ABSTRACT
The hippocampus is one of the most interesting and studied brain regions because of its involvement in memory functions and its vulnerability in pathological conditions, such as neurodegenerative processes. In the recent years, the increasing availability of Magnetic Resonance Imaging (MRI) scanners that operate at ultra‐high field (UHF), that is, with static magnetic field strength ≥7T, has opened new research perspectives. Compared to conventional high‐field scanners, these systems can provide new contrasts, increased signal‐to‐noise ratio and higher spatial resolution, thus they may improve the visualization of very small structures of the brain, such as the hippocampal subfields. Studying the morphometry of the hippocampus is crucial in neuroimaging research because changes in volume and thickness of hippocampal subregions may be relevant in the early assessment of pathological cognitive decline and Alzheimer's Disease (AD). The present review provides an overview of the manual, semi‐automated and fully automated methods that allow the assessment of hippocampal subfield morphometry at UHF MRI, focusing on the different hippocampal segmentation produced. © 2017 The Authors Hippocampus Published by Wiley Periodicals, Inc.
Keywords: ultra‐high field MRI, hippocampal subfields, morphometry, segmentation, quantification
INTRODUCTION
The hippocampal formation (HF), centerpiece of the medial temporal lobe (MTL), is a complex brain region of enormous interest in research on aging, memory, psychiatric disorders and neurological pathologies such as most notably Alzheimer's disease (AD). The study of volumetry and morphometry of the hippocampus has an important role for neuroimaging, because this region is a focus of structural changes in AD (Laakso et al., 1998; Apostolova et al., 2006), in epilepsy (Cendes et al., 1993; Iglesias et al., 2015), and also in normal aging (Petersen et al., 2000; Frisoni et al., 2008).
The HF is composed of several intricate and packed subregions (see Fig. 1) that include the cornu ammonis fields (CA1, CA2, CA3, and CA4), the dentate gyrus (DG) and the subiculum (Duvernoy, 2005). These subregions are known to be involved selectively, non‐uniformly and in complex progression in different psychiatric and neurological disorders (Arnold et al., 1995; Braak and Braak, 1995; Simiæ et al., 1997; De Lanerolle et al., 2003; West et al., 2004; Lucassen et al., 2006; Small et al., 2011), making the information about their structural properties and changes highly desirable to achieve. Unfortunately, each hippocampal subfield measures a very small volume, and the limited spatial resolution obtained by MR systems with magnetic fields lower than 3T does not allow detecting small volumetric differences and subtle changes within these structures.
Figure 1.
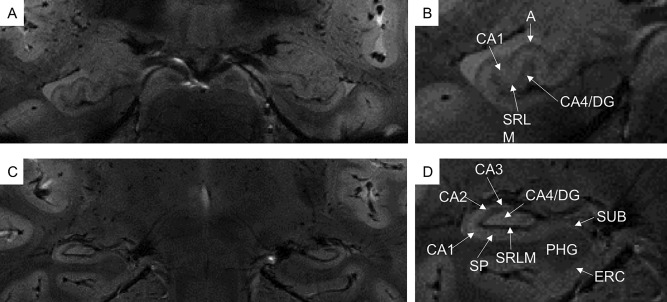
In vivo coronal T2*‐weighted images at 7T of hippocampal head (A,B) and body (C,D). Very small hippocampal structures are visible, such as cornu ammonis (CA1‐4), dentate gyrus (DG) and subiculum (SUB). In the cornu ammonis, especially in CA1, it is possible to distinguish between the stratum pyramidale (SP) and the composite of the strata radiatum, lacunosum and moleculare and the vestigial hippocampal sulcus (SRLM). A = alveus, ERC = entorhinal cortex, PHG = para‐hippocampal gyrus.
During the last years, several studies have demonstrated that 7T MR scanners may overcome these limitations. The high‐resolution structural imaging and the excellent image contrast achieved using ultra‐high field (UHF) MR systems allow the accurate depiction of several small brain structures, and the submillimetric resolution imaging achievable at UHF opens new perspectives in clinical research studies (Van Der Kolk et al., 2013; Kraff et al., 2015; van der Zwaag et al., 2016). In addition to the investigation of basal ganglia and brainstem (Cosottini et al., 2014), one of the main research field at 7T is focused on the hippocampus. The high‐resolution imaging is useful in the visualization of hippocampal subfields (Thomas et al., 2008) and in the search for in vivo radiological biomarkers of disease (Parekh et al., 2015). With UHF magnetic resonance imaging (MRI), the quantification of subtle differences in hippocampal strata, such as strata lacunosum‐moleculare and pyramidale of the CA1 (Kerchner et al., 2010), has been proved to be possible. Moreover, the ability of UHF to reveal and visualize noninvasively the endfolial pathway in the HF (Parekh et al., 2015) opens new directions in future studies on hippocampal functions and alterations in neurodegeneration. More than 20 protocols for labelling hippocampal subfields have been already published (de Flores et al., 2015a; Yushkevich et al., 2015a), confirming the increasing interest in the neuroimaging community for in vivo interrogation on the structural and functional disease‐related properties of HF subregions. A number of high‐resolution sequences centered on the MTL have been developed to acquire oblique coronal images, oriented perpendicular to the longitudinal axis of the hippocampus. The submillimetric resolution and the image contrast allow the visualization of internal details of the hippocampus, and the development of anatomical landmarks and geometrical rules to perform manual delineation of HF subfields. Despite subfield segmentation is based on morphological boundaries derived from anatomical atlases, the anatomy of human hippocampus is rather complex and the boundaries between adjacent subfields may be difficult to visualize in vivo. This leads to a certain disagreement in the way HF substructures are identified in the neuroanatomy literature.
