Abstract
Two of the primary issues with characterizing the variability of raw materials used in mammalian cell culture, such as wheat hydrolysate, is that the analyses of these materials can be time consuming, and the results of the analyses are not straightforward to interpret. To solve these issues, spectroscopy can be combined with chemometrics to provide a quick, robust and easy to understand methodology for the characterization of raw materials; which will improve cell culture performance by providing an assessment of the impact that a given raw material will have on final product quality. In this study, four spectroscopic technologies: near infrared spectroscopy, middle infrared spectroscopy, Raman spectroscopy, and fluorescence spectroscopy were used in conjunction with principal component analysis to characterize the variability of wheat hydrolysates, and to provide evidence that the classification of good and bad lots of raw material is possible. Then, the same spectroscopic platforms are combined with partial least squares regressions to quantitatively predict two cell culture critical quality attributes (CQA): integrated viable cell density and IgG titer. The results showed that near infrared (NIR) spectroscopy and fluorescence spectroscopy are capable of characterizing the wheat hydrolysate's chemical structure, with NIR performing slightly better; and that they can be used to estimate the raw materials’ impact on the CQAs. These results were justified by demonstrating that of all the components present in the wheat hydrolysates, six amino acids: arginine, glycine, phenylalanine, tyrosine, isoleucine and threonine; and five trace elements: copper, phosphorus, molybdenum, arsenic and aluminum, had a large, statistically significant effect on the CQAs, and that NIR and fluorescence spectroscopy performed the best for characterizing the important amino acids. It was also found that the trace elements of interest were not characterized well by any of the spectral technologies used; however, the trace elements were also shown to have a less significant effect on the CQAs than the amino acids. © 2017 The Authors Biotechnology Progress published by Wiley Periodicals, Inc. on behalf of American Institute of Chemical Engineers, 33:1127–1138, 2017
Keywords: raw material characterization, spectroscopy, multivariate data analysis, principal component analysis, partial least squares regressions, near infrared, middle infrared, raman, fluorescence, cell culture, biology, bioinformatics
Introduction
In the biopharmaceutical industry, a majority of therapeutic proteins are produced in mammalian cell cultures. Mammalian cells have the capacity for proper protein folding, assembly and post‐translational modification, making them the optimal choice for producing recombinant proteins with clinical applications.1 Furthermore, specific mammalian cell lines, such as those derived from Chinese hamster ovary (CHO) cell lines are preferred.2 In fact, as recently as 2007, nearly 70% of therapeutic recombinant proteins were produced in CHO cells,3 with products including blood factors, hormones, growth factors, monoclonal antibodies and others,4 whose annual revenue can exceed $99 billion.2
Reaching the level of production required by the biopharmaceutical industry has required that improvements be made in protein production. In mammalian cell cultures specifically, refinement of vector construction, biomarker selection and advances in gene‐targeting have led to higher cell line productivity.5 Other researchers have also shown that optimizing nutrient and byproduct concentrations in the bioreactor can lead to significantly higher yields.6
Further improvements in cell line productivity have been accomplished by cell culture media optimization.7, 8, 9, 10 However, improving cell culture performance through media optimization has encountered hurdles. For instance, it is no longer encouraged to use animal‐derived supplements for cell culture media7, 8, 9, 10 because serum is expensive and a source of contamination, such as viruses.11 The push to use animal‐free media has led to proteins from plant sources, such as wheat hydrolysate, being used as supplements in cell culture media.9
Furthermore, the FDA has offered guidelines to improve product quality and consistency in the biopharmaceutical industry. The FDA guidance on process analytical technology (PAT),12 advises that on‐line process monitoring equipment be used to have real‐time product quality control. Chemometric techniques, such as principal component analysis13, 14 (PCA) and partial least squares regressions (PLS),13 have been shown to an important tool in PAT14 due to their ability to ease the analysis of large data sets generated from on‐line bioprocess sensors. PAT has been used in the biopharmaceutical industry to characterize process performance,15 allowing for corrective action to be taken if process performance was poor. Furthermore, PAT systems for cell culture involving spectroscopy and PLS have been shown to be able to predict final product quality based on process conditions.16
Motivated by the above considerations, extending PAT to include the characterization of raw materials should be the logical next step. The same PAT paradigm described above, combining spectroscopy with chemometrics, has been used in the food17, 18 and petrochemical19 industries to characterize raw material variability, and to predict final product quality based only on information about the raw materials. The paradigm is robust enough to span multiple industries, and has seen use in the biopharmaceutical industry for characterizing raw material components such as soy hydrolysate,20, 21 as well. Due to the fact that cell culture raw materials are complex mixtures22 it is usually too difficult to identify and quantify every compound present.23 However, spectroscopy, such as Raman,23, 24 middle infrared,25, 26 near infrared27 – 29 and fluorescence,30 has proven to be advantageous when combined with chemometrics for characterizing complex mixtures like those present in raw materials. Furthermore, prior work has also shown the importance of assessing the performance of multiple spectroscopic platforms when determining the impact of a new raw material on cell culture performance.20, 31 This work is building upon these principles.
It is important to understand that the different chemical structures of the complex mixtures present in raw materials means that no single spectral technology will capture all of the relevant information for every raw material. It is these differences that have caused researchers to investigate a myriad of hydrolysates to use as raw materials.32 Therefore, implementing a spectral technology to characterize wheat hydrolysate based on the results from characterizing soy, or any other hydrolysates, would have low prospects for success. Until now, there is no study investigating the use of spectroscopy and chemometrics as a PAT paradigm for the characterization of wheat hydrolysate as a raw material for cell culture processes, or as a paradigm for the characterization of wheat hydrolysate for any purpose. It is for this reason that four spectroscopic platforms: Fluorescence, Middle‐Infrared, Raman and Near‐Infrared, are combined with the chemometric techniques of PCA and PLS; first, to characterize the variability present in the wheat hydrolysates; and second, to predict final product quality using only spectral data of the wheat hydrolysate used as a raw material. This is especially important, as it has been shown that cell culture performance benefits when wheat hydrolysate is used as a raw material.32
Knowing which of the spectral technologies is the best one to use for estimating the final product quality is not enough; it is important to understand why the technology was the best. The amino acid profiles of hydrolysates used to supplement CHO cell cultures have been shown to be a good indicator of cell culture performance.33, 34 Similarly, the trace element profiles have also been shown to have a strong impact on cell culture performance.35 Therefore, even though the aim of this paper is to show that the use of spectroscopy and chemometrics is an appropriate paradigm for the characterization of wheat hydrolysate, the trace element and amino acid profiles are used to justify the spectroscopic results.
Materials and methods
Samples, bioassays, cell line, and raw materials
Wheat hydrolysates were used to supplement cell culture media for 15 different batches where the cells were being cultured to express Immunoglobulin G (IgG); the wheat hydrolysate dosage was 15 g/L for all cultures. Only one vendor for the media (CD CHO Fusion, CHO DHFR; Sigma‐Aldrich, St. Louis, MO) and supplemental wheat hydrolysates was used in order to eliminate any variability that would be introduced by using different vendors. In order to eliminate variability caused by using different cell‐lines, only a single cell‐line was used for the cell cultures (CHO‐K1 Zn GS KO clone #53).
