Abstract
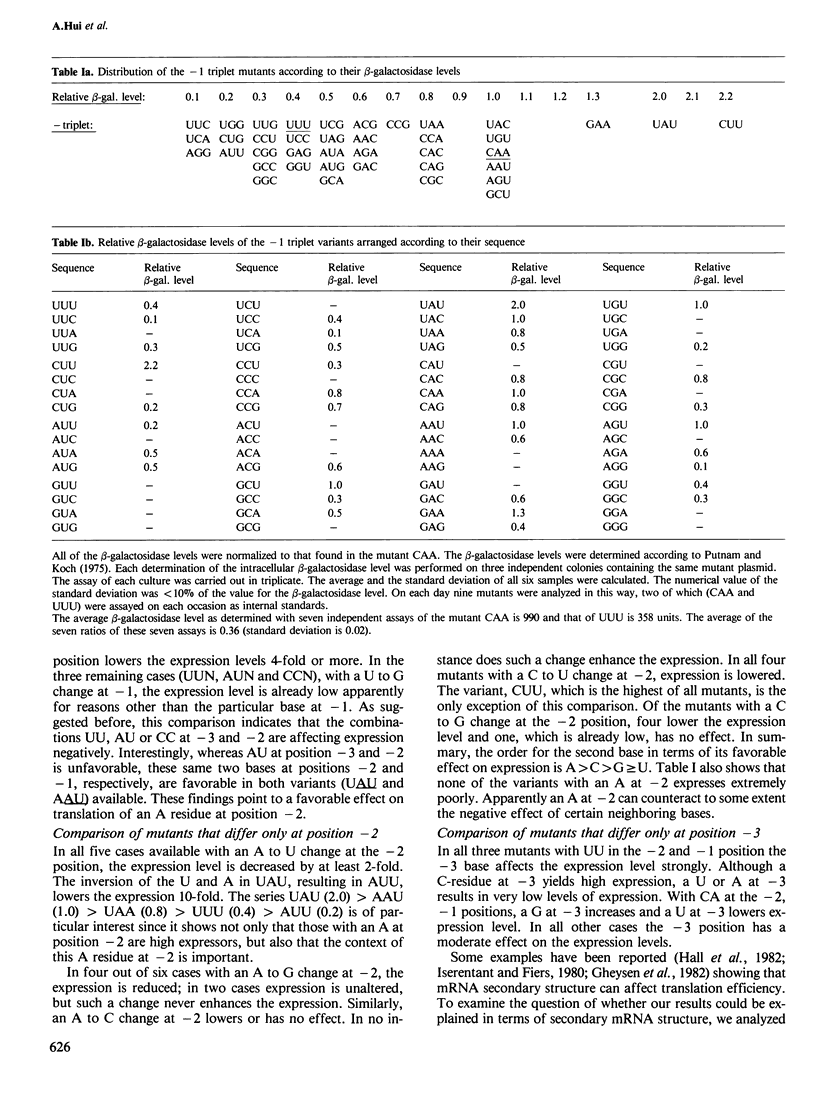
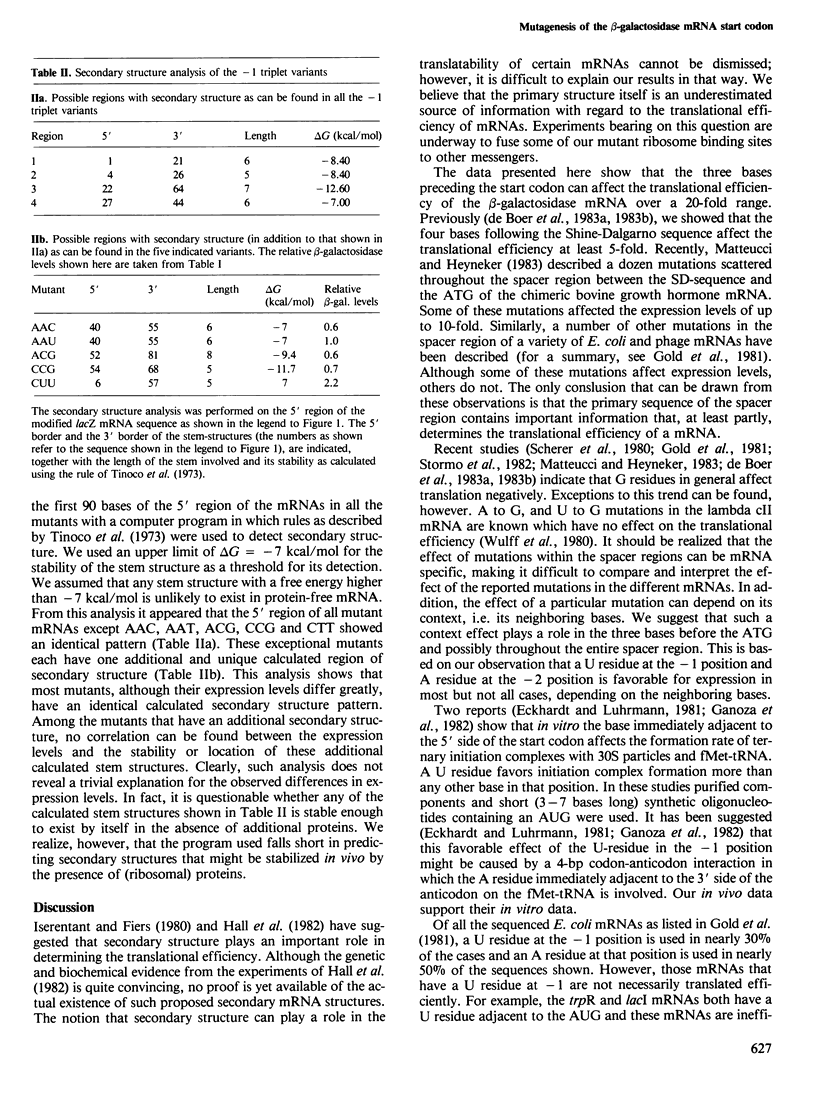
The effect on the translation efficiency of various mutations in the three bases (the -1 triplet) that precede the AUG start codon of the beta-galactosidase mRNA in Escherichia coli was studied. Of the 39 mutants examined, the level of expression varies over a 20-fold range. The most favorable combinations of bases in the -1 triplet are UAU and CUU. The expression levels in the mutants with UUC, UCA or AGG as the -1 triplet are 20-fold lower than those with UAU or CUU. In general, a U residue immediately preceding the start codon is more favorable for expression than any other base; furthermore, an A residue at the -2 position enhances the translation efficiency in most instances. In both cases, however, the degree of enhancement depends on its context, i.e. the neighboring bases. Although the rules derived from this study are complex, the results show that mutations in any of the three bases preceding the start codon can strongly affect the translational efficiency of the beta-galactosidase mRNA.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea R., Horn T. Synthesis of oligonucleotides on cellulose by a phosphotriester method. Nucleic Acids Res. 1980 May 24;8(10):2331–2348. doi: 10.1093/nar/8.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt H., Lührmann R. Recognition by initiator transfer ribonucleic acid of a uridine 5' adjacent to the AUG codon: different conformational states of formylatable methionine-accepting transfer ribonucleic acid at the ribosomal peptidyl site. Biochemistry. 1981 Apr 14;20(8):2075–2080. doi: 10.1021/bi00511a002. [DOI] [PubMed] [Google Scholar]
- Ganoza M. C., Sullivan P., Cunningham C., Hader P., Kofoid E. C., Neilson T. Effect of bases contiguous to AUG on translation initiation. J Biol Chem. 1982 Jul 25;257(14):8228–8232. [PubMed] [Google Scholar]
- Gheysen D., Iserentant D., Derom C., Fiers W. Systematic alteration of the nucleotide sequence preceding the translation initiation codon and the effects on bacterial expression of the cloned SV40 small-t antigen gene. Gene. 1982 Jan;17(1):55–63. doi: 10.1016/0378-1119(82)90100-7. [DOI] [PubMed] [Google Scholar]
