google Scholar]
- Jin R.; Jureller J. E.; Kim H. Y.; Scherer N. F. Correlating Second Harmonic Optical Responses of Single Ag Nanoparticles with Morphology. J. Am. Chem. Soc. 2005, 127 (36), 12482–12483. 10.1021/ja0537169. [DOI] [PubMed] [Google Scholar]
- Taylor A. B.; Siddiquee A. M.; Chon J. W. M. Below Melting Point Photothermal Reshaping of Single Gold Nanorods Driven by Surface Diffusion. ACS Nano 2014, 8 (12), 12071–12079. 10.1021/nn5055283. [DOI] [PubMed] [Google Scholar]
- Siddiquee A. M.; Taylor A. B.; Syed S.; Lim G. H.; Lim B.; Chon J. W. M. Measurement of Plasmon-Mediated Two-Photon Luminescence Action Cross Sections of Single Gold Bipyramids, Dumbbells, and Hemispherically Capped Cylindrical Nanorods. J. Phys. Chem. C 2015, 119 (51), 28536–28543. 10.1021/acs.jpcc.5b08214. [DOI] [Google Scholar]
- Stiles R. L.; Willets K. A.; Sherry L. J.; Roden J. M.; Van Duyne R. P. Investigating Tip-Nanoparticle Interactions in Spatially Correlated Total Internal Reflection Plasmon Spectroscopy and Atomic Force Microscopy. J. Phys. Chem. C 2008, 112 (31), 11696–11701. 10.1021/jp802066b. [DOI] [Google Scholar]
- Song Y.; Nallathamby P. D.; Huang T.; Elsayed-ali H. E.; Xu X. N. Correlation and Characterization of Three-Dimensional Morphologically Dependent Localized Surface Plasmon Resonance Spectra of Single Silver Nanoparticles Using Dark-Field Optical Microscopy and Spectroscopy and Atomic Force Microscopy. J. Phys. Chem. C 2010, 114, 74–81. 10.1021/jp9083019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bek A.; Jansen R.; Ringler M.; Mayilo S.; Klar T. A.; Feldmann J. Fluorescence Enhancement in Hot Spots of AFM-Designed Gold Nanoparticle Sandwiches. Nano Lett. 2008, 8 (2), 485–490. 10.1021/nl072602n. [DOI] [PubMed] [Google Scholar]
- Sivun D.; Vidal C.; Munkhbat B.; Arnold N.; Klar T. A.; Hrelescu C. Anticorrelation of Photoluminescence from Gold Nanoparticle Dimers with Hot-Spot Intensity. Nano Lett. 2016, 16 (11), 7203–7209. 10.1021/acs.nanolett.6b03562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenderink A. F. Single-Photon Nanoantennas. ACS Photonics 2017, 4 (4), 710–722. 10.1021/acsphotonics.7b00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauch M.; Toma K.; Toma M.; Zhang Q.; Dostalek J. Plasmon-Enhanced Fluorescence Biosensors: A Review. Plasmonics 2014, 9, 781–799. 10.1007/s11468-013-9660-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Cushing S. K.; Wu N. Plasmon-Enhanced Optical Sensors: A Review. Analyst 2015, 140 (2), 386–406. 10.1039/C4AN01079E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvill D.; Centeno A.; Xie F. Plasmonic Fluorescence Enhancement by Metal Nanostructures: Shaping the Future of Bionanotechnology. Phys. Chem. Chem. Phys. 2013, 15, 15709–15726. 10.1039/c3cp50415h. [DOI] [PubMed] [Google Scholar]
- Kern A. M.; Zhang D.; Brecht M.; Chizhik A. I.; Failla A. V.; Wackenhut F.; Meixner A. J. Enhanced Single-Molecule Spectroscopy in Highly Confined Optical Fields: From λ/2-Fabry-Pérot Resonators to Plasmonic Nano-Antennas. Chem. Soc. Rev. 2014, 43 (4), 1263–1286. 10.1039/C3CS60357A. [DOI] [PubMed] [Google Scholar]
