Abstract
Despite the large number of noncoding RNAs and their importance in several biological processes, our understanding of RNA structure and dynamics at atomic resolution is still very limited. Like many other RNAs, the Neurospora Varkud satellite (VS) ribozyme performs its functions through dynamic exchange of multiple conformational states. More specifically, the VS ribozyme recognizes and cleaves its stem‐loop substrate via a mechanism that involves several structural transitions within its stem‐loop substrate. The recent publications of high‐resolution structures of the VS ribozyme, obtained by NMR spectroscopy and X‐ray crystallography, offer an opportunity to integrate the data and closely examine the structural and dynamic properties of this model RNA system. Notably, these investigations provide a valuable example of the divide‐and‐conquer strategy for structural and dynamic characterization of a large RNA, based on NMR structures of several individual subdomains. The success of this divide‐and‐conquer approach reflects the modularity of RNA architecture and the great care taken in identifying the independently‐folding modules. Together with previous biochemical and biophysical characterizations, the recent NMR and X‐ray studies provide a coherent picture into how the VS ribozyme recognizes its stem‐loop substrate. Such in‐depth characterization of this RNA enzyme will serve as a model for future structural and engineering studies of dynamic RNAs and may be particularly useful in planning divide‐and‐conquer investigations. WIREs RNA 2017, 8:e1421. doi: 10.1002/wrna.1421
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
Noncoding RNAs form a rich and extremely diverse class of biomolecules found in all domains of life that play critical roles in many biological processes. Although our structural and functional understanding of these RNAs is rapidly growing, there are still very few high‐resolution structures of RNA available in comparison with the large number of noncoding RNAs in biology. An important part of what we know at this time comes from the investigations of ribozymes, RNAs that catalyze specific biochemical reactions including phosphoryl transfer and peptide bond formation. Several ribozymes have been shown to function in the absence of proteins and represent ideal model systems for noncoding RNAs due to their relatively small size and easily detectable biological activity. As these ribozymes adopt diverse structures and function through distinct mechanisms, previous investigations of ribozymes have given us a broad perspective on the structural diversity and dynamic nature of RNA. The case of the Neurospora VS ribozyme is noteworthy because of its unique structure and mode of substrate recognition.
The VS ribozyme was discovered more than 25 years ago as part of the Varkud satellite (VS) RNA, a mitochondrial RNA found in certain isolates of the Neurospora filamentous fungi.1, 2 Its self‐cleavage and self‐ligation activities are essential for the replication cycle of the VS RNA, for which no other specific function has been assigned yet. In vitro, a contiguous sequence of approximately 160 nucleotides (nt) containing six helical regions (I–VI) was found sufficient for self‐cleavage (Figure 1),3, 5 and extending this sequence with an additional helical domain (VII) was found important for both efficient self‐ligation and trans cleavage within a dimeric form.6, 7 The cleavage reaction yields products with 2′,3′‐cyclic phosphate and 5′‐OH termini, as also found for other small nucleolytic ribozymes, like the hairpin, hammerhead, and glmS ribozymes.8, 9, 10 A significant number of biochemical, kinetic, and biophysical investigations have allowed extensive functional and mutational characterization of the VS ribozyme11, 12, 13, 14, 15 and have provided low‐resolution three‐dimensional models (Table 1),22, 27, 29, 31, 32, 33 revealing primary, secondary, and tertiary structures that are unique among known ribozymes. They also support a general acid–base mechanism similar to that of the hairpin ribozyme. Both ribozymes require the intimate association of two internal loops to form the active site where two nucleobases, one from each internal loop, play key roles in the catalytic mechanism.13, 14 A distinctive feature of the VS ribozyme is that one internal loop is located within the catalytic domain (the A756 loop also known as the A730 loop), whereas the other one (the G638 loop) belongs exclusively to its stem‐loop substrate, such that the cleavage mechanism can be viewed as substrate‐assisted catalysis. In the proposed cleavage mechanism, G638 acts as the general base that activates the 2′‐oxygen nucleophile, whereas A756 acts as the general acid that donates a proton to the 5′‐oxygen leaving group.35 Reported pH‐activity profiles of the cleavage reaction fit a double‐ionization model with shifted pKa values of 5.2–5.8 and 8.3–8.4 attributed to A756 and G638, respectively.35, 36, 37, 38 Moreover, the metal dependence of these shifted pKa values point to an indirect role for metal ions in the reaction chemistry.38 Efficient trans cleavage of the stem‐loop substrate is made possible through a complex mechanism for substrate recognition (Figure 1), which involves formation of a magnesium‐dependent kissing‐loop interaction (KLI), an activating conformational change from an unshifted to a shifted state, and intimate association of the G638 loop with the A756 loop to form the active site.4, 39, 40, 41
Figure 1.
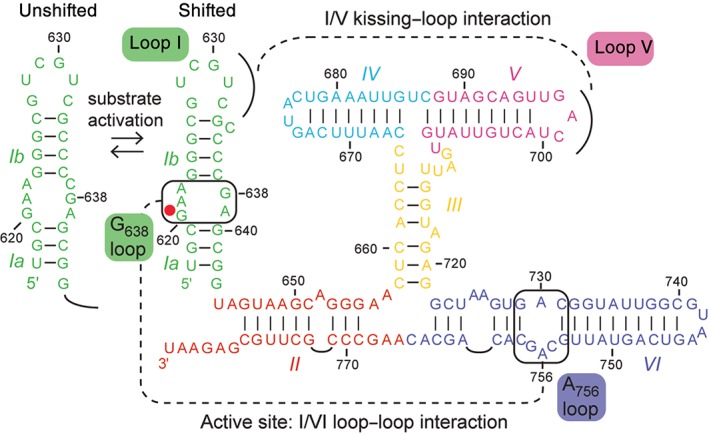
The Neurospora VS ribozyme. Primary and secondary structures of the natural cis‐cleaving VS ribozyme (residues 617–783) determined from mutagenesis and chemical probing studies.3, 4 Recognition of the substrate (stem‐loop I or SLI) involves a KLI with stem‐loop V (SLV), an activating conformation change (left) and a loop–loop interaction between the G638 and A756 internal loops to form the active site. Helical domains are color‐coded and identified with roman numerals (I–VI). The scissile phosphate is depicted by a red dot.
Table 1.
List of Three‐Dimensional Models and Structures Reported for VS Ribozyme
| Subdomain | Experimental Approach1 | PDB Code | Divalent Metal Ions in Buffer2 | Metal Ions in Structure | Reference |
|---|---|---|---|---|---|
| SLI unshifted | NMR | 1E4P | None | 16 | |
| NMR | 1HWQ | None | 17 | ||
| SLI shifted | NMR | 1OW9 | None | 18 | |
| SLV | NMR | 1TBK | None | 19 | |
| NMR | 1YN1 | ≥20 mM Mg2+ | 20 | ||
| NMR | 1YN2 | ≥20 mM Mg2+ and 20 μM Mn2+ | 4 Mn(H2O)6 2+ | 20 | |
| MD | None; 40 mM Mg2+ | 21 | |||
| I/V KLI | CP/MI/SDM | 0–15 mM Mg2+ | 22 | ||
| NMR | 2MI0 | 5 mM Mg2+ | 23 | ||
| A756 loop | NMR | 1TJZ | None | 24 | |
| NMR | 2L5Z | 5 mM Mg2+ | 25 | ||
| NMR | 2MIS | 5 mM Mg2+ and 10 μM Mn2+;4.75 mM Mg2+ and 0.25 mM Cd2+ |
3 Mg(H2O)6
2+
2 Mg(H2O)5 2+ |
26 | |
| III–IV–V junction | FRET/CGE | 3 mM Mg2+ | 27 | ||
| NMR | 2MTJ | 5 mM Mg2+ | 28 | ||
| NMR | 2MTK | 5 mM Mg2+ and 10 μM Mn2+ |
5 Mg(H2O)6
2+
1 Mg(H2O)5 2+ |
28 | |
| II–III–VI junction | FRET | 2 mM Mg2+ | 29 | ||
| NMR | 2N3Q | 5 mM Mg2+ | 30 | ||
| NMR | 2N3R | 5 mM Mg2+ and 10 μM Mn2+ | 8 Mg(H2O)6 2+ | 30 | |
| VS ribozyme (cis or trans form) | CP/MI/SDM | 0–20 mM Mg2+ | 31 | ||
| FRET/CGE | 2–3 mM Mg2+ | 27, 32 | |||
| smFRET | 35 mM Mg2+ | 32 | |||
| SAXS | 10 mM Mg2+ | 33 | |||
| NMR | 5 mM Mg2+ | 30 | |||
| X‐ray (A756G) | 4R4P | 5 mM Mg2+ | 2 Mg2+ | 34 | |
| X‐ray (G638A) | 4R4V | 5 mM Mg2+ | 9 Mg2+, 5 K+ | 34 |
The experimental approaches listed here are NMR spectroscopy (NMR), molecular dynamic simulations (MD), Föster resonance energy transfer (FRET), comparative gel electrophoresis (CGE), single‐molecule FRET (smFRET), chemical probing (CP), modification interference (MI), site‐directed mutagenesis (SDM), small‐angle X‐ray scattering (SAXS), and X‐ray crystallography (X‐ray). Only NMR and X‐ray studies have yielded high‐resolution structures reported in the pdb.