In these analyses, the day 7 integrated viable cell density (IVCD) and IgG titer were used as cell culture performance indices. IVCD is representative of overall cell culture health, where batches with high IVCD values promoted cell growth more than batches with low values; and it was measured using a Cedex automated cell counter (Roche Diagnostics Co., Mannheim, Germany). IgG titer is representative of cell culture productivity, where batches with high values generated more product than batches with low values; and it was measured using an Octet QK with Protein A tips (ForteBio, Inc., Menlo Park, CA). Duplicate measurements were taken to ensure consistency.
Amino acids
The amino acid concentrations present in the wheat hydrolysate were analyzed using the AccuQ‐Tag Ultra Derivatization Kit (Water's Corporation, Milford, MA) for high‐performance liquid chromatography. Concentrations were obtained for 20 amino acids: alanine, arginine, asparagine, aspartic acid, cysteine, glutamic acid, glutamine, glycine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, and valine.
Trace elements
The trace element concentrations present in the wheat hydolysate were analyzed using inductively coupled plasma–optical emission spectroscopy. Concentrations were obtained for 24 trace elements: aluminum, arsenic, barium, calcium, cadmium, cobalt, chromium, copper, iron, germanium, potassium, magnesium, manganese, molybdenum, sodium, nickel, phosphorus, lead, selenium, tin, strontium, titanium, zinc and zirconium.
Spectral acquisition
The chemical structure of the wheat hydrolysate used for media supplementation was examined using four different spectral technologies: 2D fluorescence spectroscopy, middle infrared spectroscopy, near infrared spectroscopy, and Raman spectroscopy.
The 2D fluorescence spectra were measured on an LS45 fluorescence spectrometer (Perkin Elmer, Inc., Waltham, MA). Prior to obtaining the spectra, the solid powder was dissolved into distilled water at 1 g/L. Then, the samples were irradiated 15 times between 250 nm and 750 nm, where the excitation wavelength was increased incrementally by 15 nm; at each increment the resulting emission spectrum was measured from 250 nm to 798.5 nm in intervals of 0.5 nm. The scanning speed was 1,000 nm/min, which was optimized through trial and error. A single spectrum was obtained for each sample. Figure 1(a) shows the unprocessed fluorescence spectrum generated from the hydrolysate used to supplement batch one. Note, only one spectrum is shown because the two‐dimensional nature of a fluorescence spectrum makes it difficult to show the spectra for all of the samples without obscuring the data. The remaining fluorescence spectra can be seen in Supporting Information Figure S1.
Figure 1.
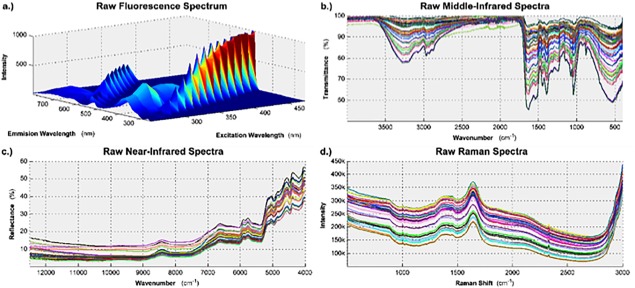
The variability of the wheat hydrolysates' chemical structure was characterized using four spectral technologies.
The raw data, before any preprocessing is applied, is shown here for: (a) fluorescence spectroscopy; (b) middle infrared spectroscopy; (c) near infrared spectroscopy; and (d) Raman spectroscopy.
To measure the middle infrared (MIR) spectra, all of the samples were loaded as a powder onto the sample stage. Then, the transmission spectra were obtained by scanning each sample from 4,000 cm−1 to 400 cm−1 in increments of 1 cm−1. Sixty‐four scans were selected to be co‐added together in order to maximize the signal‐to‐noise ratio of the resulting spectra. Triplicate spectra were obtained for each batch to ensure consistency. Figure 1(b) shows the three unprocessed MIR spectra generated from the hydrolysates used to supplement all 15 batches.
The near infrared (NIR) spectra were measured on a Bruker MPA FT‐NIR spectrophotometer (Bruker Optics, Billerica, MA). Prior to measurement, all of the samples were packed into 22 mm glass vials. Then, the reflectance spectra were obtained by scanning each sample from 12,493 cm−1 to 3,995 cm−1 in increments of 4 cm−1. 64 scans were selected to be co‐added together in order to maximize the signal‐to‐noise ratio of the resulting spectra. Triplicate spectra were obtained for each batch to ensure consistency. The spectra for the first batch were measured at the wrong resolution and were discarded. Figure 1(c) shows the three unprocessed NIR spectra generated from the hydrolysates used to supplement the remaining 14 batches.
The Raman spectra were measured on an RXN3 Raman spectrophotometer (Kaiser Optical Systems, Inc., Ann Arbor, MI) equipped with an optical fiber probe. The model RXN3 is the most optimal to use because it has the 998 nm laser, which is the least prone to cause fluorescence in the sample matrix. Prior to obtaining the spectra, the solid powder was dissolved into distilled water at 10 g/L. Then, the Raman scattering intensity was measured for Raman Shifts between 500 cm−1 to 3,000 cm−1 with a resolution of 1 cm−1; the exposure time was 30 seconds and 32 scans were co‐added together for each spectrum. Duplicate spectra were obtained for each batch to ensure consistency. Figure 1(d) shows both unprocessed Raman spectra generated from the hydrolysates used to supplement all 15 batches.
Data preprocessing and feature selection
Each of the four spectral technologies requires unique and comprehensive preprocessing techniques. This is to ensure that the multivariate analysis that follows is able to focus on the variability of the raw material as opposed to the variability induced by the measuring system.
The 2D fluorescence maps, see Figure 1(a), did not have peaks that were aligned with one another between multiple samples. This was corrected with a peak scaling and shifting algorithm that maximizes the cross‐correlation between the original signal and the target signal with aligned peaks.36 Then, the Raman and Rayleigh light scattering artifacts were removed from the spectra using polynomial interpolation.37 Figure 2(a) shows the fluorescence spectrum generated from the hydrolysate used to supplement batch one after the above preprocessing techniques were applied, the remaining fluorescence spectra can be seen in Supporting Information Figure S2. To facilitate the data analysis techniques discussed in the Multivariate Data Analysis section, each two‐dimensional fluorescence spectrum was unfolded into a one dimensional vector.37
Figure 2.
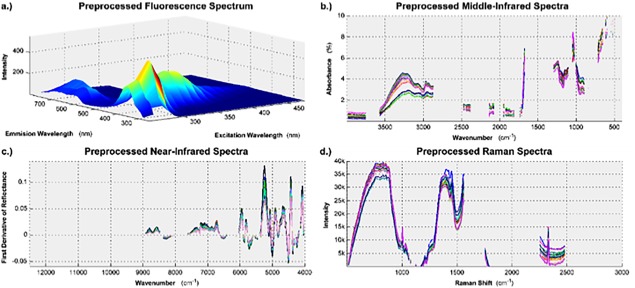
The data from each spectroscopic platform required unique preprocessing, and feature selection, to ensure that subsequent analysis was focused on sample variability, and not measurement variability.