The disease‐related modifications in the entire hippocampal morphology have been investigated through MRI, adopting several manual and automated segmentation protocols (de Flores et al., 2015a). Some methods were then quantitatively compared by the EADC‐ADNI work group (Boccardi et al., 2011). Despite the considerable improvements in MRI acquisition techniques and the great number of manual, automated and semi‐automated segmentation protocols developed, a notable between‐study variability still remains, and up to date there are not common set of rules adopted by neuroimaging groups. This heterogeneity regards the number of delineated subfields, which subregions are segmented separately or grouped together, whether the boundary delineation is focused on the hippocampus body only or on its full length, and whether the protocol concerns volumetric or strata thickness issues. An attempt to overcome the lack of consensus protocol on the segmentation of HF subfields was carried out by the Hippocampal Subfields Group (HSG, http://www.hippocampalsubfields.com). This initiative brings together researchers with the aim of characterizing the differences among existing HF subfield segmentation protocols, of identifying why they occur, and of establishing a more unified protocol to segment subfields (Dill et al., 2015).
During the last years, a number of manual and automated protocols have been developed for the segmentation of the entire hippocampus, as extensively reviewed by Konrad et al. (2009) and Dill et al. (2015). Despite the latter includes a brief section on the segmentation techniques for subfields, most of previous effort was actually focused on the segmentation of the entire hippocampal formation. Moreover, the already reviewed methods were developed and tested on data acquired with clinical field strengths, that is up to 3T. MRI data acquired at field strength below 7T have been successfully employed in a number of studies performed on large populations aimed at the identification of reliable biomarkers of pathology based on group analyses, including the noteworthy body of literature related to the ADNI initiative (Weiner et al., 2012). By contrast, UHF MRI, relying on the higher spatial resolution and signal to noise ratio than conventional magnetic fields, is expected to provide more reliable sequences able to give information about the clinical outcome of each single subject. The UHF studies are particularly aiming at the discovery of reliable biomarkers of pathology, requiring extremely high sensitivity and specificity, thus allowing moving MRI toward future personalized medicine. In particular, the presence of new image contrasts, such as susceptibility weighted imaging (SWI) technique, could be really important to identify new measures that can be correlated to AD, as first introduced by Nakada et al. (2008). In the following years, 7T imaging has been proven to be exquisitely sensitive to microscopic iron, and then it could be used to explore iron‐associated neuroinflammatory AD pathology (Zeineh et al., 2015). Additionally, the possibility to derive quantitative susceptibility maps (QSM), reflecting the presence of iron deposits, allows for a direct study of correlations between physical parameters and clinical scores of patients, as demonstrated in a study on Amyotrophic Lateral Sclerosis (Costagli et al., 2016).
The present review has the purpose of providing an overview of the state of the art of HF subfield segmentation methods working at UHF to researchers in the field aiming at exploiting the UHF potential in the assessment of HF subfield involvement in normal aging and pathological conditions. This review can be also intended as a starting point for researchers interested in improving current technical tools for HF subfield measurement or in developing new ones. Different protocols that allow the study of hippocampal subfield volumetry and morphometry using MRI data generated at UHF are presented, including manual, semi‐automated and automated methods. Several high‐resolution sequences targeting the MTL have been set up, allowing for new contrasts, increased signal‐to‐noise ratio and very high spatial resolution. These features drastically improve the capability to visualize HF subregions (Theysohn et al., 2009; Boutet et al., 2014) and to discriminate such submillimetric boundaries and hippocampal cellular strata. Even though UHF MRI may represent an important tool to investigate the early sites of AD pathology, a great deal of effort is required to combine the information obtained from standard imaging techniques, new image weightings and quantitative information achievable at UHF.
MRI ACQUISITION PROTOCOLS AND PARAMETERS
An overview of the MRI acquisition characteristics implemented in the different UHF studies targeted on hippocampal subfield segmentation is reported in Table 1. In particular, the field strength, the RF coil specification, the image weighing (T1‐w, T2‐w, T2*‐w), the acquisition sequence parameters (including TR, TE, and flip angle), the image resolution and the total acquisition time are reported.
Table 1.