- Holzmeister P.; Acuna G. P.; Grohmann D.; Tinnefeld P. Breaking the Concentration Limit of Optical Single-Molecule Detection. Chem. Soc. Rev. 2014, 43 (4), 1014–1028. 10.1039/C3CS60207A. [DOI] [PubMed] [Google Scholar]
- Punj D.; Ghenuche P.; Moparthi S. B.; de Torres J.; Grigoriev V.; Rigneault H.; Wenger J. Plasmonic Antennas and Zero-Mode Waveguides to Enhance Single Molecule Fluorescence Detection and Fluorescence Correlation Spectroscopy toward Physiological Concentrations. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2014, 6 (3), 268–282. 10.1002/wnan.1261. [DOI] [PubMed] [Google Scholar]
- Khatua S.; Paulo P. M. R.; Yuan H.; Gupta A.; Zijlstra P.; Orrit M. Resonant Plasmonic Enhancement of Single-Molecule Fluorescence by Individual Gold Nanorods. ACS Nano 2014, 8 (5), 4440–4449. 10.1021/nn406434y. [DOI] [PubMed] [Google Scholar]
- Boerigter C.; Aslam U.; Linic S. Mechanism of Charge Transfer from Plasmonic Nanostructures to Chemically Attached Materials. ACS Nano 2016, 10 (6), 6108–6115. 10.1021/acsnano.6b01846. [DOI] [PubMed] [Google Scholar]
- Hao E.; Schatz G. C. Electromagnetic Fields around Silver Nanoparticles and Dimers. J. Chem. Phys. 2004, 120 (1), 357–366. 10.1063/1.1629280. [DOI] [PubMed] [Google Scholar]
- Zhu W.; Esteban R.; Borisov A. G.; Baumberg J. J.; Nordlander P.; Lezec H. J.; Aizpurua J.; Crozier K. B. Quantum Mechanical Effects in Plasmonic Structures with Subnanometre Gaps. Nat. Commun. 2016, 7, 11495. 10.1038/ncomms11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punj D.; Regmi R.; Devilez A.; Plauchu R.; Moparthi S. B.; Stout B.; Bonod N.; Rigneault H.; Wenger J. Self-Assembled Nanoparticle Dimer Antennas for Plasmonic-Enhanced Single-Molecule Fluorescence Detection at Micromolar Concentrations. ACS Photonics 2015, 2 (8), 1099–1107. 10.1021/acsphotonics.5b00152. [DOI] [Google Scholar]
- Zhang T.; Gao N.; Li S.; Lang M. J.; Xu Q. H. Single-Particle Spectroscopic Study on Fluorescence Enhancement by Plasmon Coupled Gold Nanorod Dimers Assembled on DNA Origami. J. Phys. Chem. Lett. 2015, 6 (11), 2043–2049. 10.1021/acs.jpclett.5b00747. [DOI] [PubMed] [Google Scholar]
- Garcia De Abajo F. J. Nonlocal Effects in the Plasmons of Strongly Interacting Nanoparticles, Dimers, and Waveguides. J. Phys. Chem. C 2008, 112 (46), 17983–17987. 10.1021/jp807345h. [DOI] [Google Scholar]
- Mertens H.; Polman A. Strong Luminescence Quantum-Efficiency Enhancement near Prolate Metal Nanoparticles: Dipolar versus Higher-Order Modes. J. Appl. Phys. 2009, 105 (4), 044302. 10.1063/1.3078108. [DOI] [Google Scholar]
- Holzmeister P.; Pibiri E.; Schmied J. J.; Sen T.; Acuna G. P.; Tinnefeld P. Quantum Yield and Excitation Rate of Single Molecules close to Metallic Nanostructures. Nat. Commun. 2014, 5, 5356. 10.1038/ncomms6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuloaga J.; Nordlander P. On the Energy Shift between near-Field and Far-Field Peak Intensities in Localized Plasmon Systems. Nano Lett. 2011, 11 (3), 1280–1283. 10.1021/nl1043242. [DOI] [PubMed] [Google Scholar]
