Summary
Almost the entire seafloor is covered with sediments that can be more than 10 000 m thick and represent a vast microbial ecosystem that is a major component of Earth's element and energy cycles. Notably, a significant proportion of microbial life in marine sediments can exploit energy conserved during transformations of sulfur compounds among different redox states. Sulfur cycling, which is primarily driven by sulfate reduction, is tightly interwoven with other important element cycles (carbon, nitrogen, iron, manganese) and therefore has profound implications for both cellular‐ and ecosystem‐level processes. Sulfur‐transforming microorganisms have evolved diverse genetic, metabolic, and in some cases, peculiar phenotypic features to fill an array of ecological niches in marine sediments. Here, we review recent and selected findings on the microbial guilds that are involved in the transformation of different sulfur compounds in marine sediments and emphasise how these are interlinked and have a major influence on ecology and biogeochemistry in the seafloor. Extraordinary discoveries have increased our knowledge on microbial sulfur cycling, mainly in sulfate‐rich surface sediments, yet many questions remain regarding how sulfur redox processes may sustain the deep‐subsurface biosphere and the impact of organic sulfur compounds on the marine sulfur cycle.
Introduction
Marine sediments are dynamic environments that are shaped by interactions among biotic and abiotic processes including the redox reactions by which microorganisms harness energy (Schrenk et al., 2010). While macrofauna may exist and cause some bioturbation in the most upper sediment layers (Bertics and Ziebis, 2010), the vast diversity and biomass of life at and below the seafloor is predominantly microscopic. Sedimentary microorganisms utilise various combinations of available electron donors and acceptors for energy conservation, the combinations of which are largely under thermodynamic controls (Jørgensen, 2000) and are also highly dependent on the amounts, types and rates of their respective inputs. Together, these factors manifest in the depth stratification of marine sediments i.e. a continuum of more or less overlapping biogeochemical zones, whereby each zone is characterised by the prevailing electron acceptors (Canfield and Thamdrup, 2009). In particular, sulfate is an ubiquitous electron acceptor in marine sediments due to its high concentration in seawater (∼28 mM). Seawater diffuses into sediments and sulfate respiration is thus one of the most important microbial redox process in marine sediments once more energetically favourable electron acceptors such as oxygen, nitrate/nitrite, and iron and manganese oxides are depleted.
On a global scale, recent estimates suggest the remineralisation of up to 29% of the organic matter that is deposited to the seafloor is facilitated by sulfate‐reducing microorganisms (SRM) (Bowles et al., 2014), which are conventionally regarded as terminal components of anaerobic microbial food webs that cooperatively degrade organic matter. The activity of SRM is particularly important in organic‐rich sediments underlying the highly productive waters of continental shelves and slopes (Jørgensen, 1982; Jørgensen and Kasten, 2006). Global estimates indicate that 11.3 teramoles of sulfate are reduced to hydrogen sulfide in marine sediments every year (Bowles et al., 2014). In turn, hydrogen sulfide and other reduced sulfur compounds serve as electron donors for sulfur‐oxidising microorganisms (SOM) or are abiotically oxidised. Sulfate reduction is thereby the primary driver of the biogeochemical cycling of sulfur in marine sediments (Fig. 1). Sulfur cycling is a major determinant of the biogeochemistry and microbial ecology in sulfate‐rich surface sediments and the underlying sulfate‐methane transition zone (SMTZ). Interestingly, the discovery of cryptic sulfur cycling i.e. the rapid recycling of sulfur species at low sulfate concentrations (Holmkvist et al., 2011; Brunner et al., 2016) and continuous detection of functional genes of SRM (Leloup et al., 2007; Leloup et al., 2009; Blazejak and Schippers, 2011; Aoki et al., 2015) in sulfate‐poor sediment zones deep below the SMTZ revealed that the impact of microbial sulfur metabolism extends to biogeochemical zones that were traditionally viewed as largely fermentative and methanogenic (Bowles et al., 2014; Glombitza et al., 2016).
Figure 1.
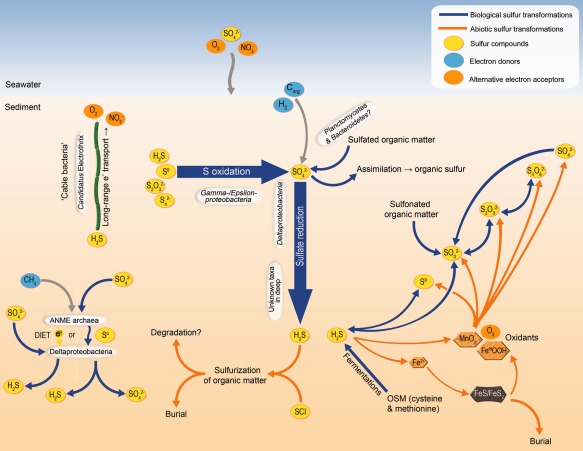
Conceptual depiction of the sulfur cycle in marine sediments, including main reactions of inorganic and organic sulfur compounds, selected taxa, sulfur oxidation via long‐range electron transport by cable bacteria, sulfate‐dependent anaerobic methane oxidation, and transformations of sulfur compounds of intermediate oxidation states (sulfur cycle intermediates, SCI). Blue lines depict biologically‐mediated sulfur transformations that can also be components of disproportionation reactions. Orange lines depict abiotic reactions. Inorganic sulfur compounds are depicted within yellow eclipses. Other electron acceptors are depicted within orange ellipses, and electron donors are depicted within blue ellipses. OSM = organo‐sulfur molecules, Corg = organic matter. DIET = direct‐interspecies electron transport. ANME = anaerobic methane‐oxidising.
Our understanding of sulfur cycling processes and the biology of microorganisms that catalyse them has improved considerably during recent years. Yet, there still remain significant questions regarding the biology of microorganisms and factors that control the turnover of sulfur compounds in marine sediments. For example, literally only the (sediment) surface of the species richness of sulfur‐transforming microorganisms in the subseafloor realm, which is one of the largest microbial biomes on Earth, has been explored. Cycling between the most oxidised (+6, sulfate) and most reduced (–2, sulfide) states of sulfur involves several inorganic sulfur compounds of intermediate oxidation states (i.e. sulfur cycle intermediates, SCI) that allow for (i) sulfur metabolite handoff to other microorganisms and (ii) shortcuts in the sulfur cycle (Fig. 1) (Jørgensen, 1990). The extent by which sulfur cycling in the different biogeochemical sediment horizons is mediated by ‘sulfur compound syntrophy’, i.e. the interspecies transfer of SCI, and the interplay between generalists that utilise diverse sulfur compounds of various oxidation states and specialists that utilise only selected sulfur compounds, is unknown. Additionally, previous research has concentrated on cycling of inorganic sulfur compounds, leaving the impact of organic sulfur compounds on life in the seafloor largely unexplored. Furthermore, individual microorganisms have evolved various, sometimes seemingly redundant enzyme systems to catalyse the many possible conversions of sulfur compounds between different oxidation states. While progress in the biochemical characterisation of these enzymes is continuously made (Denger et al., 2014; Felux et al., 2015; Santos et al., 2015; Johnston et al., 2016), some sulfur pathways and sulfur‐converting enzymes are yet to be discovered and/or functionally characterised (Milucka et al., 2012).
In this review, we highlight recent and selected findings in marine sediment sulfur cycle microbiology and address knowledge gaps that might become emphases for future research. The review especially aims to tie‐together a perspective on various microbial ecological aspects and on biogeochemical impacts of several key aspects of the sulfur cycle in marine sediments. In some points where little is known about the associated microbial ecology, we point‐out the biogeochemical aspects that suggest their probable key influences on associated microbial ecology. The review is primarily restricted to dissimilatory metabolisms used for energy conservation.
Dissimilatory sulfate reduction
Despite being of polyphyletic origin and having diverse physiological adaptations, described SRM are unified by having the same central pathway for sulfate respiration with the core enzymes ATP sulfurylase (Sat), adenylyl‐sulfate reductase (Apr), and dissimilatory (bi)sulfite reductase (Dsr) (Table 1) [for a comprehensive overview of SRM physiology, see review by (Rabus et al., 2015)]. Estimates of quantities in marine sediments suggest that SRM account for a large proportion (approx. 5–25%) of the microbial biomass in sulfate‐rich zones near the surface, and may have even higher relative abundances in the SMTZ (up to approx. 30–35%) (Leloup et al., 2007; Leloup et al., 2009). Available substrates and in situ temperatures are the key determinants of SRM community structure in sediment zones with sufficient sulfate (Robador et al., 2016). Various SRM that are ecologically relevant in these marine sediment layers belong to the class Deltaproteobacteria, whereby members of the family Desulfobacteraceae are typically one of the most important taxonomic groups in terms of abundance and activity (Fig. 1) (Robador et al., 2016).
Table 1.
Selected sulfur compound‐transforming enzymes and proteins used by microorganisms for dissimilation of sulfur compounds.
| General description | Gene/s | Enzyme/protein/complex [cofactor/'type'] a | Sulfur transformation b | Physiological function/s | Reference | UniProt ID c |
|---|---|---|---|---|---|---|
| Sulfate reduction; sulfur oxidation; other pathways in which sulfite is made available | dsrAB | Dissimilatory (bi)sulfite reductase, subunits AB [siroheme] | sulfite + [DsrC protein]‐dithiol → a [DsrC]‐trisulfide | Key enzyme in canonical sulfate/sulfite reduction; reverse function in sulfur oxidation; present in some non‐sulfate/sulfite‐reducing syntrophs | Santos et al. ( 2015) | P45574 |
| dsrC | DsrC | Co‐substrate for DsrAB in sulfite reduction | Acts as a co‐substrate for sulfite reduction by DsrAB | Santos et al. ( 2015), Oliveira et al. ( 2008), and Venceslau et al. ( 2014) | P45573 | |
| dsrMKJOP | DsrMKJOP | [DsrC]‐trisulfide → hydrogen sulfide + [DsrC protein]‐dithiol + 2 electron‐transfer quinone | Reduction of DsrC trisulfide, thereby linking cytoplasmic reduction of sulfite to energy conservation at membrane; reverse function in sulfur oxidation | Grein et al. ( 2010) | Q72CJ4 | |
| dsrN | DsrN | sirohydrochlorin + L‐glutamine → L‐glutamate + siroamide | Amidation of the siroheme cofactor of DsrAB | Lübbe et al. ( 2006) | Q9F2A0 | |
| dsrD | DsrD | Probable DNA‐binding | Probable transcriptional regulatory element | Hittel and Voordouw ( 2000) | Q46582 | |
| sat | Sulfate adenylyltransferase | sulfate + ATP + H+ ⇄ adenosine 5′‐phosphosulfate (APS) + diphosphate | Activation of sulfate with ATP to produce APS with higher redox potential than sulfate itself; also used for assimilation of sulfur from sulfate; reverse function in sulfur oxidation | Gavel et al. ( 1998) | Q72CI8 | |
| aprBA | Adenylylsulfate (APS) reductase | APS + a reduced electron acceptor ⇄ sulfite + AMP + an oxidised electron acceptor + 2 H+ | Conversion of APS to sulfite, which then acts as substrate for DsrAB/DsrC; reverse function in sulfur oxidation | Lampreia et al. ( 1994) | Q72DT2 | |
| qmoABC | Quinone‐interacting membrane‐bound oxidoreductase complex | electron transfer | Probable electron donor/transfer to Apr, linked to menaquinone pool. Appears specific for sulfate reduction pathway, but not sulfite reduction; reverse function in sulfur oxidation | Ramos et al. ( 2012) | Q7X167 | |
| Sulfite → Sulfide | dsr | Dissimilatory (bi)sulfite reductase [siroheme] | as described above | The Dsr complex and associated enzymes can also be used directly for sulfite reduction | Santos et al. ( 2015) | As above. |
| mccA/sirA | Sulfite reductase [cytochrome] | sulfite → sulfide | Direct respiratory reduction of sulfite to sulfide | Kern et al. ( 2011) | Q7MSJ8 | |
| asrABC | Anaerobic sulfite reductase [iron‐sulfur] | sulfite → sulfide | Direct respiratory reduction of sulfite to sulfide | Huang and Barrett ( 1990) | P26474 | |
| fsr | F420‐dependent sulfite reductase [siroheme] | sulfite → sulfide | Sulfite detoxification/sulfur assimilation | Johnson and Mukhopadhyay ( 2005) | Q58280 | |
| Thiosulfate → Sulfite | phsABC d | Thiosulfate reductase [molybdenum] | thiosulfate ⇄ sulfite | Respiratory reduction of thiosulfate yielding sulfite, which is further reduced (as above) to yield energy; thiosulfate disproportionation; thiosulfate oxidation | Burns and DiChristina ( 2009) | P37600 |
| Thiosulfate ⇄ Tetrathionate | tsdA | Thiosulfate dehydrogenase [diheme cytochrome] | thiosulfate ⇄ tetrathionate | Thiosulfate oxidation; tetrathionate reduction | Denkmann et al. ( 2012) | D3RVD4 |
| otr | Octoheme tetrathionate reductase [octoheme cytochrome] | tetrathionate ⇄ thiosulfate | Respiratory reduction of tetrathionate; more efficient as tetrathionate reductase than thiosulfate oxidase; possible role as nitrite reductase | Mowat et al. ( 2004) | Q8E9W8 | |