Overview of the MRI Acquisition Characteristics in the Different Studies at UHF that Focuses on Hippocampal Subfield Segmentation
| MRI studies at UHF | Field strength (T) | Coil | MRI | Weighting | Native resolution (mm3) | TR (ms) | TE (ms) | Flip angle | Total acquisition time | |
|---|---|---|---|---|---|---|---|---|---|---|
| In vivo | ||||||||||
| Chupin et al., 2009 | 7.0 | 16‐ch head | TSE | T2 |
0.612 × 0.612 × 1.2 0.612 × 0.612 × 1.1 0.5 × 0.5 × 1.3 |
4000 | 77 | 60° | 8 min | |
| Cho et al., 2010 | 7.0 | Multichannel | Spoiled FLASH | T2* | 0.35 × 0.35 × 0.35 | 50 | 25 | 10° | 14 min 27 s | |
| Wisse et al., 2012, 2014a | 7.0 | 16‐channel | TSE | T2 | 0.7 × 0.7 × 0.7 | 3158 | 467 | 120° | 10–15 min | |
| Wisse et al., 2016a, 2016b | 7.0 | 32‐channel | TSE | T2 | 0.7 × 0.7 × 0.7 | 3158 | 301 | 120° | 10 min 14 s | |
| T1 | 1.0 × 1.0 × 1.0 | 4.8 | 2.2 | 1 min 57 s | ||||||
| Boutet et al., 2014 | 7.0 | 8‐channel | GRE |
|
0.3 × 0.3 × 1.2 | 742 | 16.41‐33.22 | 65° | ||
| Parekh et al., 2015 | 7.0 | 32‐channel | 3D FIESTA | 0.4 × 0.4 × 0.4 | 8.2 | 4.1 | 25° | 40 min | ||
| Suthana et al., 2015 | 7.0 | 24‐channel | TSE | 0.35 × 0.35 × 2 | 4800 | 22 | ||||
| Santyr et al., 2016 | 7.0 | 16‐channel | Multi echo | 0.5 × 0.5 × 1.5 | 40 | 4.57 | 13° | 12 min | ||
| Kerchner et al., 2010 | 7.0 | 8‐channel | GRE |
|
0.195 × 0.195 × 2 | 250 | 15 | 20° | 9.6 min | |
| Kerchner et al., 2012 | 7.0 | 16‐channel | FSE | T2 | 0.22 × 0.22 × 1.5 | 5000–6000 | 49 | 90° | 10 min | |
| Retico et al., 2016 | 7.0 | 2ch‐Tx/32ch‐Rx head coil | GRE |
|
0.3 × 0.3 × 2 | 240 | 22 | |||
| Ex vivo | ||||||||||
| Yushkevich et al., 2009 | 9.4 | Volume | SE | 0.20 × 0.30 × 0.2 | 4000–5000 | 26 | 793‐933 min | |||
| Coras et al., 2014 | 7.0 |
Shielded gradient/shim coil Transmit/receive Birdcage |
2D‐3D T2wi | T2 | ||||||
| DTI‐EPI | ||||||||||
| Apostolova et al., 2015 | 7.0 | 16‐ch head | 0.125 × 0.125 × 0.125 | 1000 | 80.3 | 180° | 60 h | |||
| Iglesias et al., 2015 | 7.0 |
Solenoid; 4‐channel phased array; small bircage |
3D FLASH |
0.1 × 0.1 × 0.1 0.2 × 0.2 × 0.2 |
50 | 25 | 20° | |||
| Wisse et al., 2016b | 9.4 | SE | T2 | 0.2 × 0.2 × 0.2 | 4000 | 21‐22 | 13:30‐15:52 h | |||
| 0.4 × 0.4 × 2.6 | 856.72–988.72 | 17.82 | 2:29‐2:31 min | |||||||
| 0.4 × 0.4 × 2.6 | 4000 | 35 | 35:14 min | |||||||
Where no information are reported it means that it was not available in the corresponding reference.
MANUAL METHODS
Several manual methods for both mono‐dimensional and three‐dimensional measures of hippocampal subfields at UHF were developed over time (Tables 2 and 3). The volumetric assessment of hippocampal body substructures was performed in a number of studies (Boutet et al., 2014; Parekh et al., 2015; Santyr et al., 2016). Parekh and colleagues used a balanced steady‐state free precession (bSSFP) sequence for 7T whole brain images with 0.4 × 0.4 × 0.4 mm3 resolution. The final reconstructed and aligned bSSFP volumes were visualized using ITK‐SNAP (www.itksnap.org; Yushkevich et al., 2006) and segmented according to anatomical atlases and prior works (Duvernoy, 2005; Insausti and Amaral, 2008; Yushkevich et al., 2009; Zeineh et al., 2012). They segmented nine subfields: CA1‐4, stratum radiatum and stratum lacunosum‐moleculare (SRLM), alveus, fornix, subiculum and subiculum along with its molecular layer, and a hypointense structure centered in the hilus that resembled the endfolial pathway. Boutet et al. used T2*‐w images with 0.3 × 0.3 × 1.2 mm3 resolution acquired with a GRE sequence to identify hippocampal layers based on their relative amount of neuronal bodies. Manual delineation was done using the object‐based ROI module of the Anatomist software (http://www.brainvisa.info/index.html). Extending the protocol proposed by Henry et al. (2011), they segmented five subregions in the hippocampal body: alveus, SRLM, hilum, stratum pyramidale of CA and stratum pyramidale of the subiculum. They normalized the volume of each subregion to the ipsilateral hippocampal body length, obtaining measures of average cross‐sectional area. The authors demonstrated that hippocampal segmentation at 7T allows the detection of volume loss in specific layers, making it as a promising tool for AD research. Santyr et al. (2016) adapted the 4T subfield delineation protocol proposed by Mueller et al. (2007) and the atlas of Duvernoy (2005) to manually segmented five subfields (subiculum, CA1‐3, and DG + CA4) only in the hippocampal body. The segmentation was made using ITK‐SNAP software on images acquired with a multi gradient echo sequence with 0.5 × 0.5 × 1.5 mm3 resolution, and images were visually inspected for accuracy and edited for additional refinement when necessary. They apply this method in a study on hippocampal subfield volumetry at 7T for the identification of hippocampal sclerosis in temporal lobe epilepsy.