(a) Fluorescence spectrum after the peaks were aligned and light scattering artifacts were removed. (b) Middle infrared absorbance spectra after a multiplicative scatter correction algorithm and a Savitsky‐Golay smoothing filter were applied. (c) First derivative of the near infrared spectra after a Savitsky‐Golay smoothing filter was applied. (d) Raman spectra after the data was fit to a polynomial baseline.
The MIR spectra were converted from transmission spectra to absorbance spectra using Eq. (1) where %A is the percent absorbance and %T is the percent transmittance. A multiplicative scatter correction (MSC) algorithm was applied to the MIR spectra39 and then the signal was smoothed by using a third‐order Savitsky‐Golay smoothing filter based on 15 data points.40 Figure 2(b) shows the MIR spectroscopy data resulting from the applied preprocessing techniques.
| (1) |
The NIR spectra were preprocessed by smoothing with a third‐order Savitsky‐Golay filter based on 15 data points.40 Then, the first‐order derivative was taken to improve the resolution of the overlapping peaks in the 3995‐5995 cm−1 range. Figure 2(c) shows the NIR spectroscopy data resulting from the applied preprocessing techniques.
The Raman spectra were preprocessed by fitting a polynomial baseline to the spectra (fourth‐order with a window size of 2500 data points).41 Figure 2(d) shows the Raman spectroscopy data resulting from the applied preprocessing techniques.
For all spectral technologies the data was averaged for each batch, mean centered, and mean scaled prior to multivariate data analysis. After preprocessing, feature selection was performed for all spectral technologies, except for the fluorescence spectra, based on the one‐way ANOVA F‐test.42 This was done to ensure that the features used would maximize between batch variability while simultaneously minimizing within batch variability. Due to only having one fluorescence spectrum for each sample, variable importance in projection13 was used for feature selection in the fluorescence spectra. The results of the feature selection are visible in Figure 2(b–d). For all of the spectroscopic data, only the most relevant 40% was kept for subsequent analysis.
Note: All raw data is available in Supporting Information File S3.
Multivariate Data Analysis
Principal component analysis (PCA)
PCA is used in exploratory data analysis to make large, complex datasets easier to interpret visually. Each observation is projected from the original feature space onto latent variables, called principal components. Due to the collinear nature of the variables that comprise the original feature space, a relatively small number of principal components can describe the correlation structure that exists in the original feature space. Observations that are similar across many of the original variables will appear clustered together in the projection, also known as the score space. Therefore, it can be seen if there any observations, or groups of observations, that deviate from the others.
PCA is performed on a single matrix of Data, X. The data matrix has N observations (batches) and K variables (spectra wavelengths) that were measured for each observation. When projected onto the principal components, the value of each observation on these new axes are known as scores; these are represented as t 1, t 2, etc. Viewing a plot of t 1 vs. t 2 or t 1 vs. t 3 etc. is how one visualizes the data in the score space. The original variables are related to the latent variables through loading vectors, denoted as p 1, p 2, etc. Equation (2) describes mathematically the relationship between X, t i and p i. E is the residual matrix, and it contains the raw data variance not described by the first A principal components (A may have values of 1, 2, …, N‐1). More information on PCA can be found in the literature.13, 14
| (2) |
Partial least squares regression (PLS)
PLS is used to create a regression model that describes the linear functional dependence of Y on X. It differs from a traditional regression model by finding this dependence in the dimensionally reduced score space which allows for a more robust regression models to be built due to the uncorrelated noise in the raw data being left out of the projection to the score space.
In a PLS model, the X and Y blocks are both projected into the score space, as described in the PCA section, with the added constraint that the covariance between the X block scores and Y block scores be maximized. Equations (3) and (4) describe the mathematical relationship between X, Y, t i, u i p i and q i. For the Y block projection, u i represents the scores, q i represents the loadings and F is the residual matrix. More information on PLS can be found in the literature.13
| (3) |
| (4) |
The root mean square error of prediction (RMSE) is a useful statistic for comparing the relative performance of PLS models. It is calculated from Eq. (5), where y i is the value of y observed during the ith batch, is the value predicted by the model for the ith batch, and N is the number of observations a prediction is obtained for. The lower the RMSE value, the more accurate the PLS model. Therefore, when comparing the spectral technologies, the one with the lowest RMSE value performs the best. It is important to note that RMSE is scale dependent, so the RMSE for a model predicting IVCD values cannot be compared to the RMSE for a model predicting IgG titer values because of their measurements being in different units.
| (5) |
Results
Exploratory data analysis
Product Quality
There were two critical quality attributes (CQA) considered in this analysis: integrated viable cell density (IVCD) and IgG titer. They were selected as representative CQAs because they can be measured quantitatively, and a common goal for cell culture engineers is to maximize both of them. The CQAs were both measured at harvest, and Figure 3 shows a plot of IVCD vs IgG Titer for all 15 batches. It can be seen that they are positively correlated with one another; which is to be expected, because when more cells are grown, then the amount of product created should be higher. It can also be seen that some batches performed well, which are characterized by their high IVCD and IgG titer values; and other batches performed poorly, which are characterized by their low IVCD and IgG titer values. The color scheme in Figure 3 is representative of the classification assigned to each of the 15 batches by the cell culture operator. Industrial scale batches that exhibited good performance are colored in green, industrial scale batches that exhibited poor performance are colored in red and batches from the pilot plant are colored in blue. The importance of these classifications is considered in the Discussion section below.
Figure 3.
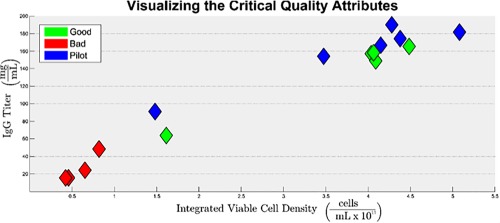
Visualization of product quality; a plot of IgG Titer vs. IVCD for all 15 batches.
Batches that performed poorly, relative to one another, are clustered in the bottom left corner of the figure and batches that performed well are clustered in the upper right corner.
Spectra
PCA was performed separately on the spectra generated from the wheat hydrolysate using each of the four spectral technologies. The resulting score spaces are shown in Figure 4. Figure 4(a) is from the fluorescence spectroscopy model, (b) is from the MIR spectroscopy model, (c) is from the NIR spectroscopy model, and (d) is from the Raman spectroscopy model. While four or five principal components were extracted for the PCA models, only two axes can be visualized simultaneously. As PCA was used for exploratory data analysis, Figure 4 shows the two components on which the various batches and their classifications can be most easily distinguished from one another visually. The Hotelling's T 2 ellipse, drawn in black in Figure 4, is the multivariate analogue of univariate confidence intervals. For all four spectral technologies, it can be seen that, at the 95% confidence level, none of batches' projections fall outside of the ellipse; therefore, none of the batches were excluded from subsequent analysis.