- Goeddel D. V., Heyneker H. L., Hozumi T., Arentzen R., Itakura K., Yansura D. G., Ross M. J., Miozzari G., Crea R., Seeburg P. H. Direct expression in Escherichia coli of a DNA sequence coding for human growth hormone. Nature. 1979 Oct 18;281(5732):544–548. doi: 10.1038/281544a0. [DOI] [PubMed] [Google Scholar]
- Goeddel D. V., Yelverton E., Ullrich A., Heyneker H. L., Miozzari G., Holmes W., Seeburg P. H., Dull T., May L., Stebbing N. Human leukocyte interferon produced by E. coli is biologically active. Nature. 1980 Oct 2;287(5781):411–416. doi: 10.1038/287411a0. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Gabay J., Débarbouillé M., Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982 Feb 18;295(5850):616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- Iserentant D., Fiers W. Secondary structure of mRNA and efficiency of translation initiation. Gene. 1980 Apr;9(1-2):1–12. doi: 10.1016/0378-1119(80)90163-8. [DOI] [PubMed] [Google Scholar]
- Kastelein R. A., Berkhout B., Overbeek G. P., van Duin J. Effect of the sequences upstream from the ribosome-binding site on the yield of protein from the cloned gene for phage MS2 coat protein. Gene. 1983 Sep;23(3):245–254. doi: 10.1016/0378-1119(83)90015-x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteucci M. D., Heyneker H. L. Targeted random mutagenesis: the use of ambiguously synthesized oligonucleotides to mutagenize sequences immediately 5' of an ATG initiation codon. Nucleic Acids Res. 1983 May 25;11(10):3113–3121. doi: 10.1093/nar/11.10.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam S. L., Koch A. L. Complications in the simplest cellular enzyme assay: lysis of Escherichia coli for the assay of beta-galactosidase. Anal Biochem. 1975 Feb;63(2):350–360. doi: 10.1016/0003-2697(75)90357-7. [DOI] [PubMed] [Google Scholar]
- Ray P. N., Pearson M. L. Evidence for post-transcriptional control of the morphogenetic genes of bacteriophage lambda. J Mol Biol. 1974 May 5;85(1):163–175. doi: 10.1016/0022-2836(74)90135-1. [DOI] [PubMed] [Google Scholar]
- Ray P. N., Pearson M. L. Functional inactivation of bacteriophage lambda morphogenetic gene in RNA. Nature. 1975 Feb 20;253(5493):647–650. doi: 10.1038/253647a0. [DOI] [PubMed] [Google Scholar]