Table 2.
Synthesis of the Hippocampal Subfield Segmentation Methods at UHF‐MRI: The Measured Subfields and Their Combinations Analyzed in the Reviewed Protocols Have Been Reported With Their Labels, Moreover, the Software and Tools Used for Segmentation are Summarized
| MRI studies at UHF | Softwares used for measurements | CA1 | CA2 | CA3 | CA4 | CA23 | CA123 | CA1:SRLM | CA1:SP | CA:SM | CA:SP | SRLM | Hilum | DarkBand | DG:GCL | DG+CA3 | DG | DG+CA4 | Sub | Sub:ML | Sub:SP | Pre | Para | DG:SM | H:Tail | H:Body | H:Head | H:Fissure | H | Alv | Fim | For | EF | HATA | ML | ERC | PRC | PHC | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Manual methods | ||||||||||||||||||||||||||||||||||||||||
| In vivo | ||||||||||||||||||||||||||||||||||||||||
| Chupin et al. 2009 | Anatomist software | x | x | x | x | x | x | |||||||||||||||||||||||||||||||||
| Cho et al. 2010 | 3D slicer; MRIcro | x | x | x | x | |||||||||||||||||||||||||||||||||||
| Wisse et al. 2012 | MeVisLab | x | x | x | x | x | x | x | ||||||||||||||||||||||||||||||||
| Wisse et al. 2014a | MeVisLab | x | x | x | x | x | x | x | x | |||||||||||||||||||||||||||||||
| Wisse et al. 2016a | MeVisLab | x | x | x | x | x | x | x | ||||||||||||||||||||||||||||||||
| Boutet et al. 2014 | Anatomist software | x | x | x | x | x | ||||||||||||||||||||||||||||||||||
| Parekh et al. 2015 | ITK‐SNAP | x | x | x | x | x | x | x | x | x | x | |||||||||||||||||||||||||||||
| Santyr et al. 2016 | ITK‐SNAP | x | x | x | x | x | ||||||||||||||||||||||||||||||||||
| Kerchner et al. 2010 | Dicom Viewer 2.0 | x | x | x | ||||||||||||||||||||||||||||||||||||
| Ex vivo | ||||||||||||||||||||||||||||||||||||||||
| Yushkevich et al. 2009 | ITK‐SNAP | x | x | x | x | x | ||||||||||||||||||||||||||||||||||
| Coras et al. 2014 | ImageJ, Image Pro‐Plus | x | x | x | x | |||||||||||||||||||||||||||||||||||
| Apostolova et al. 2015 | Multi‐Tracer, Aperio ImageScope | x | x | x | x | x | x | |||||||||||||||||||||||||||||||||
| Iglesias et al. 2015 | Freeview | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||||||||||||||||||||
| Wisse et al. 2016b | ITK‐SNAP | x | x | x | x | x | ||||||||||||||||||||||||||||||||||
| Semi‐automated methods | ||||||||||||||||||||||||||||||||||||||||
| Kerchner et al. 2012 | OsiriX 3.9; FSL‐View; Matlab | x | x | x | x | |||||||||||||||||||||||||||||||||||
| Suthana et al. 2015 | FSL | x | x | x | x | x | x | x | x | |||||||||||||||||||||||||||||||
| Retico et al. 2016 | Matlab | x | ||||||||||||||||||||||||||||||||||||||
| Automated methods | ||||||||||||||||||||||||||||||||||||||||
| Wisse et al. 2016a | ASHS | x | x | x | x | x | x | x | ||||||||||||||||||||||||||||||||
The different studies are grouped on the basis of the type of segmentation method that has been applied.
Table 3.