In terms of experimental conditions, only the concentration and type of divalent metal ions used for structure determination is provided (none = none used).
Despite the wealth of knowledge accumulated over the years on the VS ribozyme, it is only very recently that high‐resolution three‐dimensional structures have become available to provide detailed structural and dynamic insights into its mechanism (Table 1). Both a solution‐state NMR model30 and two crystal structures34 were reported recently that independently represent significant achievements given the inherently dynamic nature of this ribozyme and its tendency to form dimers and higher‐order multimers in solution. The NMR model of a trans ribozyme in complex with its stem‐loop substrate results from a divide‐and‐conquer strategy for structural characterization based on NMR structures of several individual subdomains. Two crystal structures are available for cis ribozyme variants that each form a dimer in which the substrate of one protomer interacts with the catalytic domain of the other protomer. The NMR model represents an open state of the VS ribozyme prior to formation of the active site, and complementary NMR studies have provided information on important conformational changes associated with substrate recognition and activation. However, the crystal structures capture a closed state in which the active site is formed but cleavage has not taken place, thus providing insights into the catalytic mechanism. Herein, we summarize these recent high‐resolution structural studies, highlighting the similarities and differences between the NMR and crystal structures to develop an integrated understanding of the structural and dynamic properties of this ribozyme.
A DIVIDE‐AND‐CONQUER NMR STUDY PROVIDES DETAILED STRUCTURAL AND DYNAMIC INSIGHTS
The divide‐and‐conquer approach represents a simple yet powerful strategy to structurally investigate large and dynamic biomolecular systems. It consists of determining high‐resolution structures of smaller and often more amenable subdomains to eventually reconstitute the structure of the complete biomolecule. Its recent application to characterize the complete VS ribozyme structure by solution NMR spectroscopy represents a proof‐of‐concept that this strategy can be used successfully to investigate the structure and dynamics of large functional RNAs. Prior biochemical and biophysical investigations of the VS ribozyme were instrumental in guiding the efforts. A particular challenge was that the limited number of known natural variants of the VS ribozyme did not provide sufficient sequence diversity to define the secondary structure based on covariation analysis as would normally be done for an RNA of interest. Instead, the secondary structure was delineated from careful biochemical characterization based on site‐directed mutagenesis and chemical probing studies.3 Complementary investigations helped distinguish the subdomains that are functionally required [the KLI between stem‐loop I (SLI) and stem‐loop V (SLV), the internal loops of SLI and stem‐loop VI (SLVI), the three adenosine bulges, and the two three‐way junctions] from those that are not (junction‐distal ends of stems II, IV, and VI).42, 43 They also helped delineate independent subdomains that when taken out of their natural context would remain meaningful to our structural understanding of the VS ribozyme.3, 5, 40
NMR Studies of Isolated Subdomains Relevant to Substrate Recognition
The KLI between the terminal loops of SLI and SLV is important for substrate recognition by the catalytic domain.39 Prior investigations revealed that this interaction involves three Watson–Crick (WC) base pairs and is associated with a conformational rearrangement that can be rate‐limiting, because it activates the substrate for catalysis (Figure 1).4, 39, 40, 42, 44, 45 In its free form, the substrate adopts an unshifted conformation, with a symmetric internal loop of six nt. In the bound form, the substrate adopts a shifted conformation that excludes C634 from stem Ib and reconfigures the cleavage site internal loop to an asymmetric loop of five nt.4, 40 Importantly, formation of the KLI and the associated conformation change in SLI can be reproduced when the catalytic domain is replaced by an isolated SLV, validating biophysical investigations using isolated stem‐loops.40
Unshifted and Shifted States of G638 Loop
NMR structures of the SLI substrate were determined to characterize both the unshifted (inactive) and shifted (active) states of the substrate cleavage site internal loop, termed here the G638 loop.16, 17, 18 Two individual structures were determined for the unshifted state, one of which is illustrated in Figure 2(a).16, 17 In both studies, the RNA stem‐loop used comprises the natural internal loop sequence and the adjacent base pairs from stems Ia and Ib. Both NMR structures were determined at acidic pH (pH = 5.0–6.0) and revealed the same structural elements in the G638 loop: two trans Hoogsteen/sugar edge (H/SE) G–A base pairs and a cis WC/WC A(+)•C base pair involving A622. From pH‐dependent NMR studies, pKa values of 6.1–6.4 were determined for A622, and its protonated state was found critical for stabilizing the three noncanonical base pairs of the unshifted internal loop. At higher pH, the G638 loop adopts a more open conformation that may facilitate the displacement of residue C637 into stem Ib as part of the conformational change associated with formation of the I/V KLI.17
Figure 2.
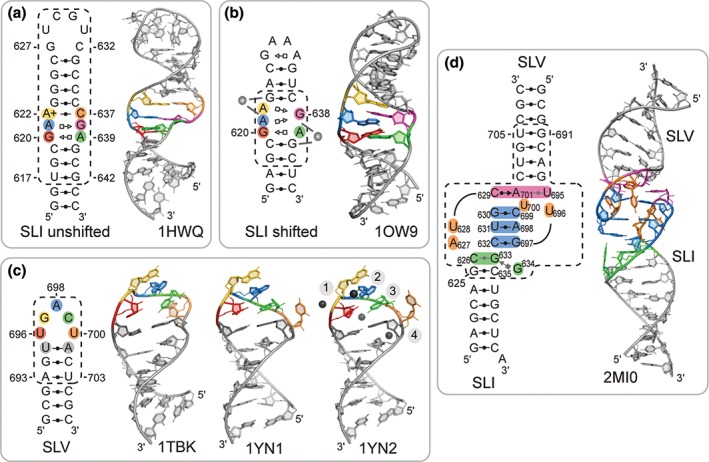
NMR structures of VS ribozyme subdomains determined to characterize substrate recognition and activation. (a) NMR structure of an SLI substrate with the G638 loop in the unshifted conformation (PDB code 1HWQ).17 (b) NMR structure of an SLI substrate with the G638 loop in the shifted conformation (PDB code 1OW9).18 The gray spheres on the structural schematics illustrate Mg2+‐binding sites near the 5′‐AG‐3′ strand of a 5′‐AG‐3′/5′‐CG‐3′ motif that are consistent with NMR chemical shift perturbations. (Adapted with permission from Ref 18. Copyright 2003 National Academy of Sciences). (c) NMR structures of the SLV subdomain of the VS ribozyme obtained in the absence of Mg2+ ions (PDB code 1TBK)19 and in the presence of Mg2+ ions (PDB code 1YN1 and 1YN2).20 The NMR structure with the PDB code 1YN2 was determined with RNA‐metal restraints to localize hexahydrated Mn2+ ions (black spheres). (Adapted with permission. Copyrights 2005 and 2006 American Chemical Society). (d) NMR structure of an SLI/SLV complex (PDB code 2MI0).23 In panels (a–d), a structural schematic is available alongside a cartoon representation of the NMR structure to illustrate the sequence and base‐pairing interactions present in the structure(s): the dashed boxes enclose the sequences derived from the VS ribozyme; specific residues are colored as in the NMR structure representations; and base pairs are depicted using the Leontis–Westhof notation46, 47 in either black (≥2 hydrogen bonds) or gray (1 hydrogen bond).
The structure of the shifted conformation of the G638 loop was also determined by NMR, as part of an RNA stem‐loop with a nonnatural stem Ib and a GNRA tetraloop that stabilize the shifted state (Figure 2(b)).18 As expected, this RNA was cleaved when incubated with a trans VS ribozyme, although at a lower rate than a wild‐type sequence given that the I/V KLI could not form due to the terminal loop modification.18 The NMR structure revealed that residues G620 and A639 form a trans H/SE G–A base pair, as found for the unshifted state (Figure 2(a) and (b)).16, 17, 18 However, residue G638 is positioned such that it can participate in a trans H/SE G–A base pair with either A621 or A622 (Figure 2(b)). Importantly, G638 is stacked on G620 with its WC edge close to the 2′‐OH of G620, in agreement with its role as general base in the cleavage reaction. Two Mg2+‐binding sites were identified in the internal loop based on chemical shift perturbations (Figure 2(b)). A detailed comparison between the NMR structures of the unshifted and shifted states of the G638 loop revealed two distinct features of the shifted state that could be important for catalysis: a Mg2+‐binding site near A622 and G623 (Figure 2(b)) and strong structural similarity with a tertiary interaction motif found in rRNA.18 In addition, a pKa value of 4.0 was estimated for residue A622 in this shifted conformation of the G638 loop, confirming that the shifted state is stable at neutral pH. Thus, NMR studies established the pH dependence of the equilibrium between the unshifted and shifted states of the SLI substrate and identify structural differences that could be important for its function.
Free Conformations of SLI and SLV Terminal Loops
U‐turn folds were initially proposed to form in the terminal loop of both SLI and SLV on the basis of their sequence, mutagenesis studies, and chemical probing experiments.39 Subsequent NMR structural studies of isolated stem‐loops have allowed in‐depth characterization of these terminal loops in their free form prior to formation of the I/V KLI.17, 19, 20, 23 NMR studies of the natural terminal loop of SLI in the absence of Mg2+ ions do not provide evidence for the proposed U‐turn fold and the adjacent G627–C632 base pair, but rather indicate that the structure of the terminal loop is mostly disordered (Figure 2(a), 1HWQ).17 Addition of Mg2+ ions did not bring significant changes in the NMR spectra. Thus, the SLI terminal loop is disordered prior to the formation of the I/V KLI both in the absence and presence of Mg2+ ions.