| ttrABC | Tetrathionate reductase [molybdenum] | tetrathionate ⇄ thiosulfate | Respiratory reduction of tetrathionate to thiosulfate; some trithionate reductase activity | Hensel et al. ( 1999) | Q9Z4S6 | |
| tetH | Tetrathionate hydrolase | tetrathionate ⇄ thiosulfate + sulfate | Tetrathionate oxidation | Kanao et al. ( 2007) | F9ZNI0 | |
| Thiosulfate → Thiocyanate | rdlA | Rhodanese‐like protein [rhodanase] | thiosulfate + cyanide ‐> thiocyanate + sulfite | Possible role in respiratory thiosulfate reduction; cyanide detoxification | Ravot et al. ( 2005) | Q6Q1E2 |
| Elemental sulfur/polysulfide transformations | psrABC | Polysulfide reductase [molybdenum] | polysulfide → sulfide | Respiratory reduction of polysulfides to sulfide | Krafft et al. ( 1992) | P31075 |
| hydDACB | Sulfhydrogenase/hydrogenase I [flavoprotein] | S° or polysulfide → sulfide | Reduction of polysulfides and/or elemental sulfur to sulfide; NADPH oxidation forming H2 when sulfur is absent; H2 oxidation | Ma et al. ( 1993) | Q8U2E5 | |
| shyCBDA/suDH | Sulfhydrogenase/hydrogenase II (a.k.a. Sulfide dehydrogenase) [flavoprotein] | S° or polysulfide → sulfide | Reduction of polysulfides and/or elemental sulfur to sulfide; NAD(P)H oxidation forming H2; H2 oxidation; ferredoxin:NADP oxidoreductase | Ma et al. ( 2000) | E7FHN9 | |
| npsr | NADH‐dependent persulfide reductase [flavoprotein/rhodanase] | S° or polysulfide → sulfide | Not clear; Reduction of range of disulfide, persulfide, and polysulfide compounds and/or elemental sulfur to sulfide | Warner et al. ( 2011) | A3QAV3 | |
| Sulfite oxidation | soeABC | Sulfite oxidising enzyme [molybdenum] | sulfite → sulfate | Sulfite oxidation to sulfate; sulfite detoxification after liberation from organo‐sulfur molecules | Lenk et al. ( 2011) | D3RNN8 |
| sorAB | Sulfite:acceptor oxidoreductase/Sulfite oxidising enzyme [molybdenum] | sulfite → sulfate | Sulfite oxidation to sulfate, sulfite may be formed during thiosulfate or sulfide oxidation; sulfite detoxificaton after liberation from organo‐sulfur molecules | Kappler et al. ( 2000) | Q9LA16 | |
| sorT | Sulfite:acceptor oxidoreductase/Sulfite oxidising enzyme [molybdenum] | sulfite → sulfate | Sulfite oxidation to sulfate, sulfite may be formed during thiosulfate or sulfide oxidation; sulfite detoxification after liberation from organo‐sulfur molecules | Wilson and Kappler ( 2009) | M4MVJ1 | |
| Sulfur oxidations | sqr | Sulfide:quinone‐oxidoreductase [flavoprotein] | sulfide → S0, + reduced quinol | Sulfide oxidation; resulting S° or , which are oxidised further by reverse‐Dsr or Sox pathways | Schütz et al. ( 1997) | Q4W5U9 |
| fccAB | Flavocytochrome c/sulfide dehydrogenases [flavoprotein/cytochrome] | sulfide → S0, | Sulfide oxidation; resulting S° or , which are oxidised further by reverse‐Dsr or Sox pathways | Dolata et al. ( 1993) | Q06529 | |
| soxXA | Sulfur‐oxidising multi‐enzyme complex, subunits XA [cytochrome] | thiosulfate or sulfite + SoxY → [SoxY protein]‐thiocysteine‐S‐sulfate | Mediates binding of thiosulfate or sulfite to a cysteine residue of SoxY | Bamford et al. ( 2002) | Q9LCV0, O33434 | |
| soxYZ | Sulfur‐oxidising multi‐enzyme complex, subunits YZ | thiosulfate or sulfite + SoxY → [SoxY protein]‐thiocysteine‐S‐sulfate | Sulfur binding/carrier to form sulfur anion adducts resulting in SoxY‐thiocysteine‐S‐sulfur | Quentmeier et al. ( 2003) | Q9LCU9, Q9LCU8 | |
| soxB | Sulfur‐oxidising multi‐enzyme complex, subunit B; Sulfate thioesterase/sulfate thiohydrolase [di‐manganese] | [SoxY protein]‐thiocysteine‐S‐sulfate → [SoxY protein]‐S‐thiocysteine + sulfate | Hydrolyses sulfonate moiety of SoxY‐thiocysteine‐S‐sulfate or SoxY‐cysteine‐S‐sulfate, releasing sulfate | Quentmeier and Friedrich ( 2001) | P72177 | |
| soxCD | Sulfur‐oxidising multi‐enzyme complex, subunits CD; Sulfane‐sulfur dehydrogenase [molybdenum/cytochrome] | [SoxY protein]‐S‐thiocysteine → a [SoxY‐protein]‐L‐cysteine‐S‐sulfate + 6 reduced c‐type cytochrome | Successive oxidation of outer sulfur of SoxY‐S‐thiocysteine, releasing 6 electrons to cytochromes | Bardischewsky et al. ( 2005) | A1B9M5, O07819 | |
| soxL | Sulfur‐oxidising multi‐enzyme complex, rhodanese‐like protein | sulfur transfer and trafficking | Possible role in transfer of SoxY‐bound sulfane sulfur to zero‐valent sulfur | Welte et al. ( 2009) | D3RVS9 | |
| rhd‐tusA‐dsrE2 | Rhodanase‐like protein‐Sulfurtransferase | sulfur transfer and trafficking | Elemental sulfur trafficking network from periplasm to cytoplasm for reverse Dsr oxidation; globules of zero‐valent sulfur may also form as intermediates during oxidation of sulfide, polysulfides, elemental sulfur, and thiosulfate to sulfate. | Stockdreher et al. ( 2012) | D3RPB9‐D3RPC0‐D3RPC1 | |
| dsrEFH | Sulfurtransferase | sulfur transfer and trafficking | Possible sulfur donor to DsrC during sulfur oxidation | Stockdreher et al. ( 2014) | O87896‐O87897‐O87898 | |
| sgpABCD | sulfur globule proteins | sulfur storage | Envelope formation and expansion of zero‐valent sulfur globules | Weissgerber et al. ( 2014), Prange et al. ( 2004) | ||
| Organo‐sulfur transformations | Sulfatase, a.k.a. sulfuric ester hydrolase e | R‐ → ROH + ; RN(H) → RNH2 + | Removal of sulfate moiety from organic molecules in order to: access organic molecule; obtain sulfur for assimilation; obtain sulfur for respiration | Barbeyron et al. ( 2016) | n.a. | |
| FGly‐sulfatase | formylglycine‐dependent sulfhydrolase, sulfatase [formylglycine] | R‐ → ROH + ; RN(H) → RNH2 + | Removal of sulfate moiety from organic molecules in order to: access organic molecule; obtain sulfur for assimilation; obtain sulfur for respiration | Hanson et al. ( 2004) | O69787 | |
| atsK | alkylsulfodioxygenase [dioxygenase] | R‐ → ROH + ; RN(H) → RNH2 + | Removal of sulfate moiety from organic molecules in order to: access organic molecule; obtain sulfur for assimilation; obtain sulfur for respiration | Kahnert and Kertesz ( 2000) | P9WKZ1 | |
| sdsA1 | alkylsulfohydrolase | R‐ → ROH + ; RN(H) → RNH2 + | Removal of sulfate moiety from organic molecules in order to: access organic molecule; obtain sulfur for assimilation; obtain sulfur for respiration | Davison et al. ( 1992) | Q9I5I9 | |
| atsA | arylsulfohydrolase | R‐ → ROH + ; RN(H) → RNH2 + | Removal of sulfate moiety from organic molecules in order to: access organic molecule; obtain sulfur for assimilation; obtain sulfur for respiration | Barbeyron et al. ( 1995) | P25549 | |
| xsc | sulfoacetaldehyde acetyltransferase | sulfoacetaldehyde → sulfite | Desulfonation of sulfoacetaldehyde in anaerobic sulfolactate degradation; part of taurine degradation pathway; sulfolactate is product of anaerobic sulfoquinovose degradation | Denger et al. ( 2009), Felux et al. ( 2015) | A3SR25 | |
| suyAB | sulfolactate sulfo‐lyase | 3‐sulfolactate → sulfite + pyruvate | Desulfonation of 3‐sulfolactate, which can be an intermediate L‐cysteate degradation. | Rein et al. ( 2005) | Q58Y43‐Q58Y44 | |
| cuyA | cysteate sulfo‐lyase | L‐cysteate → sulfite | Desulfonation of cysteate in sulfolactate degradation pathway | Denger et al. ( 2009) | A3SQG3 | |
| tauD | taurine dioxygenase | taurine → sulfite + 2‐aminoacetaldehyde + succinate + CO2 | Expressed only under conditions of sulfate starvation, aerobic | van der Ploeg et al. ( 1996) | P37610 |
This table is intended to provide an overview of enzymes catalysing key reactions in dissimilatory sulfur metabolisms described in the text and may aid in developing new marker genes for microbial ecological studies and interpreting (meta)genome/transcriptome data.
aWe provide a very brief keyword regarding cofactors in order to help distinguish different enzyme types.
bIn some cases ions and oxidizing/reducing agents are omitted for simplicity.
cUniProt IDs are mainly listed for substrate‐specific catalytic subunits.
dMolybdenum‐type oxidoreductases: Alpha subunit is catalytic, Beta subunit is electron transfer from Gamma subunit, Gamma subunit is membrane anchor.
eFour sub‐classes are listed directly below.
a.k.a = also known as.
n.a. = not available.
Prototypical of SRM in general, marine Desulfobacteraceae may use major fermentation products such as hydrogen, formate, acetate, and propionate as electron donors for sulfate respiration, but may also forage on a variety of other substrates including longer fatty acids, alkanes, and aromatic compounds, some of which allow these organisms to thrive at natural hydrocarbon seeps (Kleindienst et al., 2014; Na et al., 2015; Dorries et al., 2016b). Detailed proteomic‐genomic analyses of cultured representatives of the marine Desulfobacteraceae indicated their flexibility and ecological success is afforded by a combination of physiological mechanisms (Dorries et al., 2016a, 2016b). These include multiple oxygen defence systems, various signal transduction pathways for sensing different environmental cues such a substrate availability, as well as abundant and constitutively expressed membrane‐bound redox complexes that are important for linking electron flows from the catabolism of different substrates to respiration. Recent studies suggest that members of the uncultured Sva0081 clade of the Desulfobacteraceae are particularly abundant in coastal surface sediments around the world. They appear especially adapted to deal with oxidative stress from oxygen penetration in shallow sediments and may be exceptionally metabolically flexible, as inferred from their very large genomes (e.g. up to 9 Mb) (M. Mussmann, unpubl. data). However, Desulfobacteraceae represent just the iceberg tip of the SRM taxon diversity in marine sediments.
Other known key taxa include the families Desulfobulbaceae and Syntrophobacteraceae, and a wide diversity of bacteria related to the genus Desulfatiglans, which likely represents a previously undescribed deltaproteobacterial family. More astonishing is the extraordinary, yet undescribed diversity of SRM that is only recognised to exist from molecular ecology surveys of functional marker genes for SRM, i.e. aprBA and dsrAB (Müller et al., 2015). Marine dsrAB‐containing microorganisms are affiliated with several major uncultivated DsrAB lineages of family‐level or even higher taxon diversity, of which some (e.g. DsrAB lineages 2, 3, and 4) are almost exclusively found in marine ecosystems (Fig. 2) (Müller et al., 2015). Recent applications of single‐cell genomics and metagenomics to marine sediments have only just begun to uncover the phylogenetic identities of such dsrAB‐containing microorganisms. For instance, dsrAB sequences related to the marine‐specific DsrAB lineages 2, 3 and 4, have been uncovered in members of the Chloroflexi, which are notable for being relatively abundant in deep sediments and could therefore hint to their roles in the deep subsurface sulfur cycle (Wasmund et al., 2016). Additionally, a complete core gene set for sulfate reduction, including dsrAB from an uncultivated lineage, was recently discovered in a genome bin (SG8‐17) of a member of the phylum Gemmatimonadetes that was recovered from a sulfate‐rich estuarine sediment layer (Fig. 2) (Baker et al., 2015). Although promising, there is a long way ahead in charting the taxon diversity of marine SRM and sulfur‐transforming microorganisms by environmental genomics approaches due to the expansive microbial diversity found in marine sediments.
Figure 2.
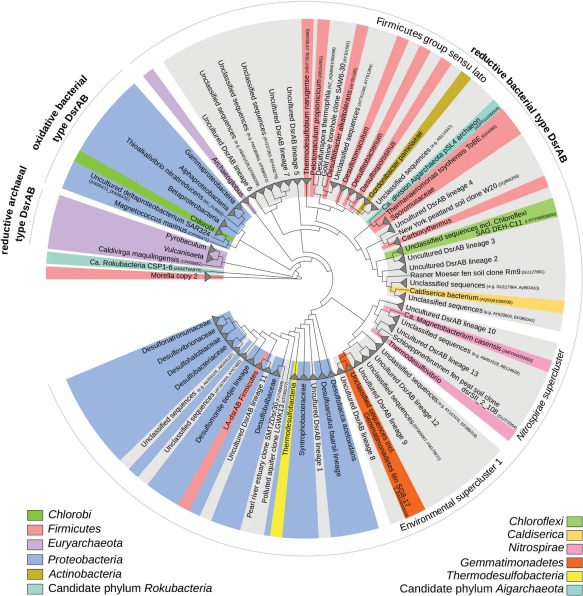
Phylogenetic tree showing the diversity of major DsrAB lineages, including sequences from the environment and isolated strains. The tree was based on a previously described DsrAB sequence set (Müller et al., 2015), including new sequences from the phyla Chloroflexi (Wasmund et al., 2016) and Gemmatimonadetes (Baker et al., 2015), and the candidate phylum Rokubacteria (Hug et al., 2016), and constructed with Fasttree (LG model of amino‐acid evolution) and an indel filter covering 530 alignment positions. Branches are unscaled. Clades with representatives from known phyla are labelled in different background colours. Desulfatiglans anilini lineage (not shown) is collapsed into the LA‐dsrAB Firmicutes group. Clades without cultured representatives are shown in grey. The three major DsrAB protein families, namely the reductive bacterial type, the oxidative bacterial type and the reductive archaeal type, are shown. The designation ‘uncultured DsrAB lineage’ depicts a stable, monophyletic lineage that consists only of environmental dsrAB sequences and has family‐level or higher taxon diversity. LA‐dsrAB, laterally acquired dsrAB. Moorella dsrAB copy 1 clustered with the LA‐dsrAB Firmicutes group. Selected accession numbers (for screen‐view only due to small font size) are given for some branches to aid in identification. See publication by Müller et al. (2015) for further information.