Synthesis of the Hippocampal Subfield Segmentation Methods at UHF‐MRI: Hippocampal Subfields That Have Been Segmented and Measured in the Different MRI Studies at UHF are Accompanied by a Brief Anatomical Explanation
| Region abbreviation | Description |
|---|---|
| CA1 | Ammon's Horn (Cornu Ammonis, CA), part 1 |
| CA2 | CA, part 2 |
| CA3 | CA, part 3 |
| CA4 | Dentate Gyrus Hilar region |
| CA23 | Combination of CA2 and CA3 |
| CA123 | Combination of CA1, CA2 and CA3 |
| CA1:SRLM | Stratum radiatum and stratum lacunosum‐moleculare of CA1 |
| CA1:SP | Stratum pyramidale of CA1 |
| CA:SM | Stratum moleculare of CA |
| CA:SP | Stratum pyramidale of CA 1‐3 |
| SRLM | Strata radiatum and stratum lacunosum moleculare of the hippocampus |
| Hilum | Stratum piramidale of CA4 and stratum granulosum and polymorphic layer of gyrus dentatus |
| DarkBand | Combination of stratum radiatum, stratum lacunosum‐moleculare and the vestigial hippocampus sulcus |
| DG:GCL | Granule Cell Layer of DG |
| DG+CA3 | Combined DG and CA3 |
| DG | Dentate gyrus |
| DG+CA4 | Combined DG and CA4 |
| Sub | Subiculum |
| Sub:ML | Subicular molecular layer |
| Sub:SP | Stratum pyramidale of subiculum |
| Pre | Presubiculum |
| Para | Parasubiculum |
| DG:SM | Stratum moleculare of dentate gyrus |
| H:Tail | Posterior hippocampus |
| H:Body | Body of the hippocampus |
| H:Head | Anterior hippocampus |
| H:Fissure | Hippocampal fissure: vestigial space between the molecular layers and the DG |
| H | Entire hippocampus |
| Alv | Alveus |
| Fim | Fimbria |
| For | Fornix |
| EF | Endfolial pathway within the hilus |
| HATA | Hippocampus‐amygdala transition area |
| ML | Molecular layer |
| ERC | Entorhinal cortex |
| PRC | Perirhinal cortex |
| PHC | Parahippocampal cortex |
The different studies are grouped on the basis of the type of segmentation method that has been applied.
The segmentation of the whole hippocampal formation was made for the first time at UHF by Chupin and colleagues in 2009. They used T2‐w images with in plane resolutions of 0.5 × 0.5 × 1.3 mm3, 0.612 × 0.612 × 1.1 mm3, and 0.612 × 0.612 × 1.2 mm3 to delineate six subparts of the hippocampus: the grey matter of CA1‐3), the stratum moleculare of CA, DG + CA4, the alveus, the fimbria and the subiculum. Successively, Cho et al. (2010) developed a manual method to divide the HF into head, body and tail on 3D T2*‐w images with 0.35 × 0.35 × 0.35 mm3 resolution (Jack et al., 1995; Pantel et al., 2000). The work of Duvernoy was used as a reference for anatomical details (Duvernoy 1998) and the manual tracings were made using two software packages: 3D slicer (http://www.slicer.org) and MRIcro (http://www.sph.sc.edu/comd/rorden/mricro.html). In this study, they compared reliability and sensitivity of the method with those of existing MRI techniques at 1.5T and 3T, clearly demonstrating the superiority of UHF in the boundary detection of the hippocampal head, body and tail. More recently, Wisse et al. performed the HF subregions segmentation along its full length in three works (Wisse et al., 2012, 2014a, 2016a). In the first study, they developed a protocol guided by two atlases (Duvernoy, 2005; Mai et al., 2008) and previous protocols of Knoops et al. (2010) and Goncharova et al. (1990) on T2‐w images with 0.7 × 0.7 × 0.7 mm3 resolution. Using such instruments, they manually outlined CA1‐3, CA4 and DG, subiculum and entorhinal cortex (ERC) and calculated both absolute subfield volumes and volumes normalized to the total intracranial volume [TIV, obtained from the T1‐w 7T sequence, using the tissue segmentation implemented in SPM8 (Ashburner and Friston, 2005)]. Subsequently they extended the segmentation protocol to include the most posterior part of the hippocampal tail (Wisse et al., 2014a), then they compared the volumetric results obtained with this manual protocol with those achieved with a fully automated procedure (Wisse et al., 2016a), as described later in this review. In the second study, analyses of covariance, adjusted for age, sex, and TIV showed that patients with AD had significantly smaller ERC, subiculum, CA1, CA3, and CA4 ± DG volumes in comparison with controls and MCI patients (Wisse et al., 2014a). Furthermore, the volume of the entire hippocampus has been evaluated as the sum of its subfields in the studies of Cho et al. (2010), Wisse et al. (2012, 2014a) and in Apostolova et al. (2015) (as reported in Tables 2 and 3, where the total hippocampal volume is labelled with “H”). Despite the great effort spent in the segmentation of hippocampal subfields, the comparison of results among different studies and the evaluation of reliability of single layers or whole hippocampal measures are hard. The comparison is mostly limited by the variability of boundary definition between subfields.
One of the first studies to assess the hippocampal body strata thickness in vivo at UHF was carried on by Kerchner et al. (2010). In this study, in addition to the measurement of the whole hippocampal body volume normalized to TIV, the authors developed a method to manually measure the thicknesses of the SRLM (CA1:SRLM) and of the stratum pyramidale of the CA1 (CA1:SP) using a 2D T2*‐w GRE sequence with an in‐plane resolution of 195 μm. On each side of the brain, they made six measurements of the thickness of the stratum at different points along the CA1 subfield, three each on two adjacent slices and then they were averaged. The same method was used for both the measures. In addition, for each stratum they computed the mean thickness from both sides then normalized by dividing for TIV1/3 (Kerchner et al., 2010), showing that such normalization did not influence data interpretation (Kerchner et al., 2012).