Figure 4.
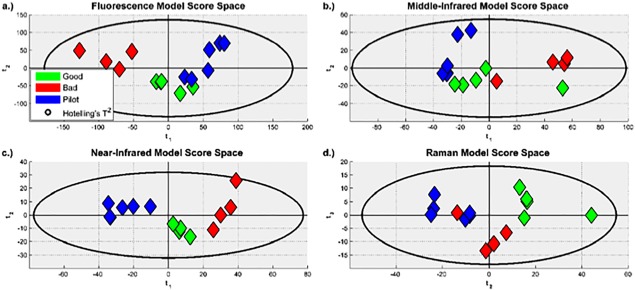
Score spaces for the PCA models built from the preprocessed: (a) Fluorescence spectra. (b) MIR spectra. (c) NIR spectra. One lot is missing because of the spectra being measured at the wrong resolution. (d) Raman spectra. It can be seen that there were no outliers; and that, for all four models, the projections are similar for batches with the same classification.
The score space for the model built from NIR spectra shows the most separation between groups, followed closely by the model built from fluorescence spectra.
The PCA model statistics are summarized in Table 1. A is the number of principal components that were extracted, as determined by 7‐fold cross‐validation; R 2(X) describes the fraction of variance present in the spectra that is explained by the A principal components and Q 2 describes the predictive fraction of variance present in the data that is explained by the A principal components. R 2 and Q 2 values of one are optimal, but are not seen in practice. The R 2(X) values seen in Table 1, which are larger than 0.98 for all four models, indicate that the multivariate data analysis techniques employed in this study accurately describe the data generated by all four of the spectral technologies under consideration. Furthermore, the Q 2 values, which are larger than 0.92 for all four models, indicate that the models will be useful for the assessment of spectra generated in the future.
Table 1.
Model Statistics for All Four PCA Models
| A | R2(X) | Q2 | |
|---|---|---|---|
| Fluorescence | 5 | 0.986 | 0.964 |
| Middle Infrared | 4 | 0.990 | 0.974 |
| Near Infrared | 4 | 0.970 | 0.926 |
| Raman | 5 | 0.987 | 0.961 |
Regression models
Predicting Product Quality from Spectral Data
Eight PLS models were created to quantitatively predict the CQA values from each spectral technology individually. Four of these models used IVCD as the Y block data, and the remaining four models used IgG titer. For both sets of four models, the X block data consisted of preprocessed spectra from one of the four technologies.
The value of each PLS model, relative to the other models that were built to predict the same CQA, was assessed by examining the predictive power of the models. For each of the four spectral technologies, an observed vs. predicted plot was generated for both IVCD, shown in Figure 5, and IgG titer, shown in Figure 6. It can be seen from Figures 5 and 6 that the CQA values predicted by the PLS models built from the fluorescence and near infrared spectra show good agreement with the CQA values observed experimentally. However, it can also be seen that the PLS models built from the middle infrared and Raman spectra do not.
Figure 5.
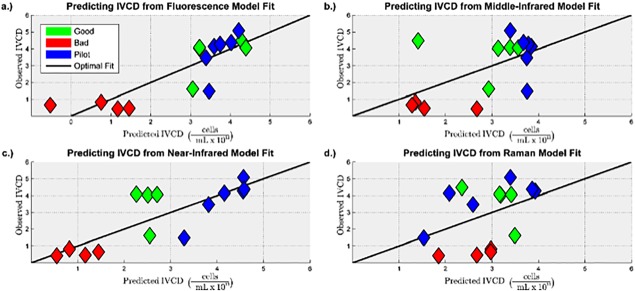
Observed vs. predicted plots for predicting IVCD for the PLS models built from the preprocessed: (a) Fluorescence spectra. (b) MIR spectra. (c) NIR spectra. (d) Raman spectra.
It can be seen that the models built from fluorescence and near infrared spectra have a lower prediction error compared to the models built from middle infrared and Raman spectra.
Figure 6.
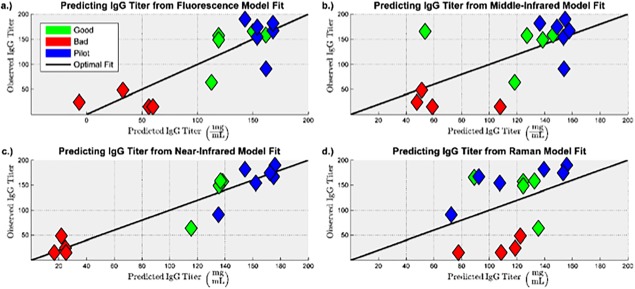
Observed vs. predicted plots for predicting IgG titer for the PLS models built from the preprocessed: (a) Fluorescence spectra. (b) MIR spectra. (c) NIR spectra. (d) Raman spectra.
It can be seen that the models built from fluorescence and near infrared spectra have a lower prediction error compared to the models built from middle infrared and Raman spectra.
The models were also assessed quantitatively from the model statistics given in Table 2. It can be seen in Table 2 that, relative to the PCA models described in Table 1, the inclusion of product quality data in the PLS models reduced the number of principal components that could be extracted before the predictive power of the model started to degrade. Furthermore, by comparing the R 2(X) values in Tables 1 and 2, it can be seen that extracting fewer principal components resulted in a smaller fraction of the variance present in the spectra being described by the PLS models. However, it can be seen that all of the PLS models have R 2(X) values greater than 0.5, which indicates that a majority of the variances present in the spectra were still captured by the PLS models.
Table 2.
Model Statistics for PLS Models that Predict Product Quality from Spectral Data
| Model | Statistics | |||||
|---|---|---|---|---|---|---|
| Y Block | X Block | A | R2(X) | R2(Y) | Q2 | RMSECV |
| IVCD | Fluorescence | 1 | 0.517 | 0.717 | 0.627 | 1.03 × 1011 |
| Middle Infrared | 1 | 0.724 | 0.326 | 0.209 | 1.50 × 1011 | |
| Near Infrared | 1 | 0.768 | 0.665 | 0.641 | 1.01 × 1011 | |
| Raman | 1 | 0.615 | 0.167 | 0.004 | 1.68 × 1011 | |
| IgG Titer | Fluorescence | 1 | 0.566 | 0.710 | 0.655 | 37.4 |
| Middle Infrared | 1 | 0.725 | 0.415 | 0.314 | 52.7 | |
| Near Infrared | 3 | 0.936 | 0.871 | 0.756 | 30.4 | |
| Raman | 1 | 0.615 | 0.150 | −7.40 × 10−5 | 63.6 | |
It is necessary for a model to capture enough of the variance present in the spectra to predict the CQA values, but it is more important for a model to adequately describe the variance present in the CQAs. It can be seen from the R 2(Y) values in Table 2 that, for both IVCD and IgG titer, the models built from the fluorescence and near infrared spectra are able to describe a majority of the variance present in the CQAs, whereas the models built from the middle infrared and Raman spectra are not. Furthermore, the Q 2 values indicate that fluorescence and near infrared spectra generated in the future can be used to predict the CQAs with reasonable accuracy. The cross‐validated root mean square error of prediction (RMSECV) provides a quantitative confirmation of the qualitative conclusions drawn from Figures 5 and 6: the models built from the Raman and middle infrared spectra have 41%–110% larger model prediction error than the models built from the fluorescence and near infrared spectra.