Our understanding of the ecology and lifestyles of SRM beyond the SMTZ and in the deep biosphere is even more limited. Sulfate depletion, decreasing nutrient availability and increasing pressure and temperature are general environmental factors that constrain the assembly of SRM communities in deeper sediment layers (Glombitza et al., 2015; Roussel et al., 2015). Biogeochemical data suggests the existence of high‐affinity SRM that are especially adapted to low sulfate concentrations in marine sediments (Tarpgaard et al., 2011). ‘Hot zones’ of cryptic sulfur cycling might provide opportunities for sulfate reduction, but SRM may also have to increasingly rely on their alternative metabolic capabilities such as fermentation due to decreased substrate availability in the deep (Glombitza et al., 2016). Metabolic versatility (Plugge et al., 2011) and longevity, enabled by cellular resistance mechanisms such as spore formation (Aüllo et al., 2013), will enable some SRM to thrive or at least survive during sediment burial (Hubert et al., 2009). Nevertheless, SRM communities present in sulfate‐rich and sulfate‐poor zones differ considerably (Leloup et al., 2009), but which SRM are active (Orsi et al., 2016), and how they make living in the deep remain key research questions.
Besides SRM ecology, recent research has also lead to unexpected discoveries regarding the biochemistry of sulfate reduction. One of the last conundrums of this pathway, namely how energy is conserved during reduction of (bi)sulfite to sulfide, was solved by showing that the DsrC protein is a co‐substrate for DsrAB (Santos et al., 2015). Furthermore, it has been an accepted fact that all SRM use the same sulfate reduction pathway. Hence, the possible existence of another pathway for sulfate reduction, which was identified by studying an anaerobic microbial consortium that oxidised methane in a sulfate‐dependent manner, came as a surprise (Milucka et al., 2012). It was shown that the methanotrophic archaea, known to lack the canonical sulfate reduction pathway, themselves reduced sulfate, albeit not to hydrogen sulfide but to elemental sulfur. The produced zero‐valent sulfur was then disproportionated to disulfide and sulfate by the associated deltaproteobacterium, which intriguingly harbour the canonical sulfate reduction pathway. It will be exciting to see how widespread and environmentally relevant this alternative pathway for dissimilatory sulfate reduction is, once its genetic and biochemical basis is unraveled and provides the necessary biomarkers for environmental surveys.
Anaerobic oxidation of methane coupled to sulfate reduction
Natural gases, mostly methane but also other alkanes such as ethane, propane, and butane (Laso‐Perez et al., 2016), originate from deep marine sediments or reservoirs in the subseafloor and diffuse upwards. Yet, teragrams of methane per year do not reach the water column because of its oxidation in anoxic sediment zones with sulfate as the dominant electron acceptor. So far, it was assumed that sulfate‐coupled methane oxidation is catalysed by an anaerobic consortium of methanotrophic archaea (ANME archaea) and sulfate‐reducing Deltaproteobacteria and that the basis of this syntrophy is the interspecies exchange of a hypothetical diffusible metabolite such as hydrogen, formate or methanethiol (Meulepas et al., 2010). A series of recent publications has now provided radically new insights into the biology of these anaerobic consortia, suggesting that they evolved diverse mechanisms for sulfate‐dependent methane oxidation (Fig. 1). As aforementioned, one such mechanism is that the methanotrophic archaea themselves perform sulfate reduction to zero‐valent sulfur (Milucka et al., 2012). The deltaproteobacterial partner would thus not be essential, rather a commensal that profits by disproportionation of the produced zero‐valent sulfur compounds. Another possible mechanism is that both partners fulfil their originally proposed function, i.e. the archaeon oxidises methane to CO2 and the deltaproteobacterium reduces sulfate to sulfide, but reducing equivalents are exchanged by direct cell‐to‐cell contacts between the partners and not via a diffusible metabolite (McGlynn et al., 2015; Wegener et al., 2015; Krukenberg et al., 2016). Electrical conductivity modelling and experimental data, including expression of large multi‐haem cytochromes by both partners, expression of type IV pili by the deltaproteobacterium, redox‐dependent staining of the intercellular matrix, and nanowire‐like structures between cells, provided evidence of direct interspecies electron transfer (DIET) (Wegener et al., 2015). Furthermore, for the first time the anaerobic methanotrophic archaea could be experimentally decoupled from syntrophic SRB by using soluble artificial single electron acceptors such as 9,10‐anthraquinone‐2,6‐disulfonate, humic acids, or Fe(III) complexes (Scheller et al., 2016). This finding is further proof for DIET and suggested a mechanism for methane oxidation coupled to insoluble iron(III) and manganese(IV) reduction, namely that methanotrophic archaea respire solid abiotic oxidants (such as metals) directly via extracellular electron transfer.
Dissimilatory reduction of sulfur cycle intermediates
As described above, the dissimilatory reduction of sulfate drives the formation of enormous quantities of reduced sulfide in marine sediments. Interestingly, these sulfides are subject to an array of biological and chemical oxidation reactions that may result not only in the reformation of sulfate, but also in a number of other SCI. These compounds may include sulfite, elemental sulfur, polysulfides, and polythionates such as thiosulfate and tetrathionate (Fig. 1). Indeed, biogeochemical studies suggest most sulfide (80–90%) is eventually reoxidised, while only 10–20% is ultimately buried, e.g. as complexes with iron (i.e. FeS, FeS2) or with organic matter after sulfurization reactions (Berner, 1982; Jørgensen, 1982; Brüchert, 1998; Sinninge Damsté et al., 1988; Ferdelman et al., 1999). Chemical oxidants of biogeochemical significance may include oxygen, nitrate, manganese‐oxides, iron‐oxides and even organics, which are present to varying degrees at different sediment sites and depths, depending on inputs and fluxes of both organics and inorganics (Burdige and Nealson, 1986; Canfield, 1989; Yao and Millero, 1996; Heitmann and Blodau, 2006; Yu et al., 2015). Intriguingly, these SCI can also be utilised as electron donors, as terminal electron acceptors for respiration, or as a both in disproportionation metabolisms (the later are described further below).
In most marine sediments examined, SCI are generally only measurable in the low micromolar ranges and do not accumulate to high concentrations because they are rapidly utilised or react upon formation, e.g. turnover times of few hours have been measured for micromolar additions of thiosulfate and tetrathionate (Jørgensen, 1990; Thamdrup et al., 1994; Zopfi et al., 2004). For other SCI such as sulfite or elemental sulfur, accessibility may hinder their utilisation, for instance, sulfite is particularly reactive with organics or elemental sulfur and it may therefore have short residence times, while elemental sulfur is stable but rather insoluble, which may also hinder its availability to microorganisms (Schauder and Kröger, 1993; Vairavamurthy et al., 1994; Zopfi et al., 2008). Only sediments with particularly high sulfate reduction rates and therefore high levels of sulfide production, e.g. those associated with particularly high organic loads or salt marsh beds, seem to exhibit accumulations of SCI (Zopfi et al., 2004). Additions of thiosulfate to sediments cause drastic decreases in the rates of sulfate reduction (Jørgensen, 1990), suggesting they draw electron‐flows away from specialist SRM to other organisms, or that SRM may switch to the use of SCI when available, or combinations thereof. Interestingly, parallel experiments have shown that tetrathionate additions reduced sulfate reduction rates to a much lesser extent, which suggests organisms fundamentally different to typical SRM may be largely responsible for tetrathionate reduction (Zopfi et al., 2004). The rapid turnover of SCI may not be surprising, since compared with sulfate, some SCI have higher redox potentials and/or more energetically favourable pathways. For instance, tetrathionate has a very high redox value of +198 ± 4 mV (Kurth et al., 2016); sulfite reduction bypasses the activation step of sulfate, which requires two ATP equivalents, and thus can lead to greater growth yields depending on the electron donor (Badziong and Thauer, 1978; Simon and Kroneck, 2013). Together, these facts mean some SCI are preferentially utilised as respiratory electron acceptors over sulfate. The available biogeochemical data clearly indicates that resident microorganisms in marine sediments are highly adapted and poised for the utilisation of various SCI.
To date, diverse microorganisms are known to be capable of, or have the genetic potential to, respire different and/or multiple SCI (Harel et al., 2016). However, this evidence is clearly far from complete, since it is reliant on information from the yet limited number of cultivated organisms or genomic information retrieved from environmental genomics. Nevertheless, in marine sediments, many cultivated Deltaproteobacteria are known to respire SCI and harbour SCI‐reductases (Krämer and Cypionka, 1989). In various tested SRM cultures, enzymes for thiosulfate and sulfite transformations were seemingly constitutively expressed (Krämer and Cypionka, 1989). This information may support the notion that SRM switch to SCI when available and why sulfate reduction rates decrease when SCI are added to sediments. Recently, various Desulfuromonadales phylotypes were highlighted as probable elemental sulfur respirers (or disproportionators) in diverse benthic habitats (Pjevac et al., 2014). Diverse members of the phyla Proteobacteria and Bacteroides, which are typically found in marine sediments are known to be capable of respiring elemental sulfur (Florentino et al., 2016).
Genes for key catalytic enzymes required for SCI transformations, such as tetrathionate‐, thiosulfate‐, polysulfide‐reductases of the complex iron‐sulfur‐molybdenum family of enzymes (Table 1) have been documented in an array of phyla, notably in the Proteobacteria and Firmicutes (Harel et al., 2016). The recent application of genome‐centric metagenomics to marine sediments has revealed potential for sulfur and possibly thiosulfate reduction in taxa completely undescribed and not previously connected with the sulfur cycle, such as the archaeal phylum Torarchaeota (Seitz et al., 2016; Lazar et al., 2017). Future research applying similar culture‐independent genomic approaches will likely illuminate a variety of microbial taxa capable of transforming SCI. Understanding the presence and expression of these genes could be of special interest in marine sediments, since they can act as molecular proxies for transformations of such cryptic biogeochemical processes, and such strategies have proven highly informative for the study of SCI transformations in the water column of oxygen minimum zones, where tracing the actual chemical transformations is difficult (Canfield, 2010). Interestingly, although not studied in marine sediments, enzymes with previously undescribed physiological functions have recently been shown to be capable of reducing SCI, e.g. sulfite, thiosulfate and tetrathionate reductases of the cytochrome family (Table 1) (Kern et al., 2011; Simon et al., 2011; Liu et al., 2013; Kurth et al., 2016). It is therefore very probable that the ability to respire SCI is even more widespread than previously conceived.
Sulfur oxidation
While anaerobic microorganisms respiring sulfur compounds can prevail deep into the marine subsurface, SOM on the other hand are generally restricted to upper sediment layers where electron acceptors with high enough redox potentials for coupling with sulfur oxidation, such as oxygen or nitrate, are available. The biological oxidation of reduced sulfur compounds competes with chemical reactions, in particular with the iron‐driven oxidation of sulfide to FeS or FeS2 (Jørgensen and Nelson, 2004) (Fig. 1). However, the half‐life of sulfide in microbial mats is controlled by cell density and is usually lower than in oxic (and iron‐rich) seawater (Nelson et al., 1986; Poulton et al., 2004; Canfield et al., 2005). Generally, thermodynamic and kinetic considerations suggest that biological oxidation probably far exceeds chemical oxidation of sulfide in most environments (Luther et al., 2011). While microorganisms are central to sulfide oxidation in marine pelagic oxygen minimum zones (Lavik et al., 2009), the general contribution of microorganisms to total sulfur oxidation in marine sediments is still unknown. A major limiting factor for sulfur oxidation in the seafloor are the gradients of oxygen, nitrate and sulfide, which often result in spatially separated maximal concentrations of these potential electron donors and acceptors (Canfield and Thamdrup, 2009). Many SOM therefore cannot directly access their primary energy sources, i.e. sulfides and SCI, and electron acceptors at the same time. This unique evolutionary pressure challenged SOM to develop distinct strategies that are reflected in remarkable morphological adaptations attracting the attention of researchers from different disciplines.
The morphologically conspicuous, large sulfur bacteria (LSB) of the gammaproteobacterial family Beggiatoaceae have been model organisms for benthic sulfur oxidation for decades. This family comprises several, morphologically distinct genera with diverse ecological strategies that reflect adaptations to the physicochemical characteristics of a wide spectrum of marine surface sediments (Teske and Salman, 2014). The molecular and morphological characteristics of Beggiatoaceae have recently been integrated in a revised taxonomic framework (Salman et al., 2011). The LSB are typical ‘gradient organisms’ that are indicative of hypoxic and sulfidic conditions in aquatic sediments. To bridge the spatial gap between oxygen and sulfide they internally store large amounts of the alternative electron acceptor nitrate and of elemental sulfur, a central intermediate of sulfide and thiosulfate oxidation. Filamentous, motile members such as Candidatus Isobeggiatoa and Thioploca glide between oxic/suboxic and sulfidic sediment layers to replenish their nitrate and sulfur reservoirs, while the non‐filamentous Thiomargarita depend on alternating sulfidic and oxygenated resuspensions of sulfidic sediments into oxygenated seawater. Certain Thiomargarita and Thiopilula attach to rocks or moving shells, where they experience alternating conditions of high sulfide, nitrate and oxygen fluxes (Bailey et al., 2011). Genome analysis of single cells or filaments revealed canonical sulfur oxidation pathways and the capacity for denitrification and/or nitrate ammonification (MacGregor et al., 2013; Winkel et al., 2016). In anoxic sediments, marine Thioploca species that respire nitrate to ammonia and leaking nitrite can fuel the anaerobic oxidation of ammonia to N2 by associated anammox bacteria and thereby drive a significant nitrogen loss (Prokopenko et al., 2013). In contrast, nitrifiers associated with LSB mats can re‐oxidise ammonia to nitrate and may thereby recycle bioavailable nitrogen within LSB mats (Winkel et al., 2014). Interestingly, some members of marine LSB such as Candidatus Thiomargarita nelsonii may employ both the Calvin‐Bassham‐Benson and the reverse tricarboxylic acid cycle for carbon fixation. Hence, these are currently the only known free‐living bacteria equipped with two carbon fixation pathways (Winkel et al., 2016).