NMR structures of an isolated SLV RNA were determined both in the absence (Figure 2(c), 1TBK) and presence of Mg2+ ions (Figure 2(c), 1YN1 and 1YN2).19, 20 In these structures, residues U696, G697, and A698 adopt a U‐turn fold while residues U695 and A701 form a canonical base pair that closes the terminal loop. In the presence of Mg2+ ions, the terminal loop adopts a more compact structure with more characteristics of the canonical U‐turn fold and with U700 being completely excluded from the loop fold (Figure 2(c), 1YN1 and 1YN2).20 Molecular dynamic simulations support the observation that the U‐turn of SLV adopts a more compact fold in the presence of Mg2+ ions, and indicate that structural characteristics of this compact fold are already present in the structural ensemble obtained in the absence of Mg2+ ions.21 Four divalent metal‐binding sites were localized in the SLV terminal loop based on Mn2+‐induced paramagnetic relaxation enhancement (PRE) studies (Figure 2(c), 1YN2).20 Three of them (sites 1–3) correspond to divalent metal‐binding sites previously identified in crystal structures of U‐turns, and the fourth one (site 4) is associated with the unusual phosphate backbone fold near U700. As a consequence of the U‐turn fold, the WC edges of residues after the sharp turn in the phosphate backbone (G697, A698, and C699) are exposed and available for tertiary contact.19, 20
Highly Stable KLI Between SLI and SLV
To characterize the KLI, several SLI/SLV complexes were investigated by NMR spectroscopy and isothermal titration calorimetry (ITC), but only one was found suitable for NMR structure determination due to its relatively small size (43 nt), the quality of the NMR data and its high thermodynamic stability (Kd = 0.2–0.4 μM).23, 48 This complex was formed with a double‐stranded SLI variant that lacks the G638 loop, but retains a terminal loop sequence compatible with KLI formation and ribozyme activity (G627A and C634G; Figure 2(d)). The three‐dimensional structure of this SLI/SLV complex confirms formation of a U‐turn fold in each of the two terminal loops and of three canonical cis WC/WC base pairs between these loops (Figure 2(d)). In addition, this NMR structure reveals two base triples: a fairly disordered C626–G633–G634 triple and a more stable C629–A701–U695 triple that contributes a fourth base pair (cis WC/SE C629–A701) between the two loops. The structure displays continuous stacking at the KLI that extends to the adjacent stems and likely contributes to the unusually high stability of this interaction.48 Nevertheless, the interhelical angle between stem I and stem V is quite variable, ranging from 127° to 175° in the ensemble of NMR structures. In addition, there is NMR evidence that several residues of loop I (C626, A627, U628, C632, G633, and G634) and loop V (U695 and U696) undergo dynamic exchange on the µs to ms timescale, suggesting that the large range of interhelical angles in the NMR ensemble may not result from the lack of sufficient NMR restraints but rather from the inherent dynamics of the KLI. In the context of the active ribozyme, some degree of flexibility at the kissing‐loop junction likely facilitates formation of the active site between the G638 and A756 loops.
Structural Evidence of Helix Shifting in SLI
NMR studies were conducted to investigate the conformational rearrangement in SLI upon binding to SLV.23 Briefly, NMR spectra were collected for monitoring the formation of several SLI/SLV complexes in the presence of Mg2+ ions using the same SLV, but several SLI variants referred to as shiftable, preshifted, or double‐stranded. Comparison of imino proton data between the free and SLV‐bound states of these SLI variants provides direct structural evidence for helix shifting in stem Ib upon complex formation with the shiftable SLI variants, but not with the preshifted and the double‐stranded SLI variants. The NMR data also reveal that the loop‐closing base pair observed in the free SLI is destabilized upon complex formation with both the shiftable and preshifted substrates. Complementary ITC studies confirm that the shiftable substrates have a lower affinity for SLV (12.5 μM ≤ Kd ≤ 65 μM) than the preshifted and double‐stranded substrates (0.4 μM ≤ Kd ≤ 0.7 μM), meaning that the helix shift in SLI reduces the stability of the KLI by 2–3 kcal/mol.48
These extensive NMR investigations allowed us to define the conformational changes associated with formation of the I/V KLI in the natural VS ribozyme (Figure 3). Prior to binding, the SLV loop adopts a compact U‐turn fold and the SLI substrate adopts an unshifted conformation with a disordered terminal loop. Formation of the I/V KLI involves only subtle structural changes in SLV, but a disorder‐to‐order transition in loop I. The NMR data are consistent with a model in which the initial contact of this disordered loop with the stable loop V destabilizes the closing C626–G633 base pair while ordering loop I and thereby initiates the helix shift in SLI. This model is consistent with the ability of U‐turns to initiate structural changes in RNA.49 Thus, the high‐energy KLI is likely responsible for triggering the helix shift in SLI and changing the conformation of the G638 loop such that it becomes cleavable.
Figure 3.
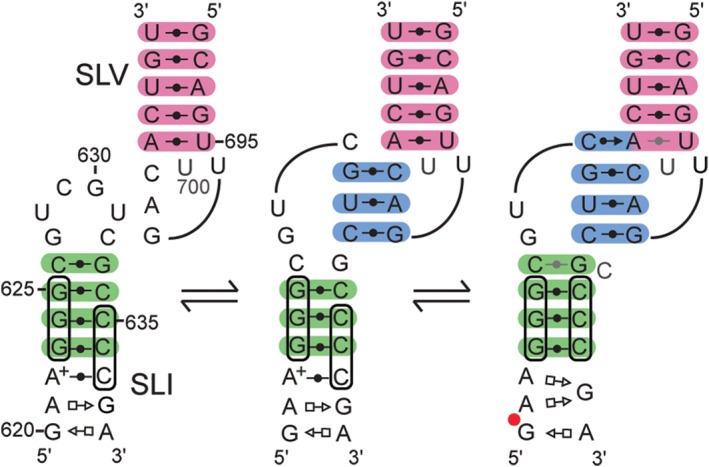
Model for substrate recognition and activation by the VS ribozyme based on NMR data.23 Structural schematics (as in Figure 2) are used to illustrate the unbound states of SLI and SLV (left), a hypothetical state in which the two loops initiate their contact (middle) and a final bound state (right). Base pairs of stem Ib, the KLI, and stem V are shaded in green, cyan, and pink, respectively.
Free Conformation of A756 Loop
For cleavage to occur, the G638 loop must interact with the A756 loop of SLVI. To investigate the conformation of the A756 loop prior to formation of the active site, NMR studies were conducted using a small SLVI subdomain containing the A756 loop and its adjacent base pairs (Figure 4).25 In preliminary NMR studies, it was found that Mg2+ ions are required for stable folding of the A756 loop and thus Mg2+ ions were included in the samples used for structure determination. The NMR structure reveals two main structural elements in the A756 loop: a cis WC/WC G–A base pair between G757 and A730 that enlarges the minor groove and an S‐turn fold involving residues G754–G757. The S‐turn motif generates a distortion of the ribose‐phosphate chain that exposes residues C755 and A756 into the enlarged minor groove with their WC edge facing the solvent. As a result, residue A756 adopts an accessible position that is compatible with its role as general acid in the catalytic mechanism.
Figure 4.
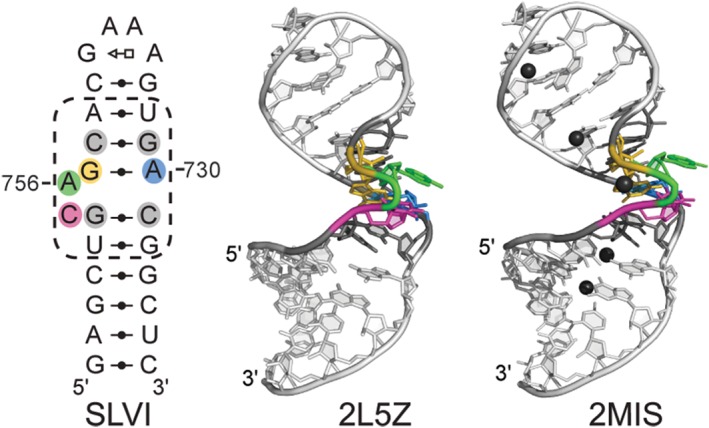
NMR structures of an SLVI subdomain containing the A756 loop. Two NMR structures were obtained in the presence of Mg2+ ions (PDB code 2L5Z and 2MIS).25, 26 The NMR structure with the PDB code 2MIS was determined with RNA‐metal restraints to localize hexahydrated Mn2+ ions (black spheres). A structural schematic (as in Figure 2) is available alongside the cartoon representation of the lowest‐energy NMR structures to provide the sequence and base‐pairing interactions present in both structures. (Adapted from articles that appeared in Oxford University Press25 and ACS publications26).