Another important application of manual HF subfield segmentation methods is the construction of high‐resolution anatomical atlases using ex vivo data. Ex vivo imaging allows using greater field strength, smaller field of view and longer acquisition time, yielding high‐contrast images with voxels about 100 times smaller in volume than in typical in vivo imaging (Yushkevich et al., 2009). Yushkevich and his group consistently identified four layers in the hippocampus: the stratum oriens and the pyramidal cell layer; the stratum radiatum, stratum lacunosum‐moleculare and the vestigial hippocampal sulcus; the stratum moleculare of the dentate gyrus; and the dentate hilus (CA4). The resulting high‐resolution computational atlas has been considered in other studies as a reference for the segmentation protocols (e.g., Parekh et al., 2015) in addition to the usually used anatomical studies (e.g., Duvernoy, 2005; Amaral and Lavenex, 2007; Insausti and Amaral, 2008). More recently, Iglesias et al. (2015) made precise delineations of 13 HF subregions in 7T ex vivo scans with extraordinary resolution. In addition to the hippocampal subfields, manual tracings for neighboring structures (e.g., amygdala, cortex) were obtained from a separate dataset of in vivo images, and they combined this information to obtain a single computational atlas with Bayesian inference. Another work on UHF ex vivo data was carried out by Apostolova et al. (2015). His group performed a manual segmentation of ex vivo temporal lobes scanned at 7T and obtained neuronal counts, β‐amyloid, and tau burden for each hippocampal traced subfield. The hippocampal labels selected for the segmentations of the studies cited above are reported in Tables 2 and 3.
The relevance of the ex‐vivo studies consists in the high precision of the resulting anatomical information and in their applicability, both for the automated segmentation of the hippocampal subregions in structural MRI images and as reliable reference for manual protocols.
Recently, aiming to investigate on hippocampal pathology along the anterior‐posterior extent in patients with temporal lobe epilepsy, Coras et al. 2014 used high‐resolution diffusion tensor imaging and coronal 2D T2‐w images of ex vivo hippocampal specimens. The values of average MRI signal intensity, areas, mean diffusivity, fractional anisotropy and histological measures were evaluated in patients and healthy subjects separately for subfields of CA1, CA2, CA3, and CA4, as identified by pathologist. The group demonstrated that the discrimination among hippocampal subfield pattern based on diffusion data could have diagnostic and prognostic value.
Finally, a recently published study by Wisse et al. (2016b) has presented a manual segmentation of ex vivo images (9.4 T, 0.2 × 0.2 × 0.2 mm3 resolution) to validate hippocampal subfield segmentation technique in vivo. The authors evaluated the ability to match information between in vivo (3T MRI scans) and ex vivo MRI in the same subjects using both rigid and deformable registration. The in vivo and ex vivo scans were registered based on manually outlined outer hippocampal boundaries. The quantitative comparison was based on SRLM thickness measurements in corresponding locations of in vivo and ex vivo scans. The registration achieved was satisfactory, with a median distance of 0.29 mm for the rigid registration, and 0.25 mm for the deformable registration, in both cases below the in vivo voxel size (0.4 × 0.4 mm2 in‐plane). Moreover, comparisons of thickness of the hippocampus and hippocampal subfields between in vivo and ex vivo pairs of images were performed.
Despite the impressive anatomical details depicted with high‐resolution images at UHF, the image analysis has still a great limitation. The majority of hippocampal subfield measurements, in fact, are based on manual method of segmentation. This is a laborious and time‐consuming process (ca. 1 h for scan) that deeply limits the amount of data that can be analyzed. Furthermore, it is affected by intra‐ and inter‐rater variability which sometimes invalidates the subfield boundary delineation and limits the following statistical analysis. Therefore, there is a need for more automation in the segmentation and measurement methods of hippocampal subfield characteristics in order to achieve more reliable and reproducible results. For these reasons, the number of semi‐automated and automated methods for HF subfield characterization is increasing, as reported in the following sections.
SEMI‐AUTOMATED METHODS FOR HF MEASURE EXTRACTION
We assigned to the category of semi‐automated methods the protocols that require manual intervention involving anatomical knowledge at the beginning, and then proceed without user intervention.
The first semi‐automated method designed to estimate HF subfield measures was published by Kerchner et al. in 2012. In this study, they visually identified hippocampal subfields based on landmarks derived from human atlases (Amaral and Lavenex, 2007; Insausti and Amaral, 2012), and combined a manual measure of DG + CA3 cross‐sectional area and entorhinal cortex (ERC) width. The semi‐automated procedure allowed measuring in vivo the widths of the CA1:SP and of the CA1:SRLM. The SRLM is an extremely thin structure whose width can be assessed only from ultra‐high resolution scans. In fact, they used 7T T2‐w fast spin echo sequence with in‐plane isotropic resolution of 222 μm to optimally view hippocampal cross‐sectional anatomy and to facilitate the quantitative study of strata thickness. In this routine, the user is asked to draw a line through the middle of the region of interest on each slice; then from a fitted spline and a set of orthogonal vectors determined along the spline, the width is measured analyzing the first derivative of the T2 signal intensity. This method was applied for the first time in 2012 and later in 2013 and 2014 to study hippocampal CA1 atrophy in AD (Kerchner and Bernstein 2013; Kerchner et al., 2014). Following the basic ideas of this method, another semi‐automated procedure that estimates the SRLM thickness has been developed and applied to 7T T2*‐w GRE sequence images with in plane resolution of 0.3 × 0.3 mm2 (Retico et al., 2016). Here, a user‐defined line following the shape of the CA1:SRLM is interpolated with a spline, and the SRLM width is calculated from the average intensity curve obtained from many profiles extracted along the vectors normal to the spline.