Another intriguing evolutionary strategy of SOM to overcome the limited access to oxidants and reductants in marine surface sediments is to partner up with motile, eukaryotic hosts. In such mutualistic, chemosynthetic symbioses, the thioautotrophic bacterium nourishes the host, which in turn provides enhanced access to resources and shelter to the symbiotic SOM. Like the filamentous Beggiatoaceae, the motile hosts can position themselves in opposing gradients of oxygen and sulfide or shuttle themselves between oxic/suboxic and sulfidic layers. This lifestyle has developed independently and multiple times between alpha‐ and gammaproteobacterial thioautotrophs and sediment‐dwelling ciliates, oligochaetes, nematodes, flatworms and bivalves [for review see (Dubilier et al., 2008)]. Some SOM hosting clams reach high biomasses underneath seagrass meadows and might substantially contribute to sulfide oxidation (van der Heide et al., 2012). Intriguingly, these symbiotic SOM are also capable of fixing N2 and possibly provide the host not only with carbon, but also with bioavailable nitrogen (Petersen et al., 2016).
Recently, an entirely new and rather astonishing mechanism for sulfur oxidation, i.e. electrogenic sulfur oxidation (e‐SOx), was discovered. In this process, multicellular filamentous bacteria bridge the oxidation of sulfide in anoxic sediment layers replete with sulfide to the reduction of oxygen or nitrate in oxic surface sediments, thereby generating and mediating electric currents in marine sediments over centimetre distances (Fig. 1). These so‐called cable bacteria are thought to conduct electrons through structures inside a common periplasm of the multicellular filament (Nielsen et al., 2010; Pfeffer et al., 2012; Marzocchi et al., 2014). The spatially separated half‐reactions of e‐SOx result in a characteristic biogeochemical profile, which has strong effects on biogeochemical cycles at aquatic sediment surfaces, for instance, by influencing iron speciation (Seitaj et al., 2015; Nielsen, 2016; Sulu‐Gambari et al., 2016). The cable bacteria belong to the family Desulfobulbaceae of the Deltaproteobacteria and are currently represented by two candidate Genera, Candidatus Electronema and Candidatus Electrothrix (Trojan et al., 2016). The cable bacteria are widespread in shallow marine sediments (Risgaard‐Petersen et al., 2012; Malkin et al., 2014; Burdorf et al., 2016). In situ mass developments of cable bacteria have been documented in a seasonally hypoxic marine basin in the Netherlands (Grevelingen) (Seitaj et al., 2015). Similar to the Beggiatoaceae, cable bacteria seem to prefer rather diffusive, undisturbed sediments with stable hydrodynamic conditions (Malkin et al., 2014; Seitaj et al., 2015). The detailed physiology and the genetic background of e‐SOx are still completely unknown and enigmatic, but first experiments with 13C‐labelled carbon suggested heterotrophic sulfide oxidation in a marine sediment (Vasquez‐Cardenas et al., 2015).
To date, the aforementioned groups of SOM have largely monopolized the attention of scientists studying benthic sulfur oxidation because of their conspicuous morphologies and fascinating lifestyles. However, these occur in high abundances only in certain habitats. In particular, the LSB and cable bacteria seem to be restricted to undisturbed sediment with stable hydrodynamic conditions, while symbiotic SOM and their hosts have been mainly found in permeable coastal sediments. Numerous 16S rRNA gene surveys of marine sediments suggest that virtually all types of marine sediments harbour a myriad of less conspicuous SOM, which probably far exceed the currently known diversity and global abundance of Beggiatoaceae, symbiotic SOM and cable bacteria. The unambiguous identification of sulfur oxidation potential in an environmental sample is obscured by the fact that SOM have diverse and complex sulfur oxidation pathways. There is currently no clearly discernible, molecular marker that is universal among all SOM. Sulfide‐quinone oxidoreductase (Sqr) and the thioesterase subunit SoxB of the thiosulfate‐oxidising multienzyme complex (SOX‐pathway) appear to be the most widespread markers among known marine SOM (Table 1), but available primers used for diversity surveys of the respective genes most likely underestimate the actual diversity in marine sediments.
Despite these limitations, the application of functional marker genes in PCR or metagenomic approaches has revealed a yet unseen diversity of SOM. Genes encoding reverse DsrAB, Sqr and SoxB have been amplified from coastal sediments and uncovered a large diversity of alpha‐, gamma‐, and epsilonproteobacterial SOM and also novel, unknown lineages (Pham et al., 2008; Lenk et al., 2011; Thomas et al., 2014). For instance, an uncultured Rhodobacteraceae (Roseobacter‐clade) member from tidal sediments harboured the reverse DSR‐pathway (Lenk et al., 2012). Intriguingly, this SOM possesses both a complete Sox‐ and the reverse DSR‐pathway, and thus a flexibility in sulfur oxidation that is unique among SOM.
Members of the epsilonproteobacterial order Campylobacterales occur in virtually all marine systems such as pelagic oxygen minimum zones, hydrothermal systems and marine sediments, typically in habitats with low oxygen/sulfide ratios. They are among the best studied SOM as they often occur in high abundances and are relatively easy to grow in the laboratory. In some highly sulfidic marine sediments, Arcobacter species oxidise sulfide to filamentous elemental sulfur forming mat‐like precipitates (Wirsen et al., 2002). Members of the Sulfurovum/Sulfurimonas‐group seem to be more competitive than other SOM in exploiting solid, elemental sulfur in marine benthic habitats, possibly due to their capability to activate cyclo‐octasulfur (S8) (Pjevac et al., 2014). Moreover, Sulfurimonas has been repeatedly found in increased frequencies in the rhizosphere of marine plants (Jensen and Kühl, 2007; Thomas et al., 2014; Cúcio et al., 2016). Interestingly, it has been hypothesised that some Sulfurimonas use cable bacteria as an electron sink by DIET when oxygen and nitrate are depleted (Vasquez‐Cardenas et al., 2015; Lovley, 2016).
Remarkably, 16S rRNA and functional gene assays have consistently detected relatives of gammaproteobacterial, thiotrophic symbionts of marine protists and invertebrates in high frequencies in marine surface sediments (Ravenschlag et al., 1999; Bowman and McCuaig, 2003; Lenk et al., 2011; Dyksma et al., 2016). Whether these are free‐living or actual symbionts of yet unidentified micro‐ or meiofaunal hosts, is not known, but is certainly an interesting avenue for future research. Moreover, 16S rRNA sequences affiliating with the genera Sedimenticola, Thiohalophilus, Thiohalorhabdus, Thiomicrospira, Thioprofundum and Thioalkalivibrio have been repeatedly detected, but in situ abundances of these SOM are still unknown. High relative cell and sequence abundances of members of the recently established family Woeseiaceae/JTB255 and of the Acidiferrobacteraceae have been detected in sediments worldwide (Bowman et al., 2005; Du et al., 2016; Dyksma et al., 2016). Genomic and isotopic tracer studies confirmed a thioautotrophic potential for members of these families (Dyksma et al., 2016; Umezawa et al., 2016; Mussmann et al., 2017). In particular, the facultative chemolithoautotrophic Woeseiaceae/JTB255 are abundant, core members of microbial communities in virtually all studied sediments worldwide, although not all members can oxidise sulfur (Du et al., 2016; Mussmann et al., 2017). Collectively, the alpha‐, gamma‐ and epsilonproteobacterial SOM may amount to average cell abundances of approximately 108 cells cm−3 in organic‐rich marine sediments (Ravenschlag et al., 2001; Bowman et al., 2005; Lenk et al., 2011; Dyksma et al., 2016). Given that biotic sulfur oxidation may by far exceed abiotic sulfur oxidation in most habitats and the relatively narrow habitat range of large sulfur bacteria, these inconspicuous SOM possibly account for a major fraction of sulfur oxidation in many marine sediments.
SOM in coastal sediments may also play a central role in marine dark carbon fixation. Recent biogeochemical modelling suggested that sulfur‐dependent carbon fixation in marine sediments could be responsible for almost half of total dark carbon fixation in the oceans (Middelburg, 2011). In line with this, recent isotopic tracer studies found that Gammaproteobacteria, including Acidiferrobacteraceae, Woeseiaceae and others, accounted for a major fraction of CO2 fixation in coastal sediments (Boschker et al., 2014; Dyksma et al., 2016). This raises the possibility that the highly abundant and widespread autotrophic SOM might play a profound, yet unconsidered role in oceanic carbon sequestration (Hawley et al., 2014; Dyksma et al., 2016).
Disproportionation of inorganic sulfur compounds
The disproportionation of SCI is performed by various anaerobes that are generally also capable of respiring with sulfur compounds. Disproportionation entails using SCI as both electron donor and acceptor, i.e. an ‘inorganic fermentation’. The biogeochemical importance of these metabolisms was demonstrated by pioneering studies applying radioisotope tracing of different sulfur atoms and demonstrated a significant proportion, i.e. 62–66% and 35–39% of thiosulfate (often the key SCI) is disproportionated in oxidised or reduced sediment layers, respectively (Bak and Cypionka, 1987; Fossing and Jørgensen, 1990; Jørgensen, 1990). While data regarding the biogeochemical impact of the disproportionation of other SCI in marine sediments is still limited, disproportionation of elemental sulfur has been demonstrated (Thamdrup et al., 1993). Disproportionation of elemental sulfur must, however, be coupled to scavenging of the resulting sulfide, for example by metal oxides, in order to keep its concentration low and make disproportionation energetically favourable. This may limit its importance to restricted biogeochemical zones containing such scavengers. As described above, disproportionation of zero‐valent sulfur might be the physiology that links the deltaproteobacterial partner to the anaerobic methanotrophic archaea. Although still disputed, recent works have intriguingly proposed that distinct sulfur isotope signatures recorded in ancient marine sediment deposits (3.4 billion‐years‐old) are evidence that sulfur‐disproportionating metabolisms existed and possibly preceded sulfate‐reducing metabolism (Philippot et al., 2007; Wacey et al., 2011). Numbers of SCI‐disproportionating microorganisms in marine sediments have been estimated by most‐probable number counts, which showed numbers of up 107 cells per cm3 sediment capable of disproportionating thiosulfate (Jørgensen and Bak, 1991). Important to note, is that this does not necessarily reflect the in situ activity of the populations, because many of these populations are possibly SRM that can switch to disproportionating metabolisms when alternative electron donors/acceptors are lacking.
Many typical sulfur compound‐dissimilating anaerobes have been shown to also disproportionate SCI. These include some members of the deltaproteobacterial family Desulfobulbaceae (e.g. Desulfocapsa) and the deltaproteobacterial genera Desulfovibrio and Desulfomonile, the facultative anaerobic gammaproteobacterium Pantoea agglomerans, and few members of the phlya Thermodesulfobacteria and Firmicutes (Mohn and Tiedje, 1990; Finster et al., 1998; Jackson and McInerney, 2000; Obraztsova et al., 2002; Mardanov et al., 2016). Generally, our knowledge of SCI‐disproportionating microorganisms is purely based on cultivated strains, since no functional marker genes/enzymes can be used to unambiguously distinguish these metabolisms from other sulfur dissimilation pathways. Nevertheless, Desulfocapsa‐related bacteria can often be detected in marine sediments from various sites and appear to be prevalent in association with seagrasses, possibly reflecting higher amounts of SCI measured in such environments (Sun et al., 2015; Cúcio et al., 2016). In situ incubations combined with molecular surveys of microbial communities colonizing elemental sulfur particles suggested various Desulfobulbaceae may disproportionate elemental sulfur (Pjevac et al., 2014). Intriguingly, some bacteria (e.g. Desulfocapsa sulfexigens, Thermosulfurimonas dismutans) appear to be specialised SCI disproportionators. This is somewhat surprising considering genome analyses of these organisms revealed genes encoding for the canonical sulfate reduction pathway, although they cannot perform sulfate reduction (Finster et al., 2013; Mardanov et al., 2016). Nevertheless, for the disproportionation of thiosulfate and sulfite, biochemical analyses suggested they use pre‐existing enzyme systems typically used for reducing other sulfur compounds, while enzymes involved in elemental sulfur transformations are yet to be revealed (Finster, 2008). Together, these findings highlight the versatility of individual microorganisms to use various sulfur compounds and in differing ways under varied environmental conditions. In this way, the disproportionation of SCI could be an especially useful strategy to switch to when other electron donors and acceptors are limited. Additionally, this must be taken into account when interpreting the presence/expression of functional genes, which are often taken as straightforward indications for, e.g. sulfate reduction, since the individual canonical enzymes in sulfate reduction can be used for various dissimilating metabolisms.