Discussion
Exploratory data analysis
Figure 3 shows that there were batches characterized by high cell growth and high productivity, and that there were batches characterized by low cell growth and low productivity. In practice, cell culture engineers should strive to operate in the upper right quadrant of Figure 3. However, it was important that the training data used to build the regression models include underperforming batches as well, so as to ensure that the models can be used to assess if a future batch will perform poorly.
It can also be seen in Figure 3 that the data points are clustered together in a way that aligns with the classification assigned to each batch by the cell culture operator. The four red data points in the bottom left quadrant of Figure 3 represent batches that are classified as bad. Similarly, the green data points that are clustered together in the top right quadrant of Figure 3 represent the batches that are classified as good, and the cluster of blue data points represent batches from the pilot plant, with some overlap between these two clusters.
The score space projections shown in Figure 4 utilize the same color scheme. It can be seen that the model built from near infrared spectra shows the clearest distinction between the clusters for each of the three classifications, followed closely by the model built from fluorescence spectra. The models built from middle infrared and Raman spectra also show that each of the three classifications are clustered together, but that there is a small degree of overlap between them, which implies that they may not be as useful as the models built from near infrared and fluorescence spectra.
Suggesting that the models built from near infrared and fluorescence spectra are more useful may appear to contradict the PCA models' statistics given in Table 1, where the models built from middle infrared and Raman spectra have the largest R 2(X) and Q 2 values. However, each of the four spectral technologies describe various aspects of the wheat hydrolysate's chemical structure, and the aspects described by one technology are not necessarily the same as the aspects described by another. Therefore, even though the inter‐batch variability that was seen in the wheat hydrolysate's chemical structure is better described by the models built from middle infrared and Raman spectra; the CQAs are more related to the aspects of the chemical structure that are described by the models built from near infrared and fluorescence spectra.
Regression models
Predicting Product Quality from Spectral Data
The PLS model statistics shown in Table 2 also validate the conclusions drawn from the PCA models. The R 2(Y) values for the models built from the middle infrared and Raman spectra are significantly lower than the values for the models built from the near infrared and fluorescence spectra. They indicate that, at best, 42% and 33% of the variability that was seen in the IgG titer and IVCD values, respectively, had any relationship with the aspects of the wheat hydrolysate's chemical structure that are described by the middle infrared spectra. They also indicate that, at best, 17% and 15% of the variability that is seen in the IgG titer and IVCD values, respectively, had any relationship with the aspects described by the Raman spectra.
Impact of Wheat Hydrolysate Composition on Product Quality
Thus far, it has been shown that the CQAs are correlated with the spectra generated from the wheat hydrolysates. In order for this to be true, specific components in the wheat hydrolysate must have impacted the cell culture performance. As amino acid and trace element concentrations are known to have an impact on cell growth and productivity, the amino acid and trace element profiles were measured for the wheat hydrolysates that were added to each batch.
Four new PLS models were created to establish the relationship between the CQAs and the amino acid and trace element profiles of the wheat hydrolysates. The results from these models are summarized in Table 3. The large R 2(X) values indicate that the amino acid and trace element profiles are described well by the PLS models; and the large R 2(Y) values indicate that the IVCD and IgG titer profiles are also well described by the models. The Q 2 values indicate that up to 64% and 58% of the variability that is seen in the IgG titer and IVCD measurements, respectively, was caused by the variations in the amino acid concentrations of the wheat hydrolysates. They also indicate that up to 45% and 39% of the variability that is seen in the IgG titer and IVCD measurements, respectively, was caused by variations in the trace element profiles. The RMSECV values indicate that the amino acid profile had a more consistently measureable effect on the CQAs than the trace element profile did.
Table 3.
Model Statistics for PLS Models that Predict Product Quality from Hydrolyste Composition
| Model | Statistics | |||||
|---|---|---|---|---|---|---|
| Y Block | X Block | A | R2(X) | R2(Y) | Q2 | RMSECV |
| IVCD | Amino Acids | 2 | 0.788 | 0.722 | 0.577 | 1.09 × 1011 |
| Trace Elements | 2 | 0.708 | 0.707 | 0.394 | 1.32 × 1011 | |
| IgG Titer | Amino Acids | 2 | 0.764 | 0.785 | 0.640 | 38.18 |
| Trace Elements | 2 | 0.706 | 0.758 | 0.445 | 48.40 | |
It is important to note that the models only imply that there is a correlation between the CQAs and the amino acid and trace element profiles. However, it is known that the relationship is causal due to the CQAs indirect dependence on the raw materials via cellular metabolism.
The most important aspect of determining the effect that the wheat hydrolysate composition had on the product quality is identifying the specific amino acids and trace elements that had a large, statistically significant effect on the CQAs. A given component is statistically significant when its effect is distinguishable from noise in measurement, which is concluded at the 5% significance level when the component's model coefficient's 95% confidence interval does not contain 0. Furthermore, the model coefficient's sign indicates whether a given component had a positive, or negative, effect on the CQAs. The size of the effect that a given component had on the CQAs is determined from the variable importance in projection (VIP) metric, where a larger VIP value indicates a larger effect.
As two principal components were extracted for each of the PLS models, a component is considered to be statistically significant only when it's model coefficients 95% confidence interval doesn't contain zero for both principal components one and two. Furthermore, only the components whose VIP values are larger than one were taken to have had a large enough effect for further consideration. These requirements may have been more strict than necessary; resulting in some amino acids or trace elements being excluded from the subsequent analysis that should possibly have been included. However, they ensured that only the most consistently significant components, with the largest effects, would be used to justify the spectral model results.
Each of the components' VIP values and model coefficients, with their respective 95% confidence intervals, are shown in Figure 7 for the models built to predict IVCD. The model coefficients that are statistically significant, and the VIP values that are greater than one, are all colored in green; the model coefficients that are not statistically significant, and the VIP values that are less than one, are colored in red. It can be seen that there are five amino acids present in the wheat hydrolysates that meet the requirements for additional consideration, all of which had a negative impact on IVCD. In order of decreasing importance, they are: arginine, phenylalanine, tyrosine, isoleucine and threonine. It can also be seen in Figure 7 that there are three trace elements that had a positive effect on IVCD that should be considered further: copper, phosphorus and molybdenum; and that there are two trace elements that had a negative effect on IVCD that should be considered further: arsenic and aluminum.
Figure 7.