A dual NMR approach was developed to localize divalent metal ion‐binding sites within this SLVI RNA.26 Mn2+‐induced paramagnetic relaxation effect (PRE) studies were used to probe almost all readily detectable 1H, 13C, and 15N atoms in SLVI, whereas Cd2+‐induced chemical‐shift perturbation (CSP) of RNAs with single phosphorothioate modifications were carried out to specifically probe the contribution of phosphate groups in divalent metal coordination. Using this dual NMR approach, five divalent metal‐binding sites were identified (Figure 4), including one at the phosphate cluster formed by the S‐turn of the A756 loop. Divalent metal ion binding at this site is likely necessary to neutralize the densely‐packed electronegative charges at the S‐turn and may explain the metal‐ion dependence of the A756 loop structure. Extrapolating to the potential role of this metal ion at the active site, it may both stabilize the S‐turn and play an indirect role in the chemistry of the reaction. Thus, NMR studies of the free conformation of the A756 loop provide insights into the formation of the active site and of a potentially important divalent metal ion for catalysis.
NMR Studies of Isolated Subdomains That Define Global Architecture of trans Ribozyme
Architecture of III–IV–V Junction
The high‐resolution NMR structure of a 47‐nt RNA subdomain containing the III–IV–V junction was determined in the presence of Mg2+ ions (Figure 5(a)).28 It shows a family C topology of three‐way junctions, with coaxial stacking of stems III and IV and side‐by‐side arrangement of stems III and V. A U‐turn fold allows backbone reversal between stems III and V and positions the residues after the turn (G711 and A712) to form minor groove base triples and a ribose zipper with stem IV.28 The junction core is stabilized by a continuous stack of four base triples, leading to coaxial stacking of stems III and IV. NMR studies indicate that Mg2+ ions are not essential for folding of the junction core. However, the position of six Mg2+ ions was determined using Mn2+‐induced PRE, revealing two Mg2+ ions within the junction core that likely help stabilize the relative orientation of stems III and V. The NMR structure of the III–IV–V junction provided the first evidence, to our knowledge, of a family C three‐way junction that does not form a long‐range tertiary interaction between the side‐by‐side helices (stems III and V).28, 50 The extensive stack of base triples allows for formation of a stable junction core in the absence of a distal tertiary interaction. In contrast, less stable three‐way junction cores may depend on a distal interaction to stabilize the global folding of the junction, as found for the II–III–VI junction (see below). Given the intrinsic stability of the III–IV–V junction core and lack of a distal tertiary interaction, SLV is free to interact with the SLI substrate to form the I/V KLI. In addition, there is a certain degree of variability in the orientation of SLV with respect to stems III and IV in the NMR ensemble, which is consistent with the III–IV–V junction forming a slightly dynamic hinge. Thus, the III–IV–V junction may assist formation of the active site by allowing the bound SLI substrate to explore the conformational space for optimum docking with the A756 loop.28
Figure 5.
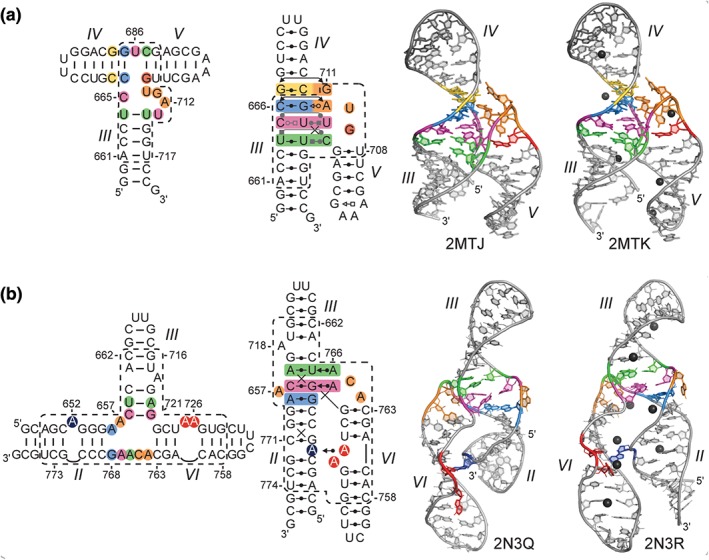
NMR structures of the VS ribozyme (a) III–IV–V(Adapted with permission from Ref 28. Copyrights 2014 American Chemical Society). and (b) II–III–VI30 three‐way junctions. From left to right: proposed secondary structures of the RNA subdomain used for NMR studies; schematic representation (as in Figure 2) of the tertiary structure derived from the NMR structures; cartoon representation of the lowest‐energy NMR structure determined in the presence of Mg2+ ions; and of the lowest‐energy NMR structure determined in the presence of Mg2+ ions and using RNA‐metal restraints to localize hydrated Mg2+ ions (black spheres).
Critical Tertiary Interaction Within II–III–VI Junction
The NMR structure of the II–III–VI junction was determined as part of a 62‐nt RNA that incorporates the core residues and adjacent stems with their A‐rich bulges, as previous mutational studies had shown the importance of these bulges for the catalytic activity of the VS ribozyme.27, 29, 51 The NMR structure determined in the presence of Mg2+ ions shows a family C topology of three‐way junctions, with coaxial stacking of stems II and III, and side‐by‐side arrangement of stems II and VI (Figure 5(b)).30 An ACAA turn (A764–A767) allows backbone reversal between stems II and VI and positions the two residues after the turn (A766 and A767) to form base triples (A–U–A and C–G–A) in the minor groove of stem III. The core of the II–III–VI junction is stabilized by these two minor groove base triples and a noncanonical cis WC/WC A656–G768 base pair, which together form a three‐layer stack between stems II and III. The NMR structure also reveals a long‐range tertiary interaction between the bulges of stems II and VI, where A652 protrudes from stem II to interact with stem VI, stacking between C760 and A761 and forming a cis SE/WC A652–A725 base pair. Interestingly, several Mg2+‐binding sites were identified from Mn2+‐induced PRE, including two at the junction core and three at the bulge–bulge interaction that are likely needed to counteract the electronegative charges that are brought in proximity of these sites. Complementary NMR studies established that Mg2+ ions and the bulge–bulge interaction are both required for the stable folding of the II–III–VI junction.
Model of VS Ribozyme Based on NMR Studies of Isolated Subdomains
Global Architecture of VS Ribozyme Based on NMR Studies: An Open State
NMR studies of VS ribozyme subdomains have allowed the structural characterization of all nonhelical regions of a minimal VS ribozyme competent for cleavage activity (Figure 6(a)). Based on the high‐resolution NMR structures of subdomains, a three‐dimensional model of a trans VS ribozyme bound to a preshifted SLI substrate was built by fragment assembly.30 This NMR‐based model and related structural schematics provided novel insights into the global structure of the VS ribozyme (Figure 6(b)–(d)). Given the specific topology of the two three‐way junctions (Figure 5), where stem II stacks on stem III (II–III–VI junction) and stem III stacks on stem IV (III–IV–V junction), the continuous stack of stems II, III, and IV defines the main frame of the VS ribozyme architecture. Stem‐loops V and VI are projected alongside this main frame like one arm (SLV) and an opposite leg (SLVI). The SLI substrate associates with the trans VS ribozyme via a KLI with the SLV loop, bringing the G638 loop of SLI in the vicinity of the A756 loop of SLVI. The NMR‐based model captures an open‐state conformation in which these two internal loops are apart, and this is compatible with chemical probing and biophysical data.32, 53 In all likelihood, such an open state is populated by a dynamic ensemble of ground‐state structures. The remodeling of the G638 and A756 loops as well as the relative motion between the supporting helical domains would be necessary to allow formation of the active site and subsequent cleavage.
Figure 6.
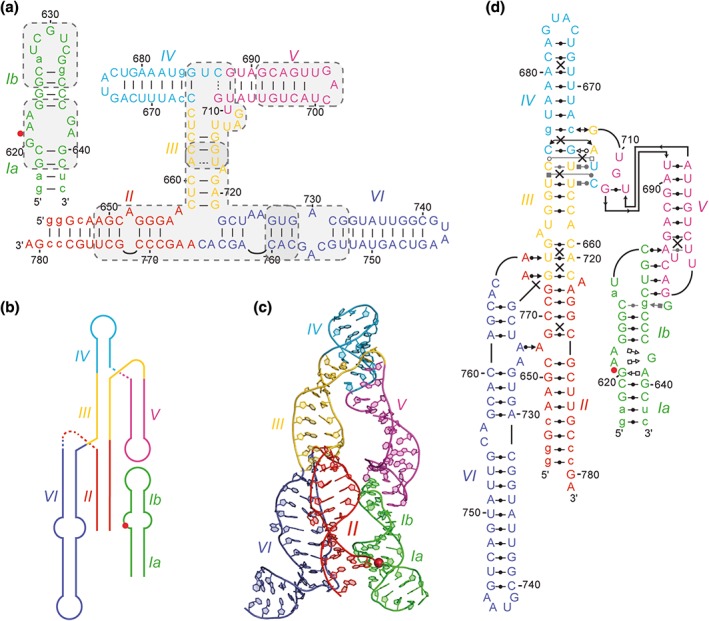
Structural overview of the open state of the VS ribozyme captured by the NMR‐based model of a trans ribozyme in complex with a preshifted stem‐loop substrate.30 (a) Proposed secondary structure of the complex,3, 4 which was previously characterized kinetically52 and used to identify important subdomains for NMR structural determination (shaded gray boxes). (b) Simplified global architecture reflecting the relative orientation of helical domains. (c) Cartoon representation of the three‐dimensional NMR model. (d) Tertiary structure schematics (as in Figure 2) based on high‐resolution NMR structures of isolated subdomains. Other legend details: In panels (a) and (d), nonnatural nucleotides are in lowercase. Also, base pairs of the proposed secondary structure are shown as a solid line when compatible with the structure shown in panel (c) and as a dashed line when not. A uniform color scheme is used for helical domains, and the scissile phosphate is depicted by a red dot or sphere.