- Zhao J.; Dieringer J. a; Zhang X.; Schatz G. C.; Van Duyne R. P. Wavelength-Scanned Surface-Enhanced Resonance Raman Excitation Spectroscopy. J. Phys. Chem. C 2008, 112 (49), 19302–19310. 10.1021/jp807837t. [DOI] [Google Scholar]
- Alonso-González P.; Albella P.; Neubrech F.; Huck C.; Chen J.; Golmar F.; Casanova F.; Hueso L. E.; Pucci A.; Aizpurua J.; Hillenbrand R. Experimental Verification of the Spectral Shift between near- and Far-Field Peak Intensities of Plasmonic Infrared Nanoantennas. Phys. Rev. Lett. 2013, 110 (20), 203902. 10.1103/PhysRevLett.110.203902. [DOI] [PubMed] [Google Scholar]
- Yuan H.; Khatua S.; Zijlstra P.; Yorulmaz M.; Orrit M. Thousand-Fold Enhancement of Single-Molecule Fluorescence near a Single Gold Nanorod. Angew. Chem., Int. Ed. 2013, 52 (4), 1217–1221. 10.1002/anie.201208125. [DOI] [PubMed] [Google Scholar]
- Punj D.; Mivelle M.; Moparthi S. B.; van Zanten T. S.; Rigneault H.; van Hulst N. F.; García-Parajó M. F.; Wenger J. A Plasmonic “Antenna-in-Box” Platform for Enhanced Single-Molecule Analysis at Micromolar Concentrations. Nat. Nanotechnol. 2013, 8 (June), 512–516. 10.1038/nnano.2013.98. [DOI] [PubMed] [Google Scholar]
- Levene M. J.; Korlach J.; Turner S. W.; Foquet M.; Craighead H. G.; Webb W. W. Zero-Mode Waveguides for Single-Molecule Analysis at High Concentrations. Science 2003, 299 (5607), 682–686. 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- Alam M. S.; Karim F.; Zhao C. Single-Molecule Detection at High Concentrations with Optical Aperture Nanoantennas. Nanoscale 2016, 8, 9480–9487. 10.1039/C6NR01645F. [DOI] [PubMed] [Google Scholar]
- Regmi R.; Al Balushi A. A.; Rigneault H.; Gordon R.; Wenger J. Nanoscale Volume Confinement and Fluorescence Enhancement with Double Nanohole Aperture. Sci. Rep. 2015, 5, 15852. 10.1038/srep15852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pibiri E.; Holzmeister P.; Lalkens B.; Acuna G. P.; Tinnefeld P. Single-Molecule Positioning in Zeromode Waveguides by DNA Origami Nanoadapters. Nano Lett. 2014, 14, 3499–3503. 10.1021/nl501064b. [DOI] [PubMed] [Google Scholar]
- Heucke S. F.; Baumann F.; Acuna G. P.; Severin P. M. D.; Stahl S. W.; Strackharn M.; Stein I. H.; Altpeter P.; Tinnefeld P.; Gaub H. E. Placing Individual Molecules in the Center of Nanoapertures. Nano Lett. 2014, 14, 391–395. 10.1021/nl401517a. [DOI] [PubMed] [Google Scholar]