The approaches briefly explained above lead to extract 1D and 2D measurements from MR images, that is, widths and areas of certain hippocampal subfields, but they do not calculate volumes. During the very last years a semi‐automated volumetric approach has been developed for studying specific human hippocampal subfield functions during associative learning. The approach was based on the use of both high‐resolution 7T fMRI and hippocampal high‐resolution structural scans (HHRs), the latter acquired with a turbo spin echo sequence (Suthana et al., 2015). In this study, the method of computational unfolding was applied to 7T HHRs for segmenting HF in CA 1–3, DG + CA4, subiculum, ERC, perirhinal cortex (PRC) and parahippocampal cortex (PHC). The manual segmentation of the white matter and CSF was mandatory in order to identify 3D grey matter of hippocampal formation and adjacent MTL regions. Using atlases by Insausti and Amaral (2012) and Duvernoy (2005), they automatically assigned the subfield labels to the hippocampal grey matter (computationally unfolded to 2D space using an iterative algorithm). In a second step, they created anatomical 3D regions of interest (ROIs) by projecting the bounded regions from 2D to 3D space, allowing final manual editing if needed. The resulting hippocampal 3D ROIs were used to measure the percent signal changes in blood oxygen level dependent (BOLD) activation in separate subfields of HF.
AUTOMATED METHODS FOR HF SUBFIELD VOLUMETRY
In the very last years a certain effort to develop and improve fully automated methods for hippocampal subfield segmentation has been spent. Automated methods are based on manually labeled atlases built on high‐resolution images or manually segmented training data inserted by the user. They only require initial configuration and setup but no anatomical initial knowledge is needed on MRI data to perform new segmentations.
Up to date, there are three automated and open‐access methods for HF subfield volumetry: Freesurfer, multiple automatically generated templates (MAGet‐Brain) and automatic segmentation of hippocampus subfields (ASHS) software packages (see Table 4).
Table 4.
Synthesis of the Hippocampal Subfield Segmentation Automated Methods
| Automated software | Author | Method | Segmented HF subfields | Validation on UHF images |
|---|---|---|---|---|
| MAGet‐Brain | Pipitone et al., 2014 | Multi‐Atlas | CA1, CA23, DG+CA4, SRLM, Sub | No |
| Freesurfer v6.0 | Iglesias et al., 2015 | Probabilistic Atlas | CA1, CA4, CA23, DG:GCL, Sub, Pre, Para, H:Tail, H:Fissure, Alv, Fim, HATA, ML | No |
| ASHS | Wisse et al., 2016a | Multi‐Atlas | CA1, CA2, CA3, CA23, DG, Sub, ERC | Yes |
The segmented subfields are reported using the abbreviations introduced and explained in Table 3.
The first method is a part of the FreeSurfer open‐source software package (Fischl, 2012, http://freesurfer.net/) and approaches the segmentation problem from a parametric perspective. This algorithm is based on a statistical atlas of hippocampal anatomy and a generative model of MRI data, where the segmentation of the observed image data is carried out by learning the hippocampal subregions from labelled training data (Van Leemput et al., 2009; Iglesias et al., 2015). The original subfield segmentation method, which was implemented in Freesurfer v5.3 and is described in detail in Van Leemput et al. (2009), is based on a probabilistic in vivo atlas generated from MR images with 0.38 × 0.38 × 0.38 mm3 resolution. Despite the high spatial resolution, it was insufficient to label the molecular layer; moreover, the protocol was designed for the hippocampal body only. This software version for hippocampal subfield segmentation has been widely criticized by other research groups (Pluta et al., 2012; Wisse et al., 2014b; de Flores et al., 2015b) and the same Freesurfer authors (http://www.freesurfer.net/fswiki/HippocampalSubfieldSegmentation; Iglesias et al., 2015). Successively, Iglesias and colleagues improved the segmentation method in 2015 with a new version (now implemented in Freesurfer v6.0 and freely available at https://surfer.nmr.mgh.harvard.edu/fswiki/HippocampalSubfields). They built a new atlas using ex vivo 7T images with spatial resolution (0.13 × 0.13 × 0.13 mm3, on average) and signal‐to‐noise ratio strongly higher than those achievable in vivo. The advantage of the resulting atlas (Iglesias et al., 2015) is its applicability within a generative Bayesian framework to directly segment in vivo MR images with various contrast properties. Freesurfer v6.0 has two main modes of operation depending on the input images. First, the segmentation can work only with standard resolution (∼1 mm) T1‐w scan available. Second, if an additional MRI volume (e.g., a T2‐w scan, proton density, or even an additional T1‐w scan) covering the hippocampus is available; it can be used to obtain a more reliable segmentation (mainly if resolution is higher than 1 mm, even if anisotropic). In this case, the only requirement is that the additional scan is coarsely aligned to the 1 mm T1‐w scan. Despite the new FreeSurfer method has been developed by using high resolution 7T ex vivo data, its applicability has been validated only on lower field strengths (T1‐w and T2‐w MRI data up to 3T). Although Iglesias et al. (2015) outline the improvements of the new atlas used in Freesurfer v6.0, a number of limitations are reported. In fact, despite the new atlas is based on ultra‐high resolution MRI, some boundaries cannot be seen in the training data, for example the interfaces between the CA fields along the pyramidal layer of the hippocampus, or the interface between the CA4 and the Granule Cell Layer of the DG (DG:GCL). Another potential limitation is that this atlas was built from manual delineations in elderly subjects only, hence it might include data of hippocampal atrophy potentially limiting its applicability in studies on younger populations (Iglesias et al., 2015). In addition, when the atlas was used to segment standard resolution (1 mm) T1‐w MR scans, the molecular layer was not visible. Because the fitting of the internal structure of the atlas relies on this prior information, volumetric results from individual subfields should be interpreted carefully.