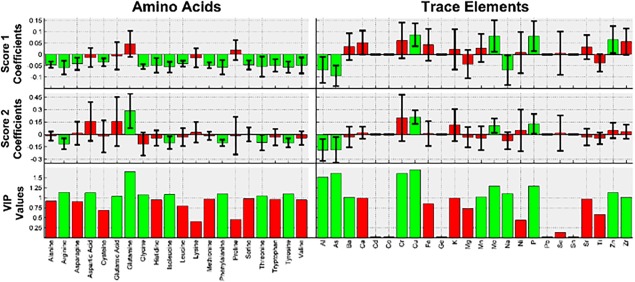
Amino acid and trace element VIP values and model coefficients, with their respective 95% confidence intervals, for the model predicting IVCD.
It can be seen that arginine, phenylalanine, tyrosine, isoleucine, threonine, copper, phosphorus, molybdenum, arsenic and aluminum had a large, statistically significant effect on cell growth.
The same data is presented in Figure 8 for the models built to predict IgG titer. Out of all of the components, five amino acids that had a negative impact on IgG titer are kept: glycine, arginine, tyrosine, isoleucine and phenylalanine; three trace elements that had a positive effect are kept: copper, molybdenum, and phosphorus; and two trace elements had a negative effect are kept: arsenic and aluminum. All three groups are listed in order of decreasing importance.
Figure 8.
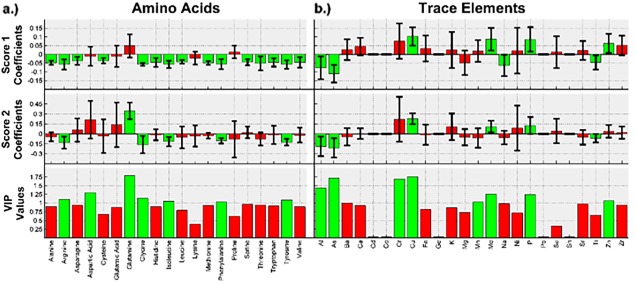
Amino acid and trace element VIP values and model coefficients, with their respective 95% confidence intervals, for the model predicting IgG titer.
It can be seen that glycine, arginine, tyrosine, isoleucine, phenylalanine, copper, molybdenum, phosphorus, arsenic and aluminum had a large, statistically significant impact on cell productivity.
Justification of the Results for Predicting Product Quality from Spectral Data
It was previously claimed that in order for the correlation that was found to exist between the CQAs and the spectra generated from the wheat hydrolysates to be valid, specific components in the wheat hydrolysate must have had an impact on the product quality. While left unstated thus far, it is also necessary that the spectra were able to describe the aspects of the wheat hydrolysate's chemical structure relevant to the components that had an impact. Several amino acids and trace elements that had a large, statistically significant effect on the critical quality attributes were identified; therefore, the spectral technology that is able to best quantify these components should also be the spectral technology that is able to predict the CQAs the most accurately.
Four new PLS models were built to evaluate the relative capability of each spectral technology to characterize the amino acids that were demonstrated to have impacted the CQAs. The results from these four models are presented in Table 4. However, as the model built from the fluorescence spectra extracted 11 principal components from a dataset that contained 15 spectra, the probability of the model being over‐fit was high. The model was confirmed to be over‐fit from a permutations plot, which is given in Supporting Information Figure S4(a). Therefore, a second model was built from the fluorescence spectra that did not use cross‐validation to determine the number of principal components to extract. In this case, two principal components were extracted; and the model is not over‐fit, as evidenced by the permutations plot given in Supporting Information Figure S4(b). The results of this model are also given in Table 4.
Table 4.
Model Statistics for PLS Models that Predict Amino Acid Concentrations from Spectral Data
| Model | Summary | RMSECV | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | R2(X) | R2(Y) | Q2 | Arginine | Glycine | Phenylalanine | Tyrosine | Isoleucine | Threonine | |
| Fluorescence a | 11 | 0.999 | 0.986 | 0.651 | 97.29 | 25.02 | 50.21 | 39.18 | 93.26 | 117.6 |
| Middle Infrared | 2 | 0.953 | 0.220 | 0.030 | 153.9 | 35.26 | 79.73 | 63.51 | 135.9 | 212.9 |
| Near Infrared | 2 | 0.899 | 0.553 | 0.354 | 111.6 | 29.28 | 60.02 | 48.88 | 102.4 | 150.1 |
| Raman | 1 | 0.615 | 0.172 | 0.031 | 154.7 | 34.91 | 77.81 | 61.57 | 130.8 | 209.8 |
| Fluorescence b | 2 | 0.778 | 0.476 | 0.176 | 133.5 | 33.99 | 68.65 | 54.32 | 125.5 | 156.0 |
Model is over‐fit.
A is determined from permutations plot in order to ensure model reproducibility.
It can be seen from Table 4 that the models built from the spectra of all four technologies are able to describe a majority of the variances that were present in the spectra. However, only the models built from the near infrared and fluorescence spectra are able to describe a large amount of the variances seen in the amino acid profiles. Furthermore, the RMSECV is lowest for the models built from the near infrared and fluorescence spectra, which indicates that these two technologies are superior when characterizing the amino acid profile of the wheat hydrolysate.
Another four PLS models were built to determine the relative performance of each spectral technology for characterizing the trace elements that were demonstrated to affect the CQAs. The results from these four models are presented in Table 5. The model built from the near infrared spectra is over‐fit, as evidenced by the permutations plot in Supporting Information Figure S5(a). Therefore, a second model was built from the near infrared spectra that extracted two principal components; and the model is not over‐fit, as evidence by the permutations plot in Supporting Information Figure S5(b). The results of this model are given in Table 5.
Table 5.
Model Statistics for PLS Models that Predict Trace Element Concentrations from Spectral Data
| Model | Summary | RMSECV | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A | R2(X) | R2(Y) | Q2 | Copper | Phosphorous | Molybdenum | Arsenic | Aluminum | |
| Fluorescence | 3 | 0.934 | 0.625 | 0.307 | 2.135 | 231.1 | 0.309 | 0.085 | 0.159 |
| Middle Infrared | 2 | 0.953 | 0.313 | 0.061 | 2.781 | 262.8 | 0.367 | 0.090 | 0.182 |
| Near Infrared a | 10 | 0.995 | 0.971 | 0.573 | 1.992 | 172.2 | 0.241 | 0.064 | 0.203 |
| Raman | 3 | 0.967 | 0.659 | 0.405 | 2.918 | 219.6 | 0.255 | 0.163 | 0.225 |
| Near Infrared b | 2 | 0.896 | 0.525 | 0.290 | 2.159 | 191.3 | 0.357 | 0.072 | 0.179 |
Model is over‐fit.
A is determined from permutations plot in order to ensure model reproducibility.
It can be seen that for the models built to predict the trace element profile, the spectra from all four technologies were described well by the model; and that for the models built from all of the spectra, except for the middle infrared spectra, a majority of the trace element profile is described by the model as well. However, it was surprising that the model built from the Raman spectra managed to perform better than the models built from the fluorescence and near infrared spectra. However, it should be noted that the trace element profile was not being characterized directly, as the trace elements do not absorb light at the frequencies that the samples were irradiated with. Rather, the ability of each spectral technology to characterize the trace element profile was more likely due to the effect that the trace elements had on other aspects of the wheat hydrolysate's chemical structure that were able to be characterized, such as the difference in bond lengths present in a glutamic acid molecule and a copper (II) glutamic acid complex.