Organo‐sulfur molecule transformations
Another important, yet largely overlooked component of the marine sediment sulfur cycle involves the utilisation and formation of organo‐sulfur molecules (OSM) (Fig. 1). OSM may include common cellular constituents such as cysteine, methionine, co‐enzymes and co‐factors, which together constitute a relatively small component of organic matter and will not be described further here, while other groups that constitute a greater proportion of organic matter include sulfonates (R‐ ) and ‘sulfated’ molecules i.e. sulfate esters/‐O‐sulfonates (R‐O‐ ). Together, OSM represent the second most important reduced sulfur pool in marine sediment environments, accounting for up to 35–80% of the reduced sulfur (Bruchert and Pratt, 1996; Passier et al., 1999). Sulfonated organics may comprise 20–40% of the organo‐sulfur in marine sediments (Vairavamurthy et al., 1994). Although no such studies have been conducted in marine sediments, studies in soils with isotopically labelled sulfur ‘tracers’ show constant fluxes of sulfur in and out of organic molecule pools, and these fluxes are thought to be primarily mediated by microorganisms (Ghani et al., 1993aa,b). The cleavage of sulfur moieties from organics by desulfonation may release oxidised sulfur compounds such as sulfite or sulfate, which can be either assimilated, converted and excreted, or utilised as electron acceptors by anaerobic microorganisms. Importantly, desulfonation may enable further catabolic degradation of the organic molecules, thereby allowing them to be used as nutrient and energy sources. This may therefore have important implications as to whether or not such organic matter is mineralised or buried. An immense diversity of sulfated organic molecules are known to exist in nature, including sulfated derivatives of major organic molecule classes such as (poly)saccharides, lipids, aminoglycans, polyaromatics, flavanoids and steroids (Barbeyron et al., 2016). A variety of microorganisms are known to desulfonate various organics via different enzymes (Table 1). For instance, members of the marine genus Rhodopirellula (phylum Planctomycetes) are desulfonating, polysaccharide‐degraders with a remarkable number (up to 110) sulfatase genes in their genomes, which appear to reflect a highly diverse substrate range (Glöckner et al., 2003; Wegner et al., 2013). Examples of sulfated polysaccharides include animal‐derived chondroitin sulfate, seaweed‐derived carrageenans, and algae‐derived fucoidan. High copy numbers of sulfatases appears to be a trait relatively widespread in the PVC‐superphylum (Planctomycetes–Verrucomicrobia–Chlamydia‐Lentisphaerae‐Omnitrophica) (Thrash et al., 2010). Intriguingly, genome comparisons among marine versus freshwater strains of the Verrucomicrobia suggest expansions of sulfatase genes in marine strains, likely reflecting the prevalence of these organic molecules in marine environments (Spring et al., 2016). Much that is known about desulfonating microorganisms in marine systems comes from aerobes, while the prevalence of these capabilities in anoxic marine sediments is essentially unknown. However, some SRM have the capacity to liberate sulfite from the small molecular weight compound taurine for anaerobic respiration (Lie et al., 1999). It is unknown if microorganisms are also able to desulfonate higher molecular weight compounds that may form during diagenetic reactions during sediment burial.
Indications for the ecological significance of OSM in marine environments is reflected in the genomes of microorganisms, whereby genes encoding enzymes for desulfonation of organics to enable further catabolism of the substrates appear to be common in pelagic and benthic marine bacteria (Glöckner et al., 2003; Woebken et al., 2007; Quaiser et al., 2011; Teeling et al., 2012). Additionally, our own examinations (unpublished data) of available subsurface Planctomycetes and Bacteroidetes genomes from tidal flat sediments (Baker et al., 2015), also uncovered that these taxa are enriched in sulfatase genes (up to 56 copies in one genome) and may be specialised for the degradation of sulfated organics in anoxic subsurface sediments.
Recent research is beginning to shed light on the importance of the diagenetic sulfurization of organics by reactive sulfides and other SCI produced by sulfur‐cycling microorganisms. Although not directly related to microbial physiology, it may have a large yet overlooked influence on ecology and biogeochemistry. Evidence from recent experiments with humic acid‐like substances at lower pH has shown reactions of sulfide with organic matter may be on par with re‐oxidation with oxygen or iron oxides (Yu et al., 2015). High resolution mass‐spectrometry‐based characterisation of dissolved organic matter in marine sediments has also directly shown a significant proportion of naturally occurring organic matter in sulfidic marine sediments can be sulfurized during early diagenesis, thereby increasing the molecular weight and diversity of the organic matter (Schmidt et al., 2017). This therefore means that the activity of SRM has a significant but little acknowledged chemical impact on the molecular structure of organic matter in marine sediments. This may also therefore pertain to the questions of whether or not certain organics are mineralised or buried, since changes in the molecular structure of organic molecules from sulfurization may mean they become unrecognisable to typical enzymes used to degrade the unmodified molecules. Interestingly, it could also be the case that some microorganisms have evolved enzymes to specially attack or dump electrons to such molecular bonds/structures, although these are completely unknown. These little studied aspects underline the highly interlinked nature of the carbon and sulfur cycles, and interesting future lines of research may relate to determining the degradability of such diagenetically‐formed OSM.
Concluding remarks
Contemporary research has revealed that remarkably diverse microorganisms have evolved to fulfil an array of niches in marine sediments by transforming sulfur compounds among various redox states (Fig. 1). It can therefore be asserted that sulfur is a major evolutionary and ecological driver of microbial life in the seafloor. From the recent deciphering of the molecular underpinnings of key biochemical pathways for fundamentally important sulfur metabolisms such as sulfate reduction, to developing understandings of the various highly interlinked biogeochemical implications associated with the sulfur cycle, we have now acquired a solid grasp about the importance of the sulfur cycle in one of the world's largest biospheres, i.e. the marine sedimentary seabed. While we have come a long way in developing this understanding, it becomes obvious that a great deal of exciting discoveries await to be made in the future that will greatly enhance our understanding of how sulfur sustains and influences microbial life in the marine seabed. We predict several major research avenues for the future are ripe for investigation. Prolific advances in linking molecular biological approaches such as strain‐resolution genome reconstruction from metagenomes, single‐cell genomics, and other molecular ‘omics’ technologies with biogeochemical and physiological processes are an obvious avenue where major progress in understanding the true diversity and revealing the metabolic capabilities of sulfur‐transforming microorganisms can be foreseen. Discovery and description of unknown sulfur‐metabolizing biochemical pathways, e.g. responsible for sulfate reduction in anaerobic methane‐oxidising archaea (Milucka et al., 2012), will be an important requirement for improved interpretation of ‘sulfur microbiomics’ data from environmental surveys. Research into the ecological and biogeochemical significance of the diverse, free‐living SOM populations will provide important perspectives on their individual environmental contributions. Additionally, exploration of SCI‐ and organo‐sulfur‐metabolizing microorganisms is needed to fully understand their diversity, ecological niches, and significance in the cycling of sulfur and other elements.
Acknowledgements
Claus Pelikan and Bela Hausmann are acknowledged for generating the DsrAB tree and highlighting sulfur enzymes, respectively. Big thanks go to all our past and present coworkers (Ivan Barisic, Stephan Duller, Diana‐Lebherz Eichinger, Stephanie Füreder, Bela Hausmann, Isabella Hinger, Albert Müller, Claus Pelikan, Michael Pester, Alexander Petritsch, Martina Putz, Doris Steger, Cecilia Wentrup) and longstanding collaborators (Bo Barker Jørgensen, Kasper Urup Kjeldsen, Andreas Schramm, Casey Hubert) on seabed sulfur microorganisms. This work was supported by the Austrian Science Fund FWF (projects P25111‐B22 to AL and P29426‐B22 to KW). The authors declare no conflict of interest
References
- Aoki, M. , Kakiuchi, R. , Yamaguchi, T. , Takai, K. , Inagaki, F. , and Imachi, H. (2015) Phylogenetic Diversity of aprA Genes in Subseafloor Sediments on the Northwestern Pacific Margin off Japan. Microbes Environ 30: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aüllo, T. , Ranchou‐Peyruse, A. , Ollivier, B. , and Magot, M. (2013) Desulfotomaculum spp. and related Gram‐positive sulfate‐reducing bacteria in deep subsurface environments. Front Microbiol 4: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badziong, W. , and Thauer, R.K. (1978) Growth yields and growth rates of Desulfovibrio vulgaris (Marburg) growing on hydrogen plus sulfate and hydrogen plus thiosulfate as the sole energy sources. Arch Microbiol 117: 209–214. [DOI] [PubMed] [Google Scholar]
- Bailey, J.V. , Salman, V. , Rouse, G.W. , Schulz‐Vogt, H.N. , Levin, L.A. , and Orphan, V.J. (2011) Dimorphism in methane seep‐dwelling ecotypes of the largest known bacteria. ISME J 5: 1926–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak, F. , and Cypionka, H. (1987) A novel type of energy‐metabolism involving fermentation of inorganic sulfur‐compounds. Nature 326: 891–892. [DOI] [PubMed] [Google Scholar]
- Baker, B.J. , Lazar, C.S. , Teske, A.P. , and Dick, G.J. (2015) Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 3: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford, V.A. , Bruno, S. , Rasmussen, T. , Appia‐Ayme, C. , Cheesman, M.R. , Berks, B.C. , and Hemmings, A.M. (2002) Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme. EMBO J 21: 5599–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeyron, T. , Potin, P. , Richard, C. , Collin, O. , and Kloareg, B. (1995) Arylsulphatase from Alteromonas carrageenovora . Microbiology 141: 2897–2904. [DOI] [PubMed] [Google Scholar]
- Barbeyron, T. , Brillet‐Gueguen, L. , Carre, W. , Carriere, C. , Caron, C. , Czjzek, M. , et al (2016) Matching the Diversity of Sulfated Biomolecules: Creation of a Classification Database for Sulfatases Reflecting Their Substrate Specificity. PLoS One 11: e0164846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardischewsky, F. , Quentmeier, A. , Rother, D. , Hellwig, P. , Kostka, S. , and Friedrich, C.G. (2005) Sulfur dehydrogenase of Paracoccus pantotrophus: the heme‐2 domain of the molybdoprotein cytochrome c complex is dispensable for catalytic activity. Biochemistry 44: 7024–7034. [DOI] [PubMed] [Google Scholar]
- Berner, R.A. (1982) Burial of organic‐carbon and pyrite sulfur in the modern ocean ‐ its geochemical and environmental significance. Am J Sci 282: 451–473. [Google Scholar]
- Bertics, V.J. , and Ziebis, W. (2010) Bioturbation and the role of microniches for sulfate reduction in coastal marine sediments. Environ Microbiol 12: 3022–3034. [DOI] [PubMed] [Google Scholar]
- Blazejak, A. , and Schippers, A. (2011) Real‐time PCR quantification and diversity analysis of the functional genes aprA and dsrA of sulfate‐reducing prokaryotes in marine sediments of the Peru continental margin and the Black Sea. Front Microbiol 2: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschker, H.T. , Vasquez‐Cardenas, D. , Bolhuis, H. , Moerdijk‐Poortvliet, T.W. , and Moodley, L. (2014) Chemoautotrophic carbon fixation rates and active bacterial communities in intertidal marine sediments. PLoS One 9: e101443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles, M.W. , Mogollon, J.M. , Kasten, S. , Zabel, M. , and Hinrichs, K.U. (2014) Global rates of marine sulfate reduction and implications for sub‐sea‐floor metabolic activities. Science 344: 889–891. [DOI] [PubMed] [Google Scholar]
- Bowman, J.P. , and McCuaig, R.D. (2003) Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl Environ Microbiol 69: 2463–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.P. , McCammon, S.A. , and Dann, A.L. (2005) Biogeographic and quantitative analyses of abundant uncultivated gamma‐proteobacterial clades from marine sediment. Microb Ecol 49: 451–460. [DOI] [PubMed] [Google Scholar]
- Brüchert, V. (1998) Early diagenesis of sulfur in estuarine sediments: The role of sedimentary humic and fulvic acids. Geochim Cosmochim Acta 62: 567–1586. [Google Scholar]
- Brüchert, V. , and Pratt, L.M. (1996) Contemporaneous early diagenetic formation of organic and inorganic sulfur in estuarine sediments from St Andrew Bay, Florida, USA. Geochim Cosmochim Acta 60: 2325–2332. [Google Scholar]