Validation of Divide‐and‐Conquer NMR Approach
The divide‐and‐conquer approach was undertaken with a good understanding of the structural elements important for structure and function of the VS ribozyme, and this facilitated the identification of RNA subdomains that can fold autonomously. To define suitable RNA subdomains for structural studies, we routinely prepared several different RNA sequences for each subdomain and performed preliminary biochemical, biophysical, and structural characterization. It was important to define an optimal RNA sequence, buffer composition, and folding conditions for which each RNA subdomain adopts a homogeneous and stable fold, and this was verified initially using native gel electrophoresis and NMR spectroscopy, and later using small‐angle X‐ray scattering (SAXS; Dagenais P and Legault, unpublished data). Imino NMR spectroscopy was particularly valuable to quickly assess conformational homogeneity and define the secondary structure of the isolated RNA subdomains. In addition, the imino NMR spectra of isolated RNA subdomains and that of the trans ribozyme were compared to validate structure determination of a given RNA subdomain (Bonneau E, Dagenais P, and Legault P, unpublished data). In some cases, NMR studies of RNA subdomain variants contributed to defining the specific residues required for folding of a given subdomain. For example, NMR studies of two variants of the isolated II–III–VI RNA subdomain revealed the importance of a bulge–bulge interaction for stable folding of this junction.30 Thus, the II–III–VI RNA subdomain used for NMR structure determination contained the bulges of stems II and VI that form this critical bulge–bulge interaction. In summary, great care was taken to make sure that RNA subdomains were well circumscribed and represented independent folding modules.
Once the high‐resolution NMR structure of an RNA subdomain was obtained, it became essential to exhaustively compare the structure with previous biochemical and biophysical data. This comparison with previous studies was particularly important to validate our NMR structure of the SLVI RNA subdomain containing the A756 loop, because no structural or functional validation of this subdomain was available prior to structure determination.25 In contrast, a previous NMR study of the A756 loop in a different structural context revealed an alternate conformation that may be relevant to the folding pathway, but that did not agree with biochemical data.24 Otherwise, the NMR structural studies of isolated subdomains are essentially consistent with a large body of biochemical and biophysical data, thereby supporting the modularity of the VS ribozyme architecture.
CRYSTAL STRUCTURES PROVIDE A DETAILED UNDERSTANDING OF THE VS RIBOZYME STRUCTURE
Overall Crystal Structure and Comparison with NMR Structures of Isolated Subdomains
Trans Recognition of a Stem‐Loop Substrate Within a Symmetric Dimer: A Closed State
Crystal structures were determined at 3.1‐Å resolution for two active‐site variants (G638A and A756G) of an extended cis VS ribozyme containing helical domain VII in addition to domains I–VI (Figure 7(a)).34 The otherwise identical RNA sequence of these two variants was optimized to improve conformational homogeneity and reduce aggregation as needed for structural studies; it incorporates a preshifted SLI substrate and a substitution in loop IV by an ACAAA pentaloop, which serves as epitope for a synthetic antibody fragment.54 Although such antibody fragment–RNA loop interaction can facilitate crystallization,54 the crystal lattice did not incorporate the antibody fragment, but was rather stabilized by crystal contacts mediated by the loop IV pentaloop itself.
Figure 7.
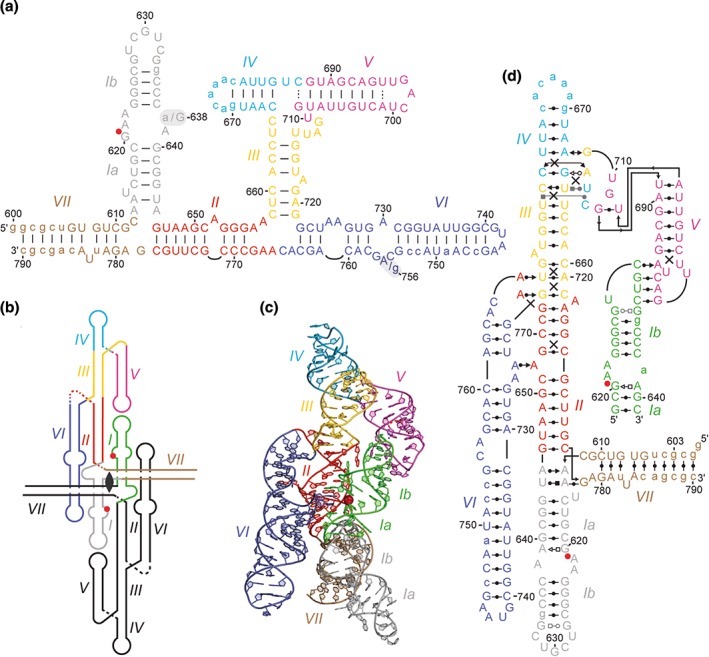
Structural overview of the closed state of the VS ribozyme captured by the crystal structure of a dimeric form.34 (a) Proposed secondary structure (as in Figure 6) of the two cis ribozyme sequences used for crystal structure determination, a G638A and an A756G variant. Of note, the secondary structure was defined for a smaller RNA sequence where the I–II–VII junction and stem VII were not present.3, 4, 34 (b) Simplified global architecture of the dimeric form reflecting the relative orientation of helical domains and showing the center of two‐fold rotational symmetry. (c) Cartoon representation of the crystal structure of the G638A variant (PDB code 4R4V) shown in a simplified trans form with one entire protomer and the SLI substrate of the partner protomer. (d) Tertiary structure schematics (as in Figure 2) of the simplified trans form. Due to constraints inherent to this type of representation, the proximity of the G638 and A756 loops is not depicted here. (Adapted with permission from Ref 34. Copyright Macmillan Publishers Ltd: Nature Chemical Biology)
The two crystal structures are very similar and both show that the VS ribozyme forms a symmetric dimer, in which the substrate of each protomer interacts with the catalytic domain of the other protomer (Figure 7(b)). This dimeric form may be relevant to efficient in vivo processing of the transcribed concatemeric form of the VS RNA, in which the upstream ribozyme catalytic domain could perform cleavage of the downstream stem‐loop substrate.55
The two central helical axes of the dimer are formed by continuous stacking of helices I–II–III–IV of one protomer on helix V of the partner protomer via the I–V KLI. The III–IV–V and II–III–VI three‐way junctions impart a parallel orientation to helices V and VI with respect to the central helices. In contrast, the I–II–VII junction directs helix VII almost perpendicular to the flat‐shaped structure formed by the side‐by‐side packing of the other helical segments. The dimeric structure can be simplified by showing only the substrate of one protomer bound to the ribozyme of the other protomer (Figure 7(c) and (d)). Interestingly, this structure captures a closed state, in which the substrate is bound via both the I/V KLI and close association of its G638 loop with the A756 loop to form a precatalytic active site.
Important Structural Features Within Crystal Structure
The global architecture of the VS ribozyme is governed by local structural features that direct its folding. With the exception of the I–II–VII junction, important structural features were previously structurally characterized by NMR. Thus, it is interesting to compare these structural features present in the crystal structures with the NMR structures of isolated subdomains.
I/V KLI
The I/V KLI within the crystal structure is very similar to the one found in the NMR structure of this subdomain, with the U‐turns of loop I and loop V directing the formation of three WC/WC base pairs and one SE/WC base pair between the two loops (Figure 8(a)). Loop V displays a compact U‐turn structure with the U700 residue bulged out and the closing A701–U695 base pair destabilized, as previously found in the NMR structure of the SLI/SLV complex (Figure 2(d)). For loop I, there is some discrepancy between the available structures (Figures 6(d) and 7(d)). In the NMR structure, there is a disordered WC/WC C626–G633 base pair that, in some models, forms a base triple with the Hoogsteen edge of the bulged out G634 (Figure 2(d)). In the X‐ray structure, it is G634 instead of G633 that forms a WC/WC base pair with C626, and G633 is involved in the H/SE G633–G627 base pair. This structural discrepancy likely results from the fact that different SLI substrates were used for X‐ray (C634G variant) and NMR (C634G/G627A variant) studies, in agreement with the notion that both the sequence and structure can be varied in this region without significantly affecting cleavage activity.4, 22
Figure 8.