The very poor performance of the models built to predict the CQAs from the Raman spectra indicates that the aspects of the chemical structure of the wheat hydrolysate that the trace elements had an effect on were not the same aspects that affected the CQAs. Therefore, if it is desirable to assess the impact that the trace element profile of a raw material has on the CQAs in the future, it is suggested to use a spectroscopic technology better suited to measuring the trace elements directly, such as x‐ray spectroscopy.
The overall implication, then, is that the amino acids present in the wheat hydrolysate, and their impact on the CQAs, are the reason, at least in part, that the models built from the near infrared and fluorescence spectra are the most effective for predicting the CQAs from the hydrolysates' spectra alone.
Conclusions
First, it should be emphasized that there is not a single spectroscopic technology that will perfectly characterize a given raw materials chemical structure. Furthermore, the aspects of the chemical structure that are characterized may not always be relevant for every product quality metric under consideration. However, the results presented here have demonstrated that if the goal is to maximize cell growth and productivity, then the most important components present in the wheat hydrolysates used to supplement cell culture media are contained within their amino acid profile. It was also demonstrated that near infrared and fluorescence spectroscopy are the best technologies to use for the characterization of wheat hydrolysates, because of their ability to characterize the amino acid profile. The authors recommend using near infrared spectroscopy because the models built from its spectra performed slightly better than the models built from Fluorescence spectra, despite the amount of data present in a fluorescence spectrum being much larger.
Facilitated by advancements in technologies such as perfusion bioreactors, the biopharmaceutical industry is moving towards continuous manufacturing. As this trend continues, the variability of the raw materials used for cell culture will have a larger and larger impact on the final product quality, relative to the in process variability, due to the process being operated at steady‐state. Therefore, the accurate and reproducible characterization of raw material quality will be of paramount importance to ensure that final product quality will be good, consistently. Furthermore, spectroscopy and chemometrics are already utilized for the real‐time monitoring of biopharmaceutical processes. Therefore, the extension of this well‐established PAT paradigm to characterize raw materials, such as wheat hydrolysates, should be the logical next step, as it will allow companies to rapidly expand the design space of their processes.
Disclaimer
This work reflects the view of the authors and should not be construed to represent official FDA's views or policies.
Conflict of Interest
The authors declare no financial or commercial conflict of interest.
Supporting information
Supporting Figures
Supporting Raw Data
Acknowledgement
The authors gratefully acknowledge scholarship support for this research from the Biopharmaceutical Product and Quality Consortium, University of Massachusetts Lowell. Additional scholarship support was funded by FDA/CDER Critical Path Project 1500. The authors appreciate SAFC for the datasets and the wheat hydrolysate samples used, and Pfizer for the partial funding of this research.
Abbreviations
- CQA
Critical Quality Attributes
- IVCD
Integrated Viable Cell Density
- PAT
Process Analytical Technology
- PCA
Principal Component Analysis
- PLS
Partial Least Squares regressions
- IgG
Immunoglobulin G
- CHO
Chinese Hamster Ovary
- MIR
Middle Infrared
- NIR
Near Infrared
- FT
Fourier Transform
- ANOVA
Analysis of Variance
- RMSECV
cross‐validated Root Mean Square Error of prediction
- VIP
Variable Importance in Projection
References
- 1. Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004; 22:1393–1398. [DOI] [PubMed] [Google Scholar]
- 2. Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, Liu X, Chen W, Xie M, Wang W, Hammond S, Andersen MR, Neff N, Passarelli B, Koh W, Fan HC, Wang J, Gui Y, Lee KH, Betenbaugh MJ, Quake SR, Famili I, Palsson BO, Wang J. The genomic sequence of the Chinese hamster ovary (CH)‐K1 cell line. Nat Biotechnol. 2011; 29:735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jayapal KP, Wlaschin KF, Yap MGS, Hu WS. Recombinant protein therapeutics from CHO cells—20 Years and counting. Chem Eng Prog. 2007; 103:40–47. [Google Scholar]
- 4. Walsh G. Biopharmaceutical benchmarks. Nat Biotechnol. 2010; 28:917–924. [DOI] [PubMed] [Google Scholar]
- 5. Andersen DC, Krummen L. Recombinant protein expression for therapeutic applications. Curr Opin Biotechnol. 2002; 13:117–123. [DOI] [PubMed] [Google Scholar]
- 6. Ozturk SS, Thrift JC, Blackie JD, Naveh D. Real‐time monitoring and control of glucose and lactate concentrations in mammalian cell perfusion reactor. Biotechnol Bioeng. 1997; 53:372–378. [DOI] [PubMed] [Google Scholar]
- 7. Zhang J, Robinson D. Development of animal‐free, protein‐free and chemically‐ defined media for NS0 cell culture. Cytotechnology. 2005; 48:59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van de Valk J, Brunner D, De Smet K, Fex Svenningsen A, Honegger P, Knudsen LE, Lindl T, Noraberg J, Price A, Scarino ML, Gstraunthaler G. Optimization of chemically defined cell culture media—Replacing fetal bovine serum in mammalian in vitro methods. Toxicol In Vitro. 2010; 24:1053–1063. [DOI] [PubMed] [Google Scholar]
- 9. Chun BK, Kim JH, Lee HJ, Chung N. Usability of size‐excluded fractions of soy protein hydrolysates for growth and viability of Chinese hamster ovary cells in protein‐ free suspension culture. Bioresour Technol. 2007; 98:1000–1005. [DOI] [PubMed] [Google Scholar]
- 10. Schroder M, Matischak K, Friedl P. Serum‐ and protein‐free media formulations for the Chinese hamster ovary cell line DUKXB11. J Biotechnol. 2004; 108:279–292. [DOI] [PubMed] [Google Scholar]
- 11. Kallel H, Jouini A, Majoul S, Samia RS. Evaluation of various serum and animal protein‐free media for the production of a veterinary rabies vaccine in BHK‐21 cells. J Biotechnol. 2002; 95:195–204. [DOI] [PubMed] [Google Scholar]
- 12. FDA . 2004. PAT‐A framework for innovative pharmaceutical development, manufacturing and quality assurance. Rockville, MD: U.S. Department of Health and Human Services Food and Drug Administration Center for Biologics Evaluation and Research. [Google Scholar]
- 13.Eriksson L, Johansson E, Kettaneh‐Wold N, Trygg J, Wikstrom C, Wold S. Multi‐ and Megavariate Data Analysis Part 1: Basic Principles and Applications. Umetrics; 2006.