- Miyake T.; Tanii T.; Sonobe H.; Akahori R.; Ueno T.; Funatsu T.; Ohdomari I.; Shimamoto N. Real-Time Imaging of Single-Molecule Fluorescence with a Zero-Mode Waveguide for the Analysis of Protein-Protein Interactions. Anal. Chem. 2008, 80 (15), 6018–6022. 10.1021/ac800726g. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Chen D.; Yue H.; Spiering M. M.; Zhao C.; Benkovic S. J.; Huang T. J. Dark-Field Illumination on Zero-Mode Waveguide/microfluidic Hybrid Chip Reveals T4 Replisomal Protein Interactions. Nano Lett. 2014, 14 (4), 1952–1960. 10.1021/nl404802f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S. M.; Triplet M. G.; Rhodes C.; Iwig S.; Tu H.; Stamou D.; Groves J. T. Monitoring the Waiting Time Sequence of Single Ras GTPase Activation Events Using Liposome Functionalized Zero-Mode Waveguides. Nano Lett. 2016, 16, 2890–2895. 10.1021/acs.nanolett.6b00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandén T.; Wyss R.; Santschi C.; Hassaïne G.; Deluz C.; Martin O. J. F.; Wennmalm S.; Vogel H. A Zeptoliter Volume Meter for Analysis of Single Protein Molecules. Nano Lett. 2012, 12 (1), 370–375. 10.1021/nl2036468. [DOI] [PubMed] [Google Scholar]
- Sobek J.; Rehrauer H.; Schauer S.; Fischer D.; Patrignani A.; Landgraf S. Single-Molecule DNA Hybridisation Studied by Using a Modified DNA Sequencer: A Comparison with Surface Plasmon Resonance Data. Methods Appl. Fluoresc. 2016, 4, a015002. 10.1088/2050-6120/4/1/015002. [DOI] [Google Scholar]
- Flusberg B. A.; Webster D. R.; Lee J. H.; Travers K. J.; Olivares E. C.; Clark T. A.; Korlach J.; Turner S. W. Direct Detection of DNA Methylation during Single-Molecule, Real-Time Sequencing. Nat. Methods 2010, 7 (6), 461–465. 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Dalal R. V.; Petrov A. N.; Tsai A.; O’Leary S. E.; Chapin K.; Cheng J.; Ewan M.; Hsiung P.-L.; Lundquist P.; Turner S. W.; Hsu D. R.; Puglisi J. D. High-Throughput Platform for Real-Time Monitoring of Biological Processes by Multicolor Single-Molecule Fluorescence. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (2), 664–669. 10.1073/pnas.1315735111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland A. D.; Van Duyne R. P. Single Silver Nanoparticles as Real-Time Optical Sensors with Zeptomole Sensitivity. Nano Lett. 2003, 3 (8), 1057–1062. 10.1021/nl034372s. [DOI] [Google Scholar]
- Raschke G.; Kowarik S.; Franzl T.; Sonnichsen C.; Klar T. A.; Feldmann J.; Nichtl A.; Kurzinger K. Biomolecular Recognition Based on Single Gold Nanoparticle Light Scattering. Nano Lett. 2003, 3 (7), 935–938. 10.1021/nl034223+. [DOI] [Google Scholar]
- Raschke G.; Brogl S.; Susha A. S.; Rogach A. L.; Klar T. A.; Feldmann J.; Fieres B.; Petkov N.; Bein T.; Nichtl A.; Kürzinger K. Gold Nanoshells Improve Single Nanoparticle Molecular Sensors. Nano Lett. 2004, 4 (10), 1853–1857. 10.1021/nl049038q. [DOI] [Google Scholar]
- Schatz G. C.; Sherry L. J.; Jin R. C.; Mirkin C. A.; Van Duyne R. P. Localized Surface Plasmon Resonance Spectroscopy of Single Silver Triangular Nanoprisms. Nano Lett. 2006, 6, 2060–2065. 10.1021/nl061286u. [DOI] [PubMed] [Google Scholar]
- Baciu C. L.; Becker J.; Janshoff A.; Sönnichsen C. Protein-Membrane Interaction Probed by Single Plasmonic Nanoparticles. Nano Lett. 2008, 8 (6), 1724–1728. 10.1021/nl080805l. [DOI] [PubMed] [Google Scholar]