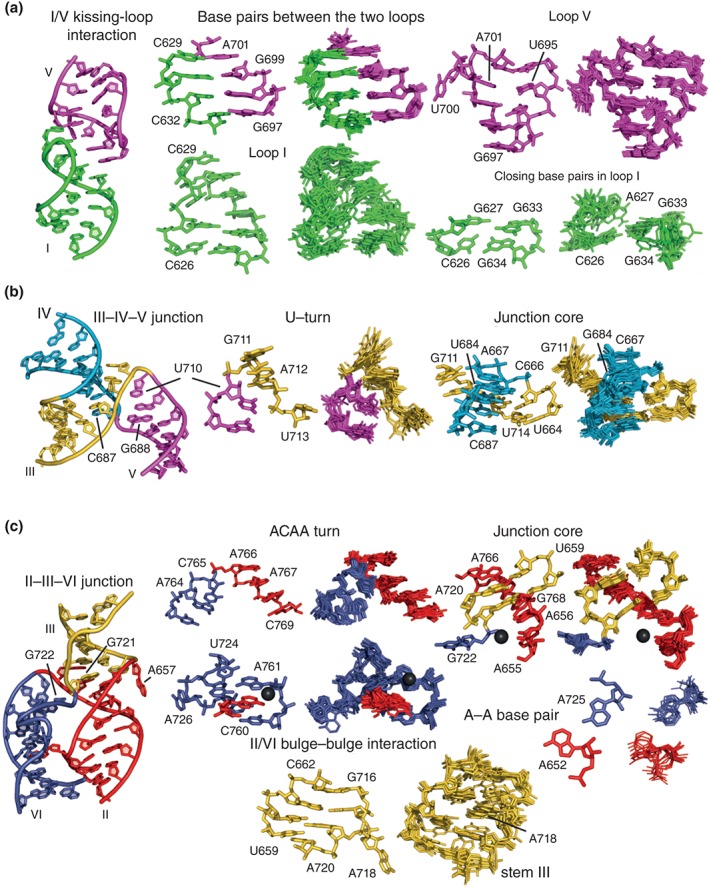
Comparative analysis of the crystal and NMR structures in terms of the structural features that direct the folding of the VS ribozyme. A general view of the crystal structure (PDB code 4R4V; left) and specific characteristic features (right) are provided for (a) the I/V kissing‐loop interaction (residues 623–637 and 692–704), (b) the III–IV–V junction (residues 661–670, 681–691, and 705–717), and (c) the II–III–VI junction (residues 649–662, 716–729, and 758–774). Each specific feature is displayed as a side‐by‐side comparison between the X‐ray crystallography structure (PDB code 4R4V) on the left and a superposition of the 10 lowest‐energy NMR structures [PDB codes 2MI0 (a), 2MTJ (b), and 2N3R (c)] on the right. Divalent metal ions that are common to both the NMR and X‐ray structures are shown as black spheres.
III–IV–V Junction
The III–IV–V junction within the crystal structure is very similar to the one found in the NMR structure of this subdomain, both adopting a family C topology with coaxial stacking of stems III and IV and close packing of stems III and V (Figure 8(b)). A U‐turn fold defined by residues U710, G711, and A712 allows backbone reversal between stems III and V as observed in the NMR ensemble, with the two typical hydrogen bonds, the sharp turn in the phosphate backbone, and continuous stacking before and after the turn. As in the NMR structure, the splayed conformation between C687 and G688 allows backbone redirection between stems IV and V. The core of the junction is stabilized by a stack of four base triples and a ribose zipper that are very similar between the crystal and NMR structures; however, there are small variations in hydrogen bond interactions that leads to different definitions of base‐pairing interactions (Figures 6 and 7).
II–III–VI Junction
The II–III–VI junction within the crystal structure is also very similar to the one found in the NMR structure of this subdomain, both adopting a family C topology with coaxial stacking of stems II and III as well as close packing of stems II and VI stabilized by a bulge–bulge interaction (Figure 8(c)). An ACAA turn defined by residues A764, C765, A766, and A767 allows backbone reversal between stems II and VI and base stacking of residues before and after the turn. Although this ACAA turn is very similar in the NMR and X‐ray structures, the NMR structure shows a slightly more compact fold with a hydrogen bond (C765 2′‐OH‐A766 N7) not present in the X‐ray structures. As observed in the NMR structure, the splayed conformation between G721 and G722 allows backbone redirection between stems III and VI, and the core of the junction is stabilized by the stack of two base triples and a WC/WC G–A base pairs. Nevertheless, there are small variations in the junction core between the crystal and NMR structures in terms of base planarity and hydrogen bond interactions (Figures 6 and 7). As in the NMR structure, a bulge–bulge interaction between stems II and VI involves A652 (stem II) stacking between C760 and A761 (stem VI) and forming a SE/WC A–A base pair with A725 (stem VI).
Despite the remarkable similarity between the structures of the II–III–VI junction core and bulge–bulge interaction, a notable difference is observed in stem III. Whereas A718 stacks on G719 within the helix in the NMR structure, it is completely bulged out in the crystal structure (Figure 8(c)). Thus, the transition of the VS ribozyme from an open state to a closed state may involve A718 shifting from an intra‐helical position to a bulged‐out state. Such conformational change, known to affect the rise and twist of flanking base pairs,56 would change the relative orientation of helices connected to the III–IV–V and II–III–VI junctions and may help bring the A638 loop in proximity of the A756 loop.
I–II–VII Junction
The I–II–VII junction is only represented in the crystal structures, where it adopts a family C topology with coaxial stacking of stems I and II and bending of stem VII toward stem I (Figure 7). This coaxial stacking of stems I and II helps orient the substrate helix outward from its own catalytic domain to interact with the catalytic domain of the partner protomer. A GCAA turn, involving G611, C612, A613, and A614, allows backbone reversal between C612 and A613 as well as base stacking of residues before and after the turn. In the core of the junction, a SE/WC A–A base pair piles up on a G–A–U base triple via cross‐stand purine stacking. Residues A614, U615, and C616 adopt an S‐turn fold that directs stacking of the U615 nucleobase on G778. This stacking is likely critical to stabilize the splayed conformation of G778 and A779, which respectively stack on stems II and VII. A superposition of the two crystal structures (not shown) reveal that residues G611, C612, U615, and A779 are less well defined, suggesting that they are relatively more dynamic than other residues of the junction. This may be relevant to the possibility of the I–II–VII junction to adopt two conformations, one that favors cleavage and one that favors ligation, as previously suggested.6
Interactions with Divalent Metal Ions
NMR studies of VS ribozyme subdomains have established that Mg2+ ions are required for folding of the A756 loop and the II–III–VI junction and help stabilize the compact fold of free SLV and the III–IV–V junction. In addition, several divalent metal‐binding sites have been identified in RNA subdomains using NMR methods (Figures 2, 4, and 5). Likewise, several metal ions have been localized from the two available crystal structures, 4R4P (2Mg2+) and 4R4V (9Mg2+ and 5K+). However, only two metal‐binding sites have been identified by both NMR spectroscopy and X‐ray crystallography methods. Interestingly, these are associated with the II–III–VI junction and are likely required for the folding of this junction. One is localized at the II–VI bulge–bulge interaction near the A652–A761 stack, such that it can counteract the electronegative repulsion from nearby phosphates (C651, A652, A761, and G762; Figure 8(c)). The other is localized in the core of the II–III–VI junction where it may stabilize a phosphate cluster (G721, A722, G655, and A656), the splayed conformation of G721 and G722 and base interactions at the junction core. Surprisingly, the Mg2+ ion localized in the S‐turn of the A756 loop by NMR methods (Figure 4) and important for folding of the free A756 loop has no equivalent in crystal structures of the VS ribozyme, even though the S‐turn displays a similar phosphate cluster in these structures.
VS Ribozyme Active Site
Mechanistic investigations of the VS ribozyme have revealed two key nucleobases, G638 and A756, responsible for general acid–base catalysis and highlighted similarities with the hairpin ribozyme, where a guanine (G8) and an adenine (A38) of two distinct internal loops also respectively serve as proton acceptor and proton donor in the cleavage reaction.13 NMR studies of individual subdomains encompassing the G638 and A756 loops of the VS ribozyme have revealed important structural features in these loops and similarities with the hairpin ribozyme active site.18, 25, 26 However, the two crystal structures of the VS ribozyme provide a detailed view of the active site architecture. They also clearly establish that the VS and hairpin ribozymes display comparable structural features at the active site despite low similarity in their sequences and tertiary structures, possibly as a result of convergent evolution.13, 14, 34
Remodeling of G638 and A756 Loops for Formation of Active Site
The NMR structures of the G638 and A756 loops were superposed to those of the crystal structures to provide insights into the local structural changes associated with formation of the active site (Figure 9(a)). For the A756 loop, the main structural characteristics of the free loop are found in the active site, namely the WC/WC G–A base pair and the S‐turn. However, formation of the active site leads to a more pronounced S‐turn and brings the bulged‐out residues (C755 and A756) from their minor groove position to a completely exposed position, away from the main body of the helix. Similarly, the largest structural change in the G638 loop upon formation of the active site is the protrusion of two adenines (A621 and A622) that were both positioned in the free G638 loop to form a G–A base pair with G638. This structural change is associated with formation of an S‐turn centered at A621 that brings the G620 2′‐OH closer to the general base G638 (from ~6.1 to 5.4 Å). Thus, the A756 and G638 internal loops are prefolded in their free form, and relatively minor conformational changes allow their less constrained residues to become completely protruded to form the active site. This is in drastic contrast with the more significant structural rearrangements that occur when the two interacting loops of the hairpin ribozyme associate to form the active site. The NMR structures of the isolated internal loops of the hairpin ribozyme57, 58 were previously compared with the crystal structure of a hairpin ribozyme–noncleavable substrate complex in which the two loops intimately associate,59 revealing important changes in base‐pairing interactions and backbone geometry upon formation of the active site.
Figure 9.