- Scherer G. F., Walkinshaw M. D., Arnott S., Morré D. J. The ribosome binding sites recognized by E. coli ribosomes have regions with signal character in both the leader and protein coding segments. Nucleic Acids Res. 1980 Sep 11;8(17):3895–3907. doi: 10.1093/nar/8.17.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard H. M., Yelverton E., Goeddel D. V. Increased synthesis in E. coli of fibroblast and leukocyte interferons through alterations in ribosome binding sites. DNA. 1982;1(2):125–131. doi: 10.1089/dna.1.1982.1.125. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A., Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A., Steege D. A. Characterization of two mRNA-rRNA complexes implicated in the initiation of protein biosynthesis. J Mol Biol. 1977 Aug 25;114(4):545–558. doi: 10.1016/0022-2836(77)90177-2. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Van Duin J., Overbeek G. P., Backendorf C. Functional recognition of phage RNA by 30-S ribosomal subunits in the absence of initiator tRNA. Eur J Biochem. 1980 Sep;110(2):593–597. doi: 10.1111/j.1432-1033.1980.tb04903.x. [DOI] [PubMed] [Google Scholar]
- Wulff D. L., Beher M., Izumi S., Beck J., Mahoney M., Shimatake H., Brady C., Court D., Rosenberg M. Structure and function of the cy control region of bacteriophage lambda. J Mol Biol. 1980 Apr;138(2):209–230. doi: 10.1016/0022-2836(80)90284-3. [DOI] [PubMed] [Google Scholar]
- de Boer H. A., Comstock L. J., Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983 Jan;80(1):21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer H. A., Hui A., Comstock L. J., Wong E., Vasser M. Portable Shine-Dalgarno regions: a system for a systematic study of defined alterations of nucleotide sequences within E. coli ribosome binding sites. DNA. 1983;2(3):231–235. doi: 10.1089/dna.1983.2.231. [DOI] [PubMed] [Google Scholar]
- van Dieijen G., Zipori P., van Prooijen W., van Duin J. Involvement of ribosomal protein S1 in the assembly of the initiation complex. Eur J Biochem. 1978 Oct 16;90(3):571–580. doi: 10.1111/j.1432-1033.1978.tb12637.x. [DOI] [PubMed] [Google Scholar]