- Brunner, B. , Arnold, G.L. , Røy, H. , Müller, I.A. , and Jørgensen, B.B. (2016) Off limits: Sulfate below the sulfate‐methane transition. Front Earth Sci 4: [Google Scholar]
- Burdige, D.J. , and Nealson, K.H. (1986) Chemical and microbiological studies of sulfide‐mediated manganese reduction. Geomicrobiol J 4: 361–387. [Google Scholar]
- Burdorf, L.D.W. , Tramper, A. , Seitaj, D. , Meire, L. , Hidalgo‐Martinez, S. , Zetsche, E.M. , et al (2016) Long‐distance electron transport occurs globally in marine sediments. Biogeosciences Discuss 2016: 1–35. [Google Scholar]
- Burns, J.L. , and DiChristina, T.J. (2009) Anaerobic respiration of elemental sulfur and thiosulfate by Shewanella oneidensis MR‐1 requires psrA, a homolog of the phsA gene of Salmonella enterica serovar typhimurium LT2. Appl Environ Microbiol 75: 5209–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield, D.E. (1989) Reactive iron in marine sediments. Geochim Cosmochim Acta 53: 619–632. [DOI] [PubMed] [Google Scholar]
- Canfield, D.E. , and Thamdrup, B. (2009) Towards a consistent classification scheme for geochemical environments, or, why we wish the term 'suboxic' would go away. Geobiology 7: 385–392. [DOI] [PubMed] [Google Scholar]
- Canfield, D.E. , Kristensen, E. , and Thamdrup, B. (2005) Aquatic Geomicrobiology. Aquat Geomicrobiol 48: 1–640. [DOI] [PubMed] [Google Scholar]
- Canfield, D.E. , Stewart, F.J. , Thamdrup, B. , De Brabandere, L. , Dalsgaard, T. , Delong, E.F. et al. (2010) A Cryptic Sulfur Cycle in Oxygen‐Minimum‐Zone Waters off the Chilean Coast. Science 330: 1375–1378. [DOI] [PubMed] [Google Scholar]
- Cúcio, C. , Engelen, A.H. , Costa, R. , and Muyzer, G. (2016) Rhizosphere microbiomes of European + seagrasses are selected by the plant, but are not species specific. Front Microbiol 7: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison, J. , Brunel, F. , Phanopoulos, A. , Prozzi, D. , and Terpstra, P. (1992) Cloning and sequencing of Pseudomonas genes determining sodium dodecyl sulfate biodegradation. Gene 114: 19–24. [DOI] [PubMed] [Google Scholar]
- Denger, K. , Mayer, J. , Buhmann, M. , Weinitschke, S. , Smits, T.H. , and Cook, A.M. (2009) Bifurcated degradative pathway of 3‐sulfolactate in Roseovarius nubinhibens ISM via sulfoacetaldehyde acetyltransferase and (S)‐cysteate sulfolyase. J Bacteriol 191: 5648–5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denger, K. , Weiss, M. , Felux, A.K. , Schneider, A. , Mayer, C. , Spiteller, D. , et al (2014) Sulphoglycolysis in Escherichia coli K‐12 closes a gap in the biogeochemical sulphur cycle. Nature 507: 114–117. [DOI] [PubMed] [Google Scholar]
- Denkmann, K. , Grein, F. , Zigann, R. , Siemen, A. , Bergmann, J. , van Helmont, S. , et al (2012) Thiosulfate dehydrogenase: a widespread unusual acidophilic c‐type cytochrome. Environ Microbiol 14: 2673–2688. [DOI] [PubMed] [Google Scholar]
- Dolata, M.M. , Van Beeumen, J.J. , Ambler, R.P. , Meyer, T.E. , and Cusanovich, M.A. (1993) Nucleotide sequence of the heme subunit of flavocytochrome c from the purple phototrophic bacterium, Chromatium vinosum. A 2.6‐kilobase pair DNA fragment contains two multiheme cytochromes, a flavoprotein, and a homolog of human ankyrin. J Biol Chem 268: 14426–14431. [PubMed] [Google Scholar]
- Dorries, M. , Wohlbrand, L. , and Rabus, R. (2016a) Differential proteomic analysis of the metabolic network of the marine sulfate‐reducer Desulfobacterium autotrophicum HRM2. Proteomics 16: 2878–2893. [DOI] [PubMed] [Google Scholar]
- Dorries, M. , Wohlbrand, L. , Kube, M. , Reinhardt, R. , and Rabus, R. (2016b) Genome and catabolic subproteomes of the marine, nutritionally versatile, sulfate‐reducing bacterium Desulfococcus multivorans DSM 2059. BMC Genomics 17: 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Z.J. , Wang, Z.J. , Zhao, J.X. , and Chen, G.J. (2016) Woeseia oceani gen. nov., sp. nov., a chemoheterotrophic member of the order Chromatiales, and proposal of Woeseiaceae fam. nov. Int J Syst Evol Microbiol 66: 107–112. [DOI] [PubMed] [Google Scholar]
- Dubilier, N. , Bergin, C. , and Lott, C. (2008) Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol 6: 725–740. [DOI] [PubMed] [Google Scholar]
- Dyksma, S. , Bischof, K. , Fuchs, B.M. , Hoffmann, K. , Meier, D. , Meyerdierks, A. , et al, (2016) Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J 10: 1939–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felux, A.K. , Spiteller, D. , Klebensberger, J. , and Schleheck, D. (2015) Entner‐Doudoroff pathway for sulfoquinovose degradation in Pseudomonas putida SQ1. Proc Natl Acad Sci USA 112: E4298–E4305. 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdelman, T.G. , Fossing, H. , Neumann, K. , and Schulz, H.D. (1999) Sulfate reduction in surface sediments of the southeast Atlantic continental margin between 15 degrees 38'S and 27 degrees 57'S (Angola and Namibia). Limnol Oceanogr 44: 650–661. [Google Scholar]
- Finster, K. (2008) Microbiological disproportionation of inorganic sulfur compounds. J Sulfur Chem 29: 281–292. [Google Scholar]
- Finster, K. , Liesack, W. , and Thamdrup, B. (1998) Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp. nov., a new anaerobic bacterium isolated from marine surface sediment. Appl Environ Microbiol 64: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finster, K.W. , Kjeldsen, K.U. , Kube, M. , Reinhardt, R. , Mussmann, M. , Amann, R. , and Schreiber, L. (2013) Complete genome sequence of Desulfocapsa sulfexigens, a marine deltaproteobacterium specialized in disproportionating inorganic sulfur compounds. Stand Genomic Sci 8: 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentino, A.P. , Weijma, J. , Stams, A.J.M. , and Sánchez‐Andrea, I. (2016) Ecophysiology and Application of Acidophilic Sulfur‐Reducing Microorganisms In Biotechnology of Extremophiles: Advances and Challenges. Rampelotto P.H. (ed.). Switzerland: Springer International Publishing. [Google Scholar]
- Fossing, H. , and Jørgensen, B.B. (1990) Oxidation and reduction of radiolabeled inorganic sulfur‐compounds in an estuarine sediment, Kysing Fjord, Denmark. Geochim Cosmochim Acta 54: 2731–2742. [Google Scholar]
- Gavel, O.Y. , Bursakov, S.A. , Calvete, J.J. , George, G.N. , Moura, J.J. , and Moura, I. (1998) ATP sulfurylases from sulfate‐reducing bacteria of the genus Desulfovibrio. A novel metalloprotein containing cobalt and zinc. Biochemistry 37: 16225–16232. [DOI] [PubMed] [Google Scholar]
- Ghani, A. , Mclaren, R.G. , and Swift, R.S. (1993a) Mobilization of Recently‐Formed Soil Organic Sulfur. Soil Biol Biochem 25: 1739–1744. [Google Scholar]
- Ghani, A. , Mclaren, R.G. , and Swift, R.S. (1993b) The incorporation and transformations of sulfur‐35 in soil: Effects of soil conditioning and glucose or sulphate additions. Soil Biol Biochem 25: 327–335. [Google Scholar]
- Glöckner, F.O. , Kube, M. , Bauer, M. , Teeling, H. , Lombardot, T. , Ludwig, W. , et al (2003) Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci USA 100: 8298–8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glombitza, C. , Jaussi, M. , Roy, H. , Seidenkrantz, M.S. , Lomstein, B.A. , and Jørgensen, B.B. (2015) Formate, acetate, and propionate as substrates for sulfate reduction in sub‐arctic sediments of Southwest Greenland. Front Microbiol 6: 846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glombitza, C. , Adhikari, R.R. , Riedinger, N. , Gilhooly, W.P. , Hinrichs, K.U. , and Inagaki, F. (2016) Microbial sulfate reduction potential in coal‐bearing sediments down to ∼2.5 km below the Seafloor off Shimokita Peninsula, Japan. Front Microbiol 7: 1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein, F. , Pereira, I.A. , and Dahl, C. (2010) Biochemical characterization of individual components of the Allochromatium vinosum DsrMKJOP transmembrane complex aids understanding of complex function in vivo . J Bacteriol 192: 6369–6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, S.R. , Best, M.D. , and Wong, C.H. (2004) Sulfatases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed Engl 43: 5736–5763. [DOI] [PubMed] [Google Scholar]
- Harel, A. , Haggblom, M.M. , Falkowski, P.G. , and Yee, N. (2016) FEMS Microbiol Ecol 92: fiw187. [DOI] [PubMed] [Google Scholar]
- Hawley, A.K. , Brewer, H.M. , Norbeck, A.D. , Pasa‐Tolic, L. , and Hallam, S.J. (2014) Metaproteomics reveals differential modes of metabolic coupling among ubiquitous oxygen minimum zone microbes. Proc Natl Acad Sci USA 111: 11395–11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitmann, T. , and Blodau, C. (2006) Oxidation and incorporation of hydrogen sulfide by dissolved organic matter. Chem Geol 235: 12–20. [Google Scholar]
- Hensel, M. , Hinsley, A.P. , Nikolaus, T. , Sawers, G. , and Berks, B.C. (1999) The genetic basis of tetrathionate respiration in Salmonella typhimurium . Mol Microbiol 32: 275–287. [DOI] [PubMed] [Google Scholar]
- Hittel, D.S. , and Voordouw, G. (2000) Overexpression, purification and immunodetection of DsrD from Desulfovibrio vulgaris Hildenborough. A Van Leeuw J Microb 77: 271–280. [DOI] [PubMed] [Google Scholar]
- Holmkvist, L. , Ferdelman, T.G. , and Jørgensen, B.B. (2011) A cryptic sulfur cycle driven by iron in the methane zone of marine sediment (Aarhus Bay, Denmark). Geochim Cosmochim Acta 75: 3581–3599. [Google Scholar]
- Huang, C.J. , and Barrett, E.L. (1990) Identification and cloning of genes involved in anaerobic sulfite reduction by Salmonella typhimurium . J Bacteriol 172: 4100–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert, C. , Loy, A. , Nickel, M. , Arnosti, C. , Baranyi, C. , Brüchert, V. , et al (2009) A constant flux of diverse thermophilic bacteria into the Cold Arctic Seabed. Science 325: 1541–1544. [DOI] [PubMed] [Google Scholar]
- Hug, L.A. , Thomas, B.C. , Sharon, I. , Brown, C.T. , Sharma, R. , Hettich, R.L. , et al (2016) Critical biogeochemical functions in the subsurface are associated with bacteria from new phyla and little studied lineages. Environ Microbiol 18: 159–173. [DOI] [PubMed] [Google Scholar]
- Jackson, B.E. , and McInerney, M.J. (2000) Thiosulfate disproportionation by Desulfotomaculum thermobenzoicum . Appl Environ Microbiol 66: 3650–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S.I. , and Kühl, M. , Priemé, A. (2007) Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina . FEMS Microbiol Ecol 62: 108–117. [DOI] [PubMed] [Google Scholar]
- Johnson, E.F. , and Mukhopadhyay, B. (2005) A new type of sulfite reductase, a novel coenzyme F420‐dependent enzyme, from the methanarchaeon Methanocaldococcus jannaschii . J Biol Chem 280: 38776–38786. [DOI] [PubMed] [Google Scholar]
- Johnston, A.W. , Green, R.T. , and Todd, J.D. (2016) Enzymatic breakage of dimethylsulfoniopropionate‐a signature molecule for life at sea. Curr Opin Chem Biol 31: 58–65. [DOI] [PubMed] [Google Scholar]
- Jørgensen, B.B. (1982) Mineralization of organic matter in the sea bed ‐ the role of sulphate reduction. Nature 296: 643–645. [Google Scholar]
- Jørgensen, B.B. (1990) A thiosulfate shunt in the sulfur cycle of marine‐sediments. Science 249: 152–154. [DOI] [PubMed] [Google Scholar]
- Jørgensen, B.B. (2000) Bacteria and marine biogeochemistry In Marine Geochemistry. Schulz H.D., and Zabel M. (eds). Berlin, Germany: Springer‐Verlag, pp. 173–208. [Google Scholar]
- Jørgensen, B.B. , and Bak, F. (1991) Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (Kattegat, Denmark). Appl Environ Microbiol 57: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen, B.B. , and Nelson, D.C. (2004) Sulfide oxidation in marine sediments: Geochemistry meets microbiology. Geol Soc Am Spec Pap 379: 63–81. [Google Scholar]
- Jørgensen, B.B. , and Kasten, S. (2006) Sulfur cycling and methane oxidation In Marine Geochemistry. Schulz H.D., and Zabel M. (eds). Berlin, Germany: Springer‐Verlag, pp. 271–309. [Google Scholar]
- Kahnert, A. , and Kertesz, M.A. (2000) Characterization of a sulfur‐regulated oxygenative alkylsulfatase from Pseudomonas putida S‐313. J Biol Chem 275: 31661–31667. [DOI] [PubMed] [Google Scholar]
- Kanao, T. , Kamimura, K. , and Sugio, T. (2007) Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans . J Biotechnol 132: 16–22. [DOI] [PubMed] [Google Scholar]
- Kappler, U. , Bennett, B. , Rethmeier, J. , Schwarz, G. , Deutzmann, R. , McEwan, A.G. , and Dahl, C. (2000) Sulfite:Cytochrome c oxidoreductase from Thiobacillus novellus. Purification, characterization, and molecular biology of a heterodimeric member of the sulfite oxidase family. J Biol Chem 275: 13202–13212. [DOI] [PubMed] [Google Scholar]
- Kern, M. , Klotz, M.G. , and Simon, J. (2011) The Wolinella succinogenes mcc gene cluster encodes an unconventional respiratory sulphite reduction system. Mol Microbiol 82: 1515–1530. [DOI] [PubMed] [Google Scholar]
- Kleindienst, S. , Herbst, F.A. , Stagars, M. , von Netzer, F. , von Bergen, M. , Seifert, J. , et al, (2014) Diverse sulfate‐reducing bacteria of the Desulfosarcina/Desulfococcus clade are the key alkane degraders at marine seeps. ISME J 8: 2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafft, T. , Bokranz, M. , Klimmek, O. , Schröder, I. , Fahrenholz, F. , Kojro, E. , and Kröger, A. (1992) Cloning and nucleotide sequence of the psrA gene of Wolinella succinogenes polysulphide reductase. Eur J Biochem 206: 503–510. [DOI] [PubMed] [Google Scholar]