- Galush W. J.; Shelby S. A.; Mulvihill M. J.; Tao A.; Yang P.; Groves J. T. A Nanocube Plasmonic Sensor for Molecular Binding on Membrane Surfaces. Nano Lett. 2009, 9 (5), 2077–2082. 10.1021/nl900513k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosman C.; Prasad J.; Neiser A.; Henkel A.; Edgar J.; Sönnichsen C. Multiplexed Plasmon Sensor for Rapid Label-Free Analyte Detection. Nano Lett. 2013, 13, 3243–3247. 10.1021/nl401354f. [DOI] [PubMed] [Google Scholar]
- Yu C.; Irudayaraj J. Multiplex Biosensor Using Gold Nanorods. Anal. Chem. 2007, 79 (2), 572–579. 10.1021/ac061730d. [DOI] [PubMed] [Google Scholar]
- Miller M. M.; Lazarides A. A. Sensitivity of Metal Nanoparticle Surface Plasmon Resonance to the Dielectric Environment. J. Phys. Chem. B 2005, 109 (46), 21556–21565. 10.1021/jp054227y. [DOI] [PubMed] [Google Scholar]
- Antosiewicz T. J.; Apell S. P.; Claudio V.; Käll M. A Simple Model for the Resonance Shift of Localized Plasmons due to Dielectric Particle Adhesion. Opt. Express 2012, 20 (1), 524–533. 10.1364/OE.20.000524. [DOI] [PubMed] [Google Scholar]
- Yang J.; Giessen H.; Lalanne P. Simple Analytical Expression for the Peak-Frequency Shifts of Plasmonic Resonances for Sensing. Nano Lett. 2015, 15, 3439–3444. 10.1021/acs.nanolett.5b00771. [DOI] [PubMed] [Google Scholar]
- Davis T. J.; Gómez D. E.; Vernon K. C. Interaction of Molecules with Localized Surface Plasmons in Metallic Nanoparticles. Phys. Rev. B: Condens. Matter Mater. Phys. 2010, 81 (4), 045432. 10.1103/PhysRevB.81.045432. [DOI] [Google Scholar]
- Lu G.; Hou L.; Zhang T.; Li W.; Liu J.; Perriat P.; Gong Q. Anisotropic Plasmonic Sensing of Individual or Coupled Gold Nanorods. J. Phys. Chem. C 2011, 115 (46), 22877–22885. 10.1021/jp2081066. [DOI] [Google Scholar]
- Claudio V.; Dahlin A. B.; Antosiewicz T. J. Single-Particle Plasmon Sensing of Discrete Molecular Events: Binding Position versus Signal Variations for Different Sensor Geometries. J. Phys. Chem. C 2014, 118 (13), 6980–6988. 10.1021/jp412219v. [DOI] [Google Scholar]
- Mayer K. M.; Hao F.; Lee S.; Nordlander P.; Hafner J. H. A Single Molecule Immunoassay by Localized Surface Plasmon Resonance. Nanotechnology 2010, 21 (25), 255503. 10.1088/0957-4484/21/25/255503. [DOI] [PubMed] [Google Scholar]
- Ament I.; Prasad J.; Henkel A.; Schmachtel S.; Sönnichsen C. Single Unlabeled Protein Detection on Individual Plasmonic Nanoparticles. Nano Lett. 2012, 12 (2), 1092–1095. 10.1021/nl204496g. [DOI] [PubMed] [Google Scholar]
- Gaiduk A.; Ruijgrok P. V.; Yorulmaz M.; Orrit M. Detection Limits in Photothermal Microscopy. Chem. Sci. 2010, 1, 343. 10.1039/c0sc00210k. [DOI] [PubMed] [Google Scholar]
- Robertson A. D.; Murphy K. P. Protein Structure and the Energetics of Protein Stability. Chem. Rev. 1997, 97 (5), 1251–1268. 10.1021/cr960383c. [DOI] [PubMed] [Google Scholar]
- Taylor R. W.; Lee T.; Scherman O. A.; Esteban R.; Aizpurua J.; Huang F. M.; Baumberg J. J.; Mahajan S. Precise Subnanometer Plasmonic Junctions for SERS within Gold Nanoparticle Assemblies Using Cucurbit [N] Uril “ Glue.. ACS Nano 2011, 5, 3878–3887. 10.1021/nn200250v. [DOI] [PubMed] [Google Scholar]