- 14. Boudreau MA, McMillan GK. New Direction in Bioprocess Modeling and Control: Maximizing Process Analytical Technology Benefits. NC, USA: Research Triangle Park; 2006. [Google Scholar]
- 15. Read EK, Park JT, Shah RB, Riely BS, Brorson KA, Rathore AS. Process analytical technology (PAT) for biopharmaceutical products: Part I. Concepts and applications. Biotechnol Bioeng. 2009; 105:276–284. [DOI] [PubMed] [Google Scholar]
- 16. Hakemeyer C, Strauss U, Werz S, Jose G, Folque F. At‐line NIR spectroscopy as effective PAT monitoring technique in Mab cultivations during process development and manufacturing. Talanta. 2012; 90:12–21. [DOI] [PubMed] [Google Scholar]
- 17. Bras LP, Bernardino SA, Lopes JA, Menezes JC. Multiblock PLS as an approach to compare and combine NIR and MIR spectra in calibrations of soy flour. Chemom Intell Lab Syst. 2005; 75:91–99. [Google Scholar]
- 18. Dupuy N, Galtier O, Dreau YL, Oinatel C, Kister J, Artaud J. Chemometric analysis of combined NIR and MIR spectra to characterize French olives. Eur J Lipid Sci Technol. 2010a; 112:463–475. [Google Scholar]
- 19. Peinder P, Visser T, Petrauskas D, Salvatori F, Soulimani F, Weckhuysen B. Partial least squares modeling of combined infrared, 1H NMR and 13C NMR spectra to predict long residue properties of crude oils. Vibrat Spectroscopy. 2009; 51:205–212. [Google Scholar]
- 20. Lee H, Christie A, Xu J, Yoon S. Data fusion‐based assessment of raw materials in mammalian cell culture. Biotechnol Bioeng. 2012; 109:2819–2828. [DOI] [PubMed] [Google Scholar]
- 21. Luo Y, Chen G. Combined approach of NMR and chemometrics for screening peptones used in the cell culture medium for the production of a recombinant therapeutic protein. Biotechnol Bioeng 2007; 97:1654–1659. [DOI] [PubMed] [Google Scholar]
- 22. Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007; 70:461–477. [DOI] [PubMed] [Google Scholar]
- 23. Li B, Ryan PW, Ray BH, Leister KJ, Sirimuthu NMS, Ryder AG. Rapid characterization and quality control of complex cell culture media solution using Raman Spectroscopy and chemometrics. Biotechnol Bioeng. 2010; 107:290–301. [DOI] [PubMed] [Google Scholar]
- 24. Herrero AM, Jimenez‐Colmenero F, Carmona P. Elucidation of structural changes in soy protein isolate upon heating by Raman spectroscopy. Int J Food Sci Technol. 2009; 44:711–717. [Google Scholar]
- 25. Hashimoto A, Yamanaka A, Kanou M, Nakanishi K, Kameoka T. Simple and rapid determination of metabolite content in plant cell culture medium using and FT‐IR/ATR method. Bioprocess Biosyst Eng. 2005; 27:115–123. [DOI] [PubMed] [Google Scholar]
- 26. Roychoudhury P, Harvey L, McNeil B. The potential of mid infrared spectroscopy (MIRS) for real time bioprocess monitoring. Anal. Chim. Acta. 2006; 571:159–166. [DOI] [PubMed] [Google Scholar]
- 27. Benoudjit N, Melgani F, Bouzgou H. Multiple regression systems for spectrophotometric data analysis. Chemom Intell Lab Syst. 2009; 95:144–149. [Google Scholar]
- 28. Rhiel M, Cohen M, Murhammer D, Arnold M. Nondestructive near‐infrared spectroscopic measurement of multiple analytes in undiluted samples of serum‐ based cell culture media. Biotechnol. Bioeng. 2002; 77:73–82. [DOI] [PubMed] [Google Scholar]
- 29. Rhiel M, Ducommun P, Bolzonella I, Marison I, von Stockar U. Real‐time in situ monitoring of freely suspended and immobilized cell cultures based on mid‐infrared spectroscopic measurements. Biotechnol. Bioeng. 2002; 77:174–185. [DOI] [PubMed] [Google Scholar]
- 30. Ryan PW, Li B, Shanahan M, Leister J, Ryder AG. Prediction of cell culture media performance using fluorescence spectroscopy. Anal Chem 2010; 82:1311–1317. [DOI] [PubMed] [Google Scholar]
- 31. Jose GE, Folque F, Menezes JC, Werz S, Strauss U, Hakemeyer C. Predicting Mab product yields from cultivation media components, using NIR and 2D‐fluorescence spectroscopies. Biotechnol. Prog 2011; 27: [DOI] [PubMed] [Google Scholar]
- 32. T Hill, S Holdread, C Hunt, K Hammett, K Chaturvedi, J Brooks. 2008. Process Optimization Using Hydrolysates: Case Studies Demonstrating Increased Performance with Hydrolysate Blends and Feed Strategies. Presented at BioProcess International Conference and Exhibition: IBC Antibody Development and Production, San Diego, CA.
- 33. G Nyberg, R Balcarcel, B Follstad, Stephanopoulos, D Wang. Metabolism of peptide amino acids by Chinese Hamster ovary cells grown in a complex medium. Biotechnol. Bioeng. 2000; 62:324–335. [PubMed] [Google Scholar]
- 34. F Franek, O Hohenwarter, H Katinger. Plant protein hydrolysates: Preparation of defined peptide fractions promoting growth and production in animal cells cultures. Biotechnol. Prog. 2008; 16:688–692. [DOI] [PubMed] [Google Scholar]
- 35. Y Huang, W Hu, Eddie Rustandi, K Chang, H Yusuf‐Makagiansar, T Ryll. Maximizing productivity of CHO cell‐based fed‐batch culture using chemically defined media conditions and typical manufacturing equipment. Biotechnol. Prog. 2010; 26:1400–1410. [DOI] [PubMed] [Google Scholar]
- 36.MATLAB. “Msalign.” Mathworks.com. Mathworks , 2013. Web. <http://www.mathworks.com/help/bioinfo/ref/msalign.html>.
- 37. Bahram M, Bro R, Stedmon C, Afkhami A. Handling of Rayleigh and Raman scatter for PARAFAC modeling of fluorescence data using interpolation. J Chemom. 2006; 20:99–105. [Google Scholar]
- 38. Gil DB, de la Pena AM, Arancibia JA, Escandar GM, Olivieri AC. Second‐order advantage achieved by unfolded‐partial least‐squares/residual bilinearization modeling of excitation‐emission fluorescence data presenting inner filter effects. Anal Chem. 2006; 2006:8051–8058. [DOI] [PubMed] [Google Scholar]
- 39. Isaksson T, Naes T. The effect of multiplicative scatter correction (MSC) and linearity improvement in NIR spectroscopy. Appl Spectrosc. 1988; 42:1273–1284. [Google Scholar]
- 40. Savitzky A, Golay M. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964; 36:1627–1639. [Google Scholar]
- 41.MATLAB. “Baselinew.” Eigenvector.com. Eigenvector, 2013. Web. <http://wiki.eigenvector.com/index.php?title=Baselinew>.
- 42. Lomax, R. "One‐Factor Analysis of Variance: Fixed‐Effects Model.” Statistical Concepts: A Second Course. Mahwah, NJ: Lawrence Erlbaum Associates, 2007. 1–46. Print. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figures
Supporting Raw Data