- Krämer, M. , and Cypionka, H. (1989) Sulfate formation via ATP sulfurylase in thiosulfate‐ and sulfite‐disproportionating bacteria. Arch Microbiol 151: 232–237. [Google Scholar]
- Krukenberg, V. , Harding, K. , Richter, M. , Glöckner, F.O. , Gruber‐Vodicka, H.R. , Adam, B. , et al (2016) Candidatus Desulfofervidus auxilii, a hydrogenotrophic sulfate‐reducing bacterium involved in the thermophilic anaerobic oxidation of methane. Environ Microbiol 18: 3073–3091. [DOI] [PubMed] [Google Scholar]
- Kurth, J.M. , Brito, J.A. , Reuter, J. , Flegler, A. , Koch, T. , Franke, T. , et al (2016) Electron accepting units of the diheme cytochrome c TsdA, a bifunctional thiosulfate dehydrogenase/tetrathionate reductase. J Biol Chem 291: 24804–24818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampreia, J. , Pereira, A.S. , and Moura, J.J.G. (1994) Adenylylsulfate reductases from sulfate‐reducing bacteria In Methods in Enzymology. Peck H.D. (ed.). Academic Press, pp. 241–260. [Google Scholar]
- Laso‐Perez, R. , Wegener, G. , Knittel, K. , Widdel, F. , Harding, K.J. , Krukenberg, V. , et al, (2016) Thermophilic archaea activate butane via alkyl‐coenzyme M formation. Nature 539: 396–401. [DOI] [PubMed] [Google Scholar]
- Lavik, G. , Stührmann, T. , Brüchert, V. , Van der Plas, A. , Mohrholz, V. , Lam, P. , et al (2009) Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature 457: 581–584. [DOI] [PubMed] [Google Scholar]
- Lazar, C.S. , Baker, B.J. , Seitz, K.W. , and Teske, A.P. (2017) Genomic reconstruction of multiple lineages of uncultured benthic archaea suggests distinct biogeochemical roles and ecological niches. ISME J. 11: 1058–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloup, J. , Loy, A. , Knab, N.J. , Borowski, C. , Wagner, M. , and Jørgensen, B.B. (2007) Diversity and abundance of sulfate‐reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ Microbiol 9: 131–142. [DOI] [PubMed] [Google Scholar]
- Leloup, J. , Fossing, H. , Kohls, K. , Holmkvist, L. , Borowski, C. , and Jørgensen, B.B. (2009) Sulfate‐reducing bacteria in marine sediment (Aarhus Bay, Denmark): abundance and diversity related to geochemical zonation. Environ Microbiol 11: 1278–1291. [DOI] [PubMed] [Google Scholar]
- Lenk, S. , Arnds, J. , Zerjatke, K. , Musat, N. , Amann, R. , and Mussmann, M. (2011) Novel groups of Gammaproteobacteria catalyse sulfur oxidation and carbon fixation in a coastal, intertidal sediment. Environ Microbiol 13: 758–774. [DOI] [PubMed] [Google Scholar]
- Lenk, S. , Moraru, C. , Hahnke, S. , Arnds, J. , Richter, M. , Kube, M. , et al (2012) Roseobacter clade bacteria are abundant in coastal sediments and encode a novel combination of sulfur oxidation genes. ISME J 6: 2178–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie, T.J. , Clawson, M.L. , Godchaux, W. , and Leadbetter, E.R. (1999) Sulfidogenesis from 2‐aminoethanesulfonate (taurine) fermentation by a morphologically unusual sulfate‐reducing bacterium, Desulforhopalus singaporensis sp. nov. Appl Environ Microbiol 65: 3328–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.W. , Denkmann, K. , Kosciow, K. , Dahl, C. , and Kelly, D.J. (2013) Tetrathionate stimulated growth of Campylobacter jejuni identifies a new type of bi‐functional tetrathionate reductase (TsdA) that is widely distributed in bacteria. Mol Microbiol 88: 173–188. [DOI] [PubMed] [Google Scholar]
- Lovley, D.R. (2016) Happy together: microbial communities that hook up to swap electrons. ISME J 11: 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübbe, Y.J. , Youn, H.S. , Timkovich, R. , and Dahl, C. (2006) Siro(haem)amide in Allochromatium vinosum and relevance of DsrL and DsrN, a homolog of cobyrinic acid a,c‐diamide synthase, for sulphur oxidation. FEMS Microbiol Lett 261: 194–202. [DOI] [PubMed] [Google Scholar]
- Luther, G.W. , Findlay, A.J. , Macdonald, D.J. , Owings, S.M. , Hanson, T.E. , Beinart, R.A. , and Girguis, P.R. (2011) Thermodynamics and kinetics of sulfide oxidation by oxygen: a look at inorganically controlled reactions and biologically mediated processes in the environment. Front Microbiol 2: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, K. , Weiss, R. , and Adams, M.W. (2000) Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J Bacteriol 182: 1864–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, K. , Schicho, R.N. , Kelly, R.M. , and Adams, M.W.W. (1993) Hydrogenase of the Hyperthermophile Pyrococcus furiosus Is an Elemental Sulfur Reductase or Sulfhydrogenase ‐ Evidence for a Sulfur‐Reducing Hydrogenase Ancestor. Proc Natl Acad Sci USA 90: 5341–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor, B.J. , Biddle, J.F. , Harbort, C. , Matthysse, A.G. , and Teske, A. (2013) Sulfide oxidation, nitrate respiration, carbon acquisition, and electron transport pathways suggested by the draft genome of a single orange Guaymas Basin Beggiatoa (Cand. Maribeggiatoa) sp. filament. Mar Genomics 11: 53–65. [DOI] [PubMed] [Google Scholar]
- Malkin, S.Y. , Rao, A.M. , Seitaj, D. , Vasquez‐Cardenas, D. , Zetsche, E.M. , Hidalgo‐Martinez, S. , et al (2014) Natural occurrence of microbial sulphur oxidation by long‐range electron transport in the seafloor. ISME J 8: 1843–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardanov, A.V. , Beletsky, A.V. , Kadnikov, V.V. , Slobodkin, A.I. , and Ravin, N.V. (2016) Genome Analysis of Thermosulfurimonas dismutans, the first thermophilic sulfur‐disproportionating bacterium of the phylum Thermodesulfobacteria . Front Microbiol 7: 950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzocchi, U. , Trojan, D. , Larsen, S. , Meyer, R.L. , Revsbech, N.P. , Schramm, A. , et al (2014) Electric coupling between distant nitrate reduction and sulfide oxidation in marine sediment. ISME J 8: 1682–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn, S.E. , Chadwick, G.L. , Kempes, C.P. , and Orphan, V.J. (2015) Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526: 531–535. [DOI] [PubMed] [Google Scholar]
- Meulepas, R.J.W. , Jagersma, C.G. , Khadem, A.F. , Stams, A.J.M. , and Lens, P.N.L. (2010) Effect of methanogenic substrates on anaerobic oxidation of methane and sulfate reduction by an anaerobic methanotrophic enrichment. Appl Microbiol Biotechnol 87: 1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelburg, J.J. (2011) Chemoautotrophy in the ocean. Geophys Res Lett 38: 1–4. [Google Scholar]
- Milucka, J. , Ferdelman, T.G. , Polerecky, L. , Franzke, D. , Wegener, G. , Schmid, M. , et al (2012) Zero‐valent sulphur is a key intermediate in marine methane oxidation. Nature 491: 541–546. [DOI] [PubMed] [Google Scholar]
- Mohn, W.W. , and Tiedje, J.M. (1990) Catabolic thiosulfate disproportionation and carbon dioxide reduction in strain DCB‐1, a reductively dechlorinating anaerobe. J Bacteriol 172: 2065–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat, C.G. , Rothery, E. , Miles, C.S. , McIver, L. , Doherty, M.K. , Drewette, K. , et al (2004) Octaheme tetrathionate reductase is a respiratory enzyme with novel heme ligation. Nat Struct Mol Biol 11: 1023–1024. [DOI] [PubMed] [Google Scholar]
- Müller, A.L. , Kjeldsen, K.U. , Rattei, T. , Pester, M. , and Loy, A. (2015) Phylogenetic and environmental diversity of DsrAB‐type dissimilatory (bi) sulfite reductases. ISME J 9: 1152–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussmann, M. , Pjevac, P. , Krüger, K. , and Dyksma, S. (2017) Genomic repertoire of the Woeseiaceae/JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J. 11: 1276–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na, H. , Lever, M.A. , Kjeldsen, K.U. , Schulz, F. , and Jørgensen, B.B. (2015) Uncultured Desulfobacteraceae and Crenarchaeotal group C3 incorporate 13C‐acetate in coastal marine sediment. Environ Microbiol Rep 7: 614–622. [DOI] [PubMed] [Google Scholar]
- Nelson, D.C. , Jorgensen, B.B. , and Revsbech, N.P. (1986) Growth Pattern and Yield of a Chemoautotrophic Beggiatoa sp. in Oxygen‐Sulfide Microgradients. Appl Environ Microbiol 52: 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, L.P. (2016) Ecology: Electrical cable bacteria save marine life. Curr Biol 26: R32–R33. [DOI] [PubMed] [Google Scholar]
- Nielsen, L.P. , Risgaard‐Petersen, N. , Fossing, H. , Christensen, P.B. , and Sayama, M. (2010) Electric currents couple spatially separated biogeochemical processes in marine sediment. Nature 463: 1071–1074. [DOI] [PubMed] [Google Scholar]
- Obraztsova, A.Y. , Francis, C.A. , and Tebo, B.M. (2002) Sulfur disproportionation by the facultative anaerobe Pantoea agglomerans SP1 as a mechanism for chromium(VI) reduction. Geomicrobiol J 19: 121–132. [Google Scholar]
- Oliveira, T.F. , Vonrhein, C. , Matias, P.M. , Venceslau, S.S. , Pereira, I.A. , and Archer, M. (2008) Purification, crystallization and preliminary crystallographic analysis of a dissimilatory DsrAB sulfite reductase in complex with DsrC. J Struct Biol 164: 236–239. [DOI] [PubMed] [Google Scholar]
- Orsi, W.D. , Jørgensen, B.B. , and Biddle, J.F. (2016) Transcriptional analysis of sulfate reducing and chemolithoautotrophic sulfur oxidizing bacteria in the deep subseafloor. Environ Microbiol Rep 8: 452–460. [DOI] [PubMed] [Google Scholar]
- Passier, H.F. , Böttcher, M.E. , and De Lange, G.J. (1999) Sulphur enrichment in organic matter of eastern Mediterranean sapropels: A study of sulphur isotope partitioning. Aquat Geochem 5: 99–118. [Google Scholar]
- Petersen, J.M. , Kemper, A. , Gruber‐Vodicka, H. , Cardini, U. , van der Geest, M. , Kleiner, M. , et al (2016) Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation. Nat Microbiol 2: 16195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer, C. , Larsen, S. , Song, J. , Dong, M. , Besenbacher, F. , Meyer, R.L. , et al (2012) Filamentous bacteria transport electrons over centimetre distances. Nature 491: 218–221. [DOI] [PubMed] [Google Scholar]
- Pham, V.H. , Yong, J.J. , Park, S.J. , Yoon, D.N. , Chung, W.H. , and Rhee, S.K. (2008) Molecular analysis of the diversity of the sulfide:quinone reductase (sqr) gene in sediment environments. Microbiology 154: 3112–3121. [DOI] [PubMed] [Google Scholar]
- Philippot, P. , Van Zuilen, M. , Lepot, K. , Thomazo, C. , Farquhar, J. , and Van Kranendonk, M.J. (2007) Early Archaean microorganisms preferred elemental sulfur, not sulfate. Science 317: 1534–1537. [DOI] [PubMed] [Google Scholar]
- Pjevac, P. , Kamyshny, A., Jr. , Dyksma, S. , and Mussmann, M. (2014) Microbial consumption of zero‐valence sulfur in marine benthic habitats. Environ Microbiol 16: 3416–3430. [DOI] [PubMed] [Google Scholar]
- Plugge, C.M. , Zhang, W.W. , Scholten, J.C.M. , and Stams, A.J.M. (2011) Metabolic flexibility of sulfate‐reducing bacteria. Front Microbiol 2: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton, S.W. , Krom, M.D. , and Raiswell, R. (2004) A revised scheme for the reactivity of iron (oxyhydr)oxide minerals towards dissolved sulfide. Geochim Cosmochim Acta 68: 3703–3715. [Google Scholar]
- Prange, A. , Engelhardt, H. , Trüper, H.G. , and Dahl, C. (2004) The role of the sulfur globule proteins of Allochromatium vinosum: mutagenesis of the sulfur globule protein genes and expression studies by real‐time RT‐PCR. Arch Microbiol 182: 165–174. [DOI] [PubMed] [Google Scholar]
- Prokopenko, M.G. , Hirst, M.B. , De Brabandere, L. , Lawrence, D.J. , Berelson, W.M. , Granger, J. , et al (2013) Nitrogen losses in anoxic marine sediments driven by Thioploca‐anammox bacterial consortia. Nature 500: 194–198. [DOI] [PubMed] [Google Scholar]
- Quaiser, A. , Zivanovic, Y. , Moreira, D. , and Lopez‐Garcia, P. (2011) Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J 5: 285–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentmeier, A. , and Friedrich, C.G. (2001) The cysteine residue of the SoxY protein as the active site of protein‐bound sulfur oxidation of Paracoccus pantotrophus GB17. FEBS Lett 503: 168–172. [DOI] [PubMed] [Google Scholar]
- Quentmeier, A. , Hellwig, P. , Bardischewsky, F. , Grelle, G. , Kraft, R. , and Friedrich, C.G. (2003) Sulfur oxidation in Paracoccus pantotrophus: interaction of the sulfur‐binding protein SoxYZ with the dimanganese SoxB protein. Biochem Biophys Res Commun 312: 1011–1018. [DOI] [PubMed] [Google Scholar]
- Rabus, R. , Venceslau, S.S. , Wöhlbrand, L. , Voordouw, G. , Wall, J.D. , and Pereira, I.A. (2015) A Post‐Genomic View of the Ecophysiology, Catabolism and Biotechnological Relevance of Sulphate‐Reducing Prokaryotes. Adv Microb Physiol 66: 55–321. [DOI] [PubMed] [Google Scholar]
- Ramos, A.R. , Keller, K.L. , Wall, J.D. , and Pereira, I.A.C. (2012) The membrane QmoABC complex interacts directly with the dissimilatory adenosine 5 ′‐phosphosulfate reductase sulfate reducing bacteria. Front Microbiol 3: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenschlag, K. , Sahm, K. , and Amann, R. (2001) Quantitative molecular analysis of the microbial community in marine Arctic sediments (Svalbard). Appl Environ Microbiol 67: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenschlag, K. , Sahm, K. , Pernthaler, J. , and Amann, R. (1999) High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol 65: 3982–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravot, G. , Casalot, L. , Ollivier, B. , Loison, G. , and Magot, M. (2005) rdlA, a new gene encoding a rhodanese‐like protein in Halanaerobium congolense and other thiosulfate‐reducing anaerobes. Res Microbiol 156: 1031–1038. [DOI] [PubMed] [Google Scholar]