- Sigle D. O.; Kasera S.; Herrmann L. O.; Palma A.; Benz F.; Mahajan S.; Baumberg J. J.; Scherman O. Observing Single Molecules Complexing with Cucurbit [7] Uril through Nanogap-SERS. J. Phys. Chem. Lett. 2016, 7, 704–710. 10.1021/acs.jpclett.5b02535. [DOI] [PubMed] [Google Scholar]
- Benz F.; Schmidt M. K.; Dreismann A.; Chikkaraddy R.; Zhang Y.; Demetriadou A.; Carnegie C.; Ohadi H.; de Nijs B.; Esteban R.; Aizpurua J.; Baumberg J. J. Single-Molecule Optomechanics in “picocavities.. Science 2016, 354 (6313), 726–729. 10.1126/science.aah5243. [DOI] [PubMed] [Google Scholar]
- Lee S. E.; Chen Q.; Bhat R.; Petkiewicz S.; Smith J. M.; Ferry V. E.; Correia A. L.; Alivisatos A. P.; Bissell M. J. Reversible Aptamer-Au Plasmon Rulers for Secreted Single Molecules. Nano Lett. 2015, 15 (7), 4564–4570. 10.1021/acs.nanolett.5b01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann T.; Patel D. J. Adaptive Recognition by Nucleic Acid Aptamers. Science 2000, 287 (5454), 820–825. 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- del Pino P.; Pelaz B.; Zhang Q.; Maffre P.; Nienhaus G. U.; Parak W. J. Protein Corona Formation around Nanoparticles – from the Past to the Future. Mater. Horiz. 2014, 1 (3), 301–313. 10.1039/C3MH00106G. [DOI] [Google Scholar]
- Johnson-Buck A.; Su X.; Giraldez M. D.; Zhao M.; Tewari M.; Walter N. G. Kinetic Fingerprinting to Identify and Count Single Nucleic Acids. Nat. Biotechnol. 2015, 33, 730–732. 10.1038/nbt.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y.; Li X.; Li H.; Zhang W. Quantifying Thiol–gold Interactions towards the Efficient Strength Control. Nat. Commun. 2014, 5, 4348. 10.1038/ncomms5348. [DOI] [PubMed] [Google Scholar]
- Chen S.; Zheng J.; Li L.; Jiang S. Strong Resistance of Phosphorylcholine Self-Assembled Monolayers to Protein Adsorption: Insights into Nonfouling Properties of Zwitterionic Materials. J. Am. Chem. Soc. 2005, 127 (41), 14473–14478. 10.1021/ja054169u. [DOI] [PubMed] [Google Scholar]
- Sheehan P. E.; Whitman L. J. Detection Limits for Nanoscale Biosensors. Nano Lett. 2005, 5 (4), 803–807. 10.1021/nl050298x. [DOI] [PubMed] [Google Scholar]
- Nair P. R.; Alam M. A. Performance Limits of Nanobiosensors. Appl. Phys. Lett. 2006, 88 (23), 233120. 10.1063/1.2211310. [DOI] [Google Scholar]
- Squires T. M.; Messinger R. J.; Manalis S. R. Making It Stick: Convection, Reaction and Diffusion in Surface-Based Biosensors. Nat. Biotechnol. 2008, 26 (4), 417–426. 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- De Angelis F.; Gentile F.; Mecarini F.; Das G.; Moretti M.; Candeloro P.; Coluccio M. L.; Cojoc G.; Accardo A.; Liberale C.; Zaccaria R. P.; Perozziello G.; Tirinato L.; Toma A.; Cuda G.; Cingolani R.; Di Fabrizio E. Breaking the Diffusion Limit with Super-Hydrophobic Delivery of Molecules to Plasmonic Nanofocusing SERS Structures. Nat. Photonics 2011, 5 (11), 682–687. 10.1038/nphoton.2011.222. [DOI] [Google Scholar]