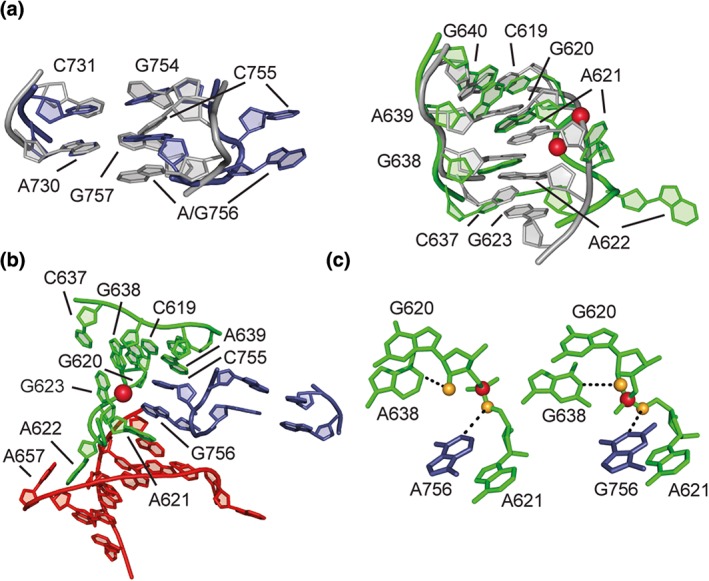
Formation of the active site in the VS ribozyme. (a) Remodeling of the A756 loop (left) and G638 loop (right) upon formation of the active site as seen from the superposition of the lowest‐energy NMR structures of the free loops [in gray; PDB codes 2L5Z (left) and 1OW9 (right)] onto those forming the active site in the crystal structure (in color; PDB code 4R4P). (b) Local environment at the active site (PDB code 4R4P). (c) Key residues at the active site of the G638A variant (left; PDB code 4R4V) and the A756G variant (right; PDB code 4R4P). The representation highlights the atoms that are expected to adopt an inline geometry in the transition state, namely the 2′‐oxygen (gold sphere), scissile P (red), and 5′‐oxygen (gold sphere) that rather form angles of 97° (left) and 129° (right).34 The N1 atoms of G638 and A638 are respectively 5.7 Å (left) and 4.9 Å (right) away from the 2′‐oxygen nucleophile (dashed line), whereas the N1 atoms of A756 and G756 are respectively 4.2 Å (left) and 4.0 Å (right) away from the 5′‐oxygen leaving group (dashed line).34 (Adapted with permission from Ref 34. Macmillan Publishers Ltd: Nature Chemical Biology)
Active Site Architecture
As originally envisioned, the G638 loop is nested in a cleft formed by the helical domains II and VI,41 and this involves extensive interloop stacking between the G638 and A756 loops as well as interactions of these loops with the minor groove of stem II (Figure 9(b)). Starting with the protruded adenines of the G638 loop, A622 stacks with the bulged out A657 from the II–III–VI junction and A621 forms an A‐minor interaction with helix II and stacks with A756 of stem VI. The protruded residues of the A756 loop, C755 and A756, are stacked on one another and sandwiched between A621 and A639 from SLI. These stacking interactions are consistent with the observation of a UV‐inducible crosslink between A756 and a 4‐thiouridine at position 621.53 As observed in the hairpin ribozyme, the residues flanking the scissile phosphate [N − 1 (G620) and N + 1 (A621)] adopt a splayed conformation that allow stacking of the key catalytic nucleobases (G638 and A756) and position their reactive groups near the scissile phosphate (Figure 9(c)). The similarity with the hairpin ribozyme extends further as the general acid (A756) is protruded from the internal loop by an S‐turn structural element and the N − 1 residue forms a SE/H base‐pair with the residue immediately 3′ of the general base (A639). In the proposed cleavage mechanism, a proton transfer occurs between the G620 2′‐OH group and the deprotonated N1 group of the G638 nucleobase, followed by a nucleophile attack of the 2′‐oxygen on the scissile phosphate to form a trigonal bipyramidal transition state, which is resolved by protonation of the 5′‐oxygen leaving group by the N1+‐H group of the A756 nucleobase.35 In many nucleolytic ribozymes, the splayed conformation of the cleavage site nucleotides is associated with the in‐line geometry of reacting groups (2′‐oxygen, P, and 5′‐oxygen) and is required in the transition state; however, such in‐line geometry was not observed in the available crystal structures of the VS ribozyme (Figure 9(c)). Also, the key nucleobases are not within hydrogen bond distances of their associated reacting groups (Figure 9(c)). Nevertheless, the crystal structures are in general agreement with the respective roles of G638 and A756 as the general base and acid in the cleavage reaction since only minor structural rearrangement may be required to bring the active site in a fully functional state.
Shifted pKas of Two Key Nucleobases in Light of Available Structures
In light of the available NMR and X‐ray structures of the VS ribozyme, it is interesting to consider the shifted pKas of 5.2–5.8 and 8.3–8.4 previously assigned to A756 and G638, respectively, based on the pH dependence of the VS ribozyme cleavage reaction.35, 36, 37, 38 From NMR studies of the isolated SLVI subdomain, a pKa value of 4.4 was attributed to A756, in agreement with its nucleobase being exposed in the open state conformation of the VS ribozyme.25 The increase of its pKa value to 5.2–5.8 could result from its proximity to the scissile phosphate in the active site. In contrast, the lowering of the pKa value of G638 by approximately one log unit compared to 5′‐GMP is more intriguing and suggests the presence of a metal ion at the active site. Modulation of the pKa values of the two key nucleobases upon changes in the ionic environment also suggests the presence of a metal ion at the active site.38 Although no metal ion is observed at the active site in the available X‐ray structures, NMR studies have revealed divalent metal ion binding sites in both the A756 and G638 loops that may remain bound in the active site, namely the metal ion at the S‐turn of the A756 loop (Figure 4) and the one associated with the 5′‐A639–G640‐3′ sequence of the G638 loop (Figure 2(b)). Alternatively, the stabilization of an S‐turn in the G638 loop upon formation of the active site (Figure 9(a); near A621) may create a favorable environment for binding of a metal ion that could stabilize the transition state. Higher resolution crystal structures are clearly needed to provide a more detailed characterization of Mg2+ ion binding sites in the VS ribozyme.
CONCLUSION
Several high‐resolution NMR and X‐ray studies reported in the past few years have provided novel insights into the dynamic structure of the Neurospora VS ribozyme. More specifically, they have allowed us to better understand: (1) the global architecture of the VS ribozyme, (2) the specific structural features that direct folding, (3) important features of the active site that direct catalysis, (4) the role of magnesium ions, and (5) the conformational changes associated with substrate recognition, activation, and docking. Several hotspots for dynamics have been identified that are likely important for the cleavage and ligation activities of this ribozyme. Follow‐up investigations are needed to capture a fully active ribozyme with an in‐line conformation at the active site and better understand the role of metal ions and dynamics in the reaction. Together with available structural data, such information will support current efforts aimed at engineering the VS ribozyme to cleave alternate substrates.52, 60
ACKNOWLEDGMENTS
The authors thank Dr. Richard A. Collins for critical reading of the manuscript. This work was supported by a grant from the Canadian Institutes of Health Research (CIHR 148801 to P.L.).
Conflict of interest: The authors have declared no conflicts of interest for this article.