- Rein, U. , Gueta, R. , Denger, K. , Ruff, J. , Hollemeyer, K. , and Cook, A.M. (2005) Dissimilation of cysteate via 3‐sulfolactate sulfo‐lyase and a sulfate exporter in Paracoccus pantotrophus NKNCYSA. Microbiology 151: 737–747. [DOI] [PubMed] [Google Scholar]
- Risgaard‐Petersen, N. , Revil, A. , Meister, P. , and Nielsen, L.P. (2012) Sulfur, iron‐, and calcium cycling associated with natural electric currents running through marine sediment. Geochim Cosmochim Acta 92: 1–13. [Google Scholar]
- Robador, A. , Muller, A.L. , Sawicka, J.E. , Berry, D. , Hubert, C.R.J. , Loy, A. , et al (2016) Activity and community structures of sulfate‐reducing microorganisms in polar, temperate and tropical marine sediments. ISME J 10: 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel, E.G. , Cragg, B.A. , Webster, G. , Sass, H. , Tang, X.H. , Williams, A.S. , et al (2015) Complex coupled metabolic and prokaryotic community responses to increasing temperatures in anaerobic marine sediments: critical temperatures and substrate changes. FEMS Microbiol Ecol 91: fiv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman, V. , Amann, R. , Girnth, A.C. , Polerecky, L. , Bailey, J.V. , Hogslund, S. , et al (2011) A single‐cell sequencing approach to the classification of large, vacuolated sulfur bacteria. Syst Appl Microbiol 34: 243–259. [DOI] [PubMed] [Google Scholar]
- Santos, A.A. , Venceslau, S.S. , Grein, F. , Leavitt, W.D. , Dahl, C. , Johnston, D.T. , and Pereira, I.A.C. (2015) A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science 350: 1541–1545. [DOI] [PubMed] [Google Scholar]
- Schauder, R. , and Kröger, A. (1993) Bacterial sulfur respiration. Archiv Microbiol 159: 491–497. [Google Scholar]
- Scheller, S. , Yu, H. , Chadwick, G.L. , McGlynn, S.E. , and Orphan, V.J. (2016) Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351: 703–707. [DOI] [PubMed] [Google Scholar]
- Schmidt, F. , Koch, B.P. , Goldhammer, T. , Elvert, M. , Witt, M. , Lin, Y.S. , et al (2017) Unraveling signatures of biogeochemical processes and the depositional setting in the molecular composition of pore water DOM across different marine environments. Geochim Cosmochim Acta. Advance online publication. [Google Scholar]
- Schrenk, M.O. , Huber, J.A. , and Edwards, K.J. (2010) Microbial provinces in the subseafloor. Ann Rev Mar Sci 2: 279–304. [DOI] [PubMed] [Google Scholar]
- Schütz, M. , Shahak, Y. , Padan, E. , and Hauska, G. (1997) Sulfide‐quinone reductase from Rhodobacter capsulatus. Purification, cloning, and expression. J Biol Chem 272: 9890–9894. [DOI] [PubMed] [Google Scholar]
- Seidel, M. , Beck, M. , Riedel, T. , Waska, H. , Suryaputra, I.G.N.A. , Schnetger, B. et al (2014) Biogeochemistry of dissolved organic matter in an anoxic intertidal creek bank. Geochim Cosmochim Acta 140: 418–434. [Google Scholar]
- Seitaj, D. , Schauer, R. , Sulu‐Gambari, F. , Hidalgo‐Martinez, S. , Malkin, S.Y. , Burdorf, L.D. , et al (2015) Cable bacteria generate a firewall against euxinia in seasonally hypoxic basins. Proc Natl Acad Sci USA 112: 13278–13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz, K.W. , Lazar, C.S. , Hinrichs, K.U. , Teske, A.P. , and Baker, B.J. (2016) Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J 10: 1696–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, J. , and Kroneck, P.M. (2013) Microbial sulfite respiration. Adv Microb Physiol 62: 45–117. [DOI] [PubMed] [Google Scholar]
- Simon, J. , Kern, M. , Hermann, B. , Einsle, O. , and Butt, J.N. (2011) Physiological function and catalytic versatility of bacterial multihaem cytochromes c involved in nitrogen and sulfur cycling. Biochem Soc Trans 39: 1864–1870. [DOI] [PubMed] [Google Scholar]
- Sinninge Damsté, J.S. , Irene, W. , Rijpstra, C. , de Leeuw, J.W. , and Schenck, P.A. (1988) Origin of organic sulphur compounds and sulphur‐containing high molecular weight substances in sediments and immature crude oils. Org Geochem 13: 593–606. [Google Scholar]
- Spring, S. , Bunk, B. , Sproer, C. , Schumann, P. , Rohde, M. , Tindall, B.J. , and Klenk, H.P. (2016) Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J 10: 2801–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdreher, Y. , Venceslau, S.S. , Josten, M. , Sahl, H.G. , Pereira, I.A. , and Dahl, C. (2012) Cytoplasmic sulfur transferases in the purple sulfur bacterium Allochromatium vinosum: evidence for sulfur transfer from DsrEFH to DsrC. PLoS One 7: e40785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdreher, Y. , Sturm, M. , Josten, M. , Sahl, H.G. , Dobler, N. , Zigann, R. , and Dahl, C. (2014) New proteins involved in sulfur trafficking in the cytoplasm of Allochromatium vinosum . J Biol Chem 289: 12390–12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulu‐Gambari, F. , Seitaj, D. , Meysman, F.J. , Schauer, R. , Polerecky, L. , and Slomp, C.P. (2016) Cable Bacteria Control Iron‐Phosphorus Dynamics in Sediments of a Coastal Hypoxic Basin. Environ Sci Technol 50: 1227–1233. [DOI] [PubMed] [Google Scholar]
- Sun, F. , Zhang, X. , Zhang, Q. , Liu, F. , Zhang, J. , and Gong, J. (2015) Seagrass (Zostera marina) colonization promotes the accumulation of diazotrophic bacteria and alters the relative abundances of specific bacterial lineages involved in benthic carbon and sulfur cycling. Appl Environ Microbiol 81: 6901–6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpgaard, I.H. , Roy, H. , and Jørgensen, B.B. (2011) Concurrent low‐ and high‐affinity sulfate reduction kinetics in marine sediment. Geochim Cosmochim Acta 75: 2997–3010. [Google Scholar]
- Teeling, H. , Fuchs, B.M. , Becher, D. , Klockow, C. , Gardebrecht, A. , Bennke, C.M. , et al (2012) Substrate‐controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 608–611. [DOI] [PubMed] [Google Scholar]
- Teske, A. , and Salman, V. (2014) The Family Beggiatoaceae In The Prokaryotes: Gammaproteobacteria. Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., and Thompson F. (eds). Berlin, Heidelberg: Springer Berlin Heidelberg, pp. 93–134. [Google Scholar]
- Thamdrup, B. , Finster, K. , Hansen, J.W. , and Bak, F. (1993) Bacterial disproportionation of elemental sulfur coupled to chemical reduction of iron or manganese. Appl Environ Microbiol 59: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamdrup, B. , Finster, K. , Fossing, H. , Hansen, J.W. , and Jørgensen, B.B. (1994) Thiosulfate and sulfite distributions in porewater of marine‐sediments related to manganese, iron, and sulfur geochemistry. Geochim Cosmochim Acta 58: 67–73. [Google Scholar]
- Thomas, F. , Giblin, A.E. , Cardon, Z.G. , and Sievert, S.M. (2014) Rhizosphere heterogeneity shapes abundance and activity of sulfur‐oxidizing bacteria in vegetated salt marsh sediments. Front Microbiol 5: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash, J.C. , Cho, J.C. , Vergin, K.L. , Morris, R.M. , and Giovannoni, S.J. (2010) Genome sequence of Lentisphaera araneosa HTCC2155T, the type species of the order Lentisphaerales in the phylum Lentisphaerae . J Bacteriol 192: 2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojan, D. , Schreiber, L. , Bjerg, J.T. , Boggild, A. , Yang, T. , Kjeldsen, K.U. , and Schramm, A. (2016) A taxonomic framework for cable bacteria and proposal of the candidate genera Electrothrix and Electronema. Syst Appl Microbiol 39: 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa, K. , Watanabe, T. , Miura, A. , Kojima, H. , and Fukui, M. (2016) The complete genome sequences of sulfur‐oxidizing Gammaproteobacteria Sulfurifustis variabilis skN76(T) and Sulfuricaulis limicola HA5(T). Stand Genomic Sci 11: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vairavamurthy, A. , Zhou, W.Q. , Eglinton, T. , and Manowitz, B. (1994) Sulfonates ‐ a Novel Class of Organic Sulfur‐Compounds in Marine‐Sediments. Geochim Cosmochim Acta 58: 4681–4687. [Google Scholar]
- van der Heide, T. , Govers, L.L. , de Fouw, J. , Olff, H. , van der Geest, M. , van Katwijk, M.M. , et al (2012) A three‐stage symbiosis forms the foundation of seagrass ecosystems. Science 336: 1432–1434. [DOI] [PubMed] [Google Scholar]
- van der Ploeg, J.R. , Weiss, M.A. , Saller, E. , Nashimoto, H. , Saito, N. , Kertesz, M.A. , and Leisinger, T. (1996) Identification of sulfate starvation‐regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J Bacteriol 178: 5438–5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez‐Cardenas, D. , van de Vossenberg, J. , Polerecky, L. , Malkin, S.Y. , Schauer, R. , Hidalgo‐Martinez, S. , et al (2015) Microbial carbon metabolism associated with electrogenic sulphur oxidation in coastal sediments. ISME J 9: 1966–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venceslau, S.S. , Stockdreher, Y. , Dahl, C. , and Pereira, I.A.C. (2014) The “bacterial heterodisulfide” DsrC is a key protein in dissimilatory sulfur metabolism. Biochim Biophys Acta Bioenerg 1837: 1148–1164. [DOI] [PubMed] [Google Scholar]
- Wacey, D. , Kilburn, M.R. , Saunders, M. , Cliff, J. , and Brasier, M.D. (2011) Microfossils of sulphur‐metabolizing cells in 3.4‐billion‐year‐old rocks of Western Australia. Nat Geosci 4: 698–702. [Google Scholar]
- Warner, M.D. , Lukose, V. , Lee, K.H. , Lopez, K. , Sazinsky, M.H. , and Crane, E.J. 3rd. (2011) Characterization of an NADH‐dependent persulfide reductase from Shewanella loihica PV‐4: implications for the mechanism of sulfur respiration via FAD‐dependent enzymes. Biochemistry 50: 194–206. [DOI] [PubMed] [Google Scholar]
- Wasmund, K. , Cooper, M. , Schreiber, L. , Lloyd, K.G. , Baker, B.J. , Petersen, D.G. , et al (2016) Single‐cell genome and group‐specific dsrAB sequencing implicate marine members of the class Dehalococcoidia (Phylum Chloroflexi) in sulfur cycling. MBio 7: e00266–e00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener, G. , Krukenberg, V. , Riedel, D. , Tegetmeyer, H.E. , and Boetius, A. (2015) Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526: 587–590. [DOI] [PubMed] [Google Scholar]
- Wegner, C.E. , Richter‐Heitmann, T. , Klindworth, A. , Klockow, C. , Richter, M. , Achstetter, T. , et al (2013) Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula . Mar Genomics 9: 51–61. [DOI] [PubMed] [Google Scholar]
- Weissgerber, T. , Sylvester, M. , Kröninger, L. , and Dahl, C. (2014) A comparative quantitative proteomic study identifies new proteins relevant for sulfur oxidation in the purple sulfur bacterium Allochromatium vinosum . Appl Environ Microbiol 80: 2279–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, C. , Hafner, S. , Kratzer, C. , Quentmeier, A. , Friedrich, C.G. , and Dahl, C. (2009) Interaction between Sox proteins of two physiologically distinct bacteria and a new protein involved in thiosulfate oxidation. FEBS Lett 583: 1281–1286. [DOI] [PubMed] [Google Scholar]
- Wilson, J.J. , and Kappler, U. (2009) Sulfite oxidation in Sinorhizobium meliloti . Biochim Biophys Acta 1787: 1516–1525. [DOI] [PubMed] [Google Scholar]
- Winkel, M. , de Beer, D. , Lavik, G. , Peplies, J. , and Mussmann, M. (2014) Close association of active nitrifiers with Beggiatoa mats covering deep‐sea hydrothermal sediments. Environ Microbiol 16: 1612–1626. [DOI] [PubMed] [Google Scholar]
- Winkel, M. , Salman‐Carvalho, V. , Woyke, T. , Richter, M. , Schulz‐Vogt, H.N. , Flood, B.E. , et al (2016) Single‐cell sequencing of Thiomargarita reveals genomic flexibility for adaptation to dynamic redox conditions. Front Microbiol 7: 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsen, C.O. , Sievert, S.M. , Cavanaugh, C.M. , Molyneaux, S.J. , Ahmad, A. , Taylor, L.T. , et al (2002) Characterization of an autotrophic sulfide‐oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl Environ Microbiol 68: 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woebken, D. , Teeling, H. , Wecker, P. , Dumitriu, A. , Kostadinov, I. , Delong, E.F. , et al (2007) Fosmids of novel marine Planctomycetes from the Namibian and Oregon coast upwelling systems and their cross‐comparison with planctomycete genomes. ISME J 1: 419–435. [DOI] [PubMed] [Google Scholar]
- Yao, W.S. , and Millero, F.J. (1996) Oxidation of hydrogen sulfide by hydrous Fe(III) oxides in seawater. Mar Chem 52: 1–16. [Google Scholar]
- Yu, Z.G. , Peiffer, S. , Göttlicher, J. , and Knorr, K.H. (2015) Electron transfer budgets and kinetics of abiotic oxidation and incorporation of aqueous sulfide by dissolved organic matter. Environ Sci Technol 49: 5441–5449. [DOI] [PubMed] [Google Scholar]
- Zopfi, J. , Ferdelman, T.G. , and Fossing, H. (2004) Distribution and fate of sulfur intermediates ‐ sulfite, tetrathionate, thiosulfate, and elemental sulfur ‐ in marine sediments In Sulfur Biogeochemistry – Past and Present. Amend J.P., Edwards K.J., and Lyons T.W. (eds). Boulder Colorado, USA: Geological Society of America, pp. 97–116. [Google Scholar]
- Zopfi, J. , Böttcher, M.E. , and Jørgensen, B.B. (2008) Biogeochemistry of sulfur and iron in Thioploca‐colonized surface sediments in the upwelling area off central Chile. Geochim Cosmochim Acta 72: 827–843. [Google Scholar]