- Barik A.; Otto L. M.; Yoo D.; Jose J.; Johnson T. W.; Oh S.-H. Dielectrophoresis-Enhanced Plasmonic Sensing with Gold Nanohole Arrays. Nano Lett. 2014, 14, 2006–2012. 10.1021/nl500149h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche J.; Albinsson D.; Fritzsche M.; Antosiewicz T. J.; Westerlund F.; Langhammer C. Single Particle Nanoplasmonic Sensing in Individual Nanofluidic Channels. Nano Lett. 2016, 16 (12), 7857–7864. 10.1021/acs.nanolett.6b04124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahie M.; Chisholm C.; Chen M. Resolved Single-Molecule Detection of Individual Species within a Mixture of Anti-Biotin Antibodies Using an Engineered Monomeric Nanopore. ACS Nano 2015, 9 (2), 1089–1098. 10.1021/nn506606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgia M. B.; Borgia A.; Best R. B.; Steward A.; Nettels D.; Wunderlich B.; Schuler B.; Clarke J. Single-Molecule Fluorescence Reveals Sequence-Specific Misfolding in Multidomain Proteins. Nature 2011, 474 (7353), 662–665. 10.1038/nature10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler B.; Eaton W. A. Protein Folding Studied by Single-Molecule FRET. Curr. Opin. Struct. Biol. 2008, 18 (1), 16–26. 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-X.; Rivas G.; Minton A. P. Macromolecular Crowding and Confinement: Biochemical, Biophysical, and Potential Physiological Consequences. Annu. Rev. Biophys. 2008, 37 (1), 375–397. 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck A.; Wurch T.; Bailly C.; Corvaia N. Strategies and Challenges for the next Generation of Therapeutic Antibodies. Nat. Rev. Immunol. 2010, 10 (5), 345–352. 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- Dobson C. M. Protein Folding and Misfolding. Nature 2003, 426, 884–890. 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- Steinmetz L. M.; Jones A. Sensing a Revolution. Mol. Syst. Biol. 2016, 12 (4), 867. 10.15252/msb.20166873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong G.; Corrie S. R.; Clark H. A. In Vivo Biosensing: Progress and Perspectives. ACS Sensors 2017, 2, 327. 10.1021/acssensors.6b00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S. Continuous Glucose Monitoring Systems: A Review. Diagnostics 2013, 3 (4), 385–412. 10.3390/diagnostics3040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. L.; Boutelle M. G. Real-Time Clinical Monitoring of Biomolecules. Annu. Rev. Anal. Chem. 2013, 6 (1), 427–453. 10.1146/annurev.anchem.111808.073648. [DOI] [PubMed] [Google Scholar]
- Khatua S.; Orrit M. Toward Single-Molecule Microscopy on a Smart Phone. ACS Nano 2013, 7 (10), 8340–8343. 10.1021/nn405167q. [DOI] [PubMed] [Google Scholar]
- Kühnemund M.; Wei Q.; Darai E.; Wang Y.; Hernández-Neuta I.; Yang Z.; Tseng D.; Ahlford A.; Mathot L.; Sjöblom T.; Ozcan A.; Nilsson M. Targeted DNA Sequencing and in Situ Mutation Analysis Using Mobile Phone Microscopy. Nat. Commun. 2017, 8, 537–540. 10.1038/ncomms13913. [DOI] [PMC free article] [PubMed] [Google Scholar]