REFERENCES
- 1. Saville BJ, Collins RA. A site‐specific self‐cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell 1990, 61:685–696. [DOI] [PubMed] [Google Scholar]
- 2. Saville BL, Collins RA. RNA‐mediated ligation of self‐cleavage products of a Neurospora mitochondrial plasmid transcript. Proc Natl Acad Sci USA 1991, 88:8826–8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beattie TL, Olive JE, Collins RA. A secondary‐structure model for the self‐cleaving region of Neurospora VS RNA. Proc Natl Acad Sci USA 1995, 92:4686–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersen A, Collins RA. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol Cell 2000, 5:469–478. [DOI] [PubMed] [Google Scholar]
- 5. Guo HCT, De Abreu DM, Tillier ERM, Saville BJ, Olive JE, Collins RA. Nucleotide sequence requirements for self‐cleavage of Neurospora VS RNA. J Mol Biol 1993, 232:351–361. [DOI] [PubMed] [Google Scholar]
- 6. Jones FD, Ryder SP, Strobel SA. An efficient ligation reaction promoted by a Varkud satellite ribozyme with extended 5′‐ an 3′‐termini. Nucleic Acids Res 2001, 29:5115–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ouellet J, Byrne M, Lilley DM. Formation of an active site in trans by interaction of two complete Varkud Satellite ribozymes. RNA 2009, 15:1822–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lilley DMJ. The hairpin and Varkud satellite ribozymes In: Lilley DMJ, Eckstein F, eds. Ribozymes and RNA Catalysis. Cambridge: Royal Society of Chemistry; 2008, 66–91. [Google Scholar]
- 9. Cochrane JC, Strobel SA. Catalytic strategies of self‐cleaving ribozymes. Acc Chem Res 2008, 41:1027–1035. [DOI] [PubMed] [Google Scholar]
- 10. Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet 2007, 8:776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collins RA. The Neurospora Varkud satellite ribozyme. Biochem Soc Trans Rev 2002, 30:1122–1126. [DOI] [PubMed] [Google Scholar]
- 12. Lilley DM. The Varkud satellite ribozyme. RNA 2004, 10:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lilley DM. Catalysis by the nucleolytic ribozymes. Biochem Soc Trans 2011, 39:641–646. [DOI] [PubMed] [Google Scholar]
- 14. Wilson TJ, Lilley DM. Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid–base catalysis? A critical assessment of available experimental data. RNA 2011, 17:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lilley DM. Mechanisms of RNA catalysis. Philos Trans R Soc Lond B Biol Sci 2011, 366:2910–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Michiels PJ, Schouten CHJ, Hilbers CW, Heus HA. Structure of the ribozyme substrate hairpin of Neurospora VS RNA: a close look at the cleavage site. RNA 2000, 6:1821–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Flinders J, Dieckmann T. A pH controlled conformational switch in the cleavage site of the VS ribozyme substrate RNA. J Mol Biol 2001, 308:665–679. [DOI] [PubMed] [Google Scholar]
- 18. Hoffmann B, Mitchell GT, Gendron P, Major F, Andersen AA, Collins RA, Legault P. NMR structure of the active conformation of the Varkud satellite ribozyme cleavage site. Proc Natl Acad Sci USA 2003, 100:7003–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Campbell DO, Legault P. NMR structure of the Varkud satellite ribozyme stem‐loop V RNA and magnesium‐ion binding from chemical‐shift mapping. Biochemistry 2005, 44:4157–4170. [DOI] [PubMed] [Google Scholar]
- 20. Campbell DO, Bouchard P, Desjardins G, Legault P. NMR structure of Varkud satellite ribozyme stem‐loop V in the presence of magnesium ions and localization of metal‐binding sites. Biochemistry 2006, 45:10591–10605. [DOI] [PubMed] [Google Scholar]
- 21. Bergonzo C, Hall KB, Cheatham TE 3rd.. Stem‐loop V of Varkud satellite RNA exhibits characteristics of the Mg(2+) bound structure in the presence of monovalent ions. J Phys Chem B 2015, 119:12355–12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bouchard P, Lacroix‐Labonté J, Desjardins G, Lampron P, Lisi V, Lemieux S, Major F, Legault P. Role of SLV in SLI substrate recognition by the Neurospora VS ribozyme. RNA 2008, 14:736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bouchard P, Legault P. Structural insights into substrate recognition by the Neurospora Varkud satellite ribozyme: importance of U‐turns at the kissing‐loop junction. Biochemistry 2014, 53:258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flinders J, Dieckmann T. The solution structure of the VS ribozyme active site loop reveals a dynamic “hot‐spot”. J Mol Biol 2004, 341:935–949. [DOI] [PubMed] [Google Scholar]
- 25. Desjardins G, Bonneau E, Girard N, Boisbouvier J, Legault P. NMR structure of the A730 loop of the Neurospora VS ribozyme: insights into the formation of the active site. Nucleic Acids Res 2011, 39:4427–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bonneau E, Legault P. NMR localization of divalent cations at the active site of the Neurospora VS ribozyme provides insights into RNA‐metal‐ion interactions. Biochemistry 2014, 53:579–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lafontaine DA, Norman DG, Lilley DM. The global structure of the VS ribozyme. EMBO J 2002, 21:2461–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bonneau E, Legault P. Nuclear magnetic resonance structure of the III‐IV‐V three‐way junction from the Varkud Satellite ribozyme and identification of magnesium‐binding sites using paramagnetic relaxation enhancement. Biochemistry 2014, 53:6264–6275. [DOI] [PubMed] [Google Scholar]
- 29. Lafontaine DA, Norman DG, Lilley DM. Structure, folding and activity of the VS ribozyme: importance of the 2‐3‐6 helical junction. EMBO J 2001, 20:1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bonneau E, Girard N, Lemieux S, Legault P. The NMR structure of the II‐III‐VI three‐way junction from the Neurospora VS ribozyme reveals a critical tertiary interaction and provides new insights into the global ribozyme structure. RNA 2015, 21:1621–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hiley SL, Collins RA. Rapid formation of a solvent‐inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J 2001, 20:5461–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pereira MJ, Nikolova EN, Hiley SL, Jaikaran D, Collins RA, Walter NG. Single VS ribozyme molecules reveal dynamic and hierarchical folding toward catalysis. J Mol Biol 2008, 382:496–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley DM. The complete VS ribozyme in solution studied by small‐angle X‐ray scattering. Structure 2008, 16:1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suslov NB, DasGupta S, Huang H, Fuller JR, Lilley DM, Rice PA, Piccirilli JA. Crystal structure of the Varkud satellite ribozyme. Nat Chem Biol 2015, 11:840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilson TJ, Li NS, Lu J, Frederiksen JK, Piccirilli JA, Lilley DM. Nucleobase‐mediated general acid–base catalysis in the Varkud satellite ribozyme. Proc Natl Acad Sci USA 2010, 107:11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith MD, Collins RA. Evidence for proton transfer in the rate‐limiting step of a fast‐cleaving Varkud satellite ribozyme. Proc Natl Acad Sci USA 2007, 104:5818–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilson TJ, McLeod AC, Lilley DM. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J 2007, 26:2489–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith MD, Mehdizadeh R, Olive JE, Collins RA. The ionic environment determines ribozyme cleavage rate by modulation of nucleobase pKa . RNA 2008, 14:1942–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rastogi T, Beattie TL, Olive JE, Collins RA. A long‐range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J 1996, 15:2820–2825. [PMC free article] [PubMed] [Google Scholar]
- 40. Andersen AA, Collins RA. Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme. Proc Natl Acad Sci USA 2001, 98:7730–7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sood VD, Collins RA. Identification of the catalytic subdomain of the VS ribozyme and evidence for remarkable sequence tolerance in the active site loop. J Mol Biol 2002, 320:443–454. [DOI] [PubMed] [Google Scholar]
- 42. Rastogi T, Collins RA. Smaller, faster ribozymes reveal the catalytic core of Neurospora VS RNA. J Mol Biol 1998, 277:215–224. [DOI] [PubMed] [Google Scholar]
- 43. Beattie TL, Collins RA. Identification of functional domains in the self‐cleaving Neurospora VS ribozyme using damage selection. J Mol Biol 1997, 267:830–840. [DOI] [PubMed] [Google Scholar]
- 44. Zamel R, Collins RA. Rearrangement of substrate secondary structure facilitates binding to the Neurospora VS ribozyme. J Mol Biol 2002, 324:903–915. [DOI] [PubMed] [Google Scholar]
- 45. Zamel R, Poon A, Jaikaran D, Andersen A, Olive J, De Abreu D, Collins RA. Exceptionally fast self‐cleavage by a Neurospora Varkud satellite ribozyme. Proc Natl Acad Sci USA 2004, 101:1467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA 2001, 7:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abu Almakarem AS, Petrov AI, Stombaugh J, Zirbel CL, Leontis NB. Comprehensive survey and geometric classification of base triples in RNA structures. Nucleic Acids Res 2012, 40:1407–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bouchard P, Legault P. A remarkably stable kissing‐loop interaction defines substrate recognition by the Neurospora VS Ribozyme. RNA 2014, 20:1451–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Franch T, Petersen M, Wagner EGH, Jacobsen JP, Gerdes K. Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U‐turn loop structure. J Mol Biol 1999, 294:1115–1125. [DOI] [PubMed] [Google Scholar]
- 50. de la Pena M, Dufour D, Gallego J. Three‐way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold. RNA 2009, 15:1949–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lafontaine DA, Wilson TJ, Norman DG, Lilley DM. The A730 loop is an important component of the active site of the VS ribozyme. J Mol Biol 2001, 312:663–674. [DOI] [PubMed] [Google Scholar]
- 52. Lacroix‐Labonté J, Girard N, Lemieux S, Legault P. Helix‐length compensation studies reveal the adaptability of the VS ribozyme architecture. Nucleic Acids Res 2012, 40:2284–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hiley SL, Sood VD, Fan J, Collins RA. 4‐Thio‐U cross‐linking identifies the active site of the VS ribozyme. EMBO J 2002, 21:4691–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Koldobskaya Y, Duguid EM, Shechner DM, Suslov NB, Ye J, Sidhu SS, Bartel DP, Koide S, Kossiakoff AA, Piccirilli JA. A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat Struct Mol Biol 2011, 18:100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Poon AH, Olive JE, McLaren M, Collins RA. Identification of separate structural features that affect rate and cation concentration dependence of self‐cleavage by the Neurospora VS ribozyme. Biochemistry 2006, 45:13394–13400. [DOI] [PubMed] [Google Scholar]
- 56. Barthel A, Zacharias M. Conformational transitions in RNA single uridine and adenosine bulge structures: a molecular dynamics free energy simulation study. Biophys J 2006, 90:2450–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cai Z, Tinoco IJ. Solution structure of loop A from the hairpin ribozyme from Tobacco ringspot virus satellite. Biochemistry 1996, 35:6026–6036. [DOI] [PubMed] [Google Scholar]
- 58. Butcher SE, Allain FH, Feigon J. Solution structure of the loop B domain from the hairpin ribozyme. Nat Struct Biol 1999, 6:212–216. [DOI] [PubMed] [Google Scholar]
- 59. Rupert PB, Ferré‐D'Amaré AR. Crystal structure of a haripin ribozyme‐inhibitor complex with implications for catalysis. Nature 2001, 410:780–786. [DOI] [PubMed] [Google Scholar]
- 60. Lacroix‐Labonté J, Girard N, Dagenais P, Legault P. Rational engineering of the Neurospora VS ribozyme to allow substrate recognition via different kissing‐loop interactions. Nucleic Acids Res 2016, 44:6924–6934. [DOI] [PMC free article] [PubMed] [Google Scholar]