Second, Pipitone et al. in 2014 developed MAGeT‐Brain that acts as a multi‐atlas method for the segmentation of the whole hippocampus and its subfields (CA1, CA2‐3, DG + CA4, SRLM, and subiculum) based on a priori knowledge. This technique requires in input a set of manually segmented images (known as the atlas library) to generate a much larger group of automatically segmented images (known as template library). The segmentations are propagated to a subset of the template library (created by sampling either randomly or representatively from the subject images) via image registration and then the labels are combined into a single one by label fusion. When only a single atlas is used, the multi‐atlas segmentation method degenerates into model‐based segmentation technique: labels are propagated from the atlas to the target, without any label fusion step. Although the authors have demonstrated a reliable reproduction of hippocampal subfield segmentations in standard 1.5–3T T1‐w images, the method has never been validated at higher field strengths.
A third automated method has very recently been applied to 7T MRI data (Wisse et al., 2016a). ASHS consists in a combination of multi‐atlas algorithm (non‐parametric and registration‐based) and machine learning techniques. The first version of the method needed prior knowledge to run, hence it was semi‐automated; moreover, it was designed to be used only on high‐resolution anisotropic T2‐w images and was based on the VACIND atlas (Yushkevich et al., 2010, Pluta et al., 2012). The most recent version is fully automated (i.e., it requires no manual initialization), labels hippocampal subfields over a greater anterior‐posterior extent, and delineates the perirhinal cortex which is further subdivided into Brodmann areas 35 and 36 (Upenn PMC atlas, Yushkevich et al., 2015b). ASHS consists of a training pipeline and a segmentation pipeline. The first procedure generates an atlas package requiring in input a set of manually labeled dedicated T2‐w scans and standard whole‐brain T1‐w images. The ASHS segmentation pipeline allows the automated segmentation of hippocampal subfields on the T2‐w MRI scan of a new subject, using the T1‐w MRI as an additional input. Moreover, it allows both volume and thickness analyses. Compared to semi‐automated methods, a great advantage of this software is the possibility to run a completely automated procedure using one of the freely available atlases developed by the authors of ASHS (https://sites.google.com/site/hipposubfields/). Another point of force is that it can be easily re‐trained on a set of scans acquired with a different MRI sequence and labeled with a different segmentation protocol.
To the best of our knowledge, ASHS is the unique fully automated software for HF subfield volumetry that has been applied to UHF MRI in vivo data. This study was recently made by Wisse et al. (2016a) and used ASHS with the aim of comparing automated segmentations of CA1, CA2, CA3, CA2‐3, DG, subiculum, and ERC with the corresponding manual segmentations. As shown in Table 1, their protocol included 0.7 × 0.7 × 0.7 mm3 3D T2‐w TSE and 1.0 × 1.0 × 1.0 mm3 T1‐w sequences and both the acquisitions were used for the segmentation. In this work, a high and competitive accuracy emerged for most subfield segmentations with respect to the labor‐intensive manual segmentation, and their results indicated that there might be a benefit in using 7T data for the segmentation of hippocampal subfields.
VARIABILITY AND RELIABILITY ASSESSMENT OF SEGMENTATION PROTOCOLS
All the protocols described above are targeted to the measurement of either volumes or thicknesses of subtle regions of the hippocampus. Despite the use of UHF MR systems allows the accurate depiction of such small brain substructures, a number of uncertainties emerge from the use a nonstandardized set of sequences and image acquisition protocols (see Table 1), from the application of large variety of segmentation methods (see Tables 2 and 3) to identify and measure the HF subfields.
As reported in the study by Yushkevich et al. (2015a), that quantitatively compared 21 segmentation protocols applied both at 3T and 7T images, significant variability exists among the protocols in terms of what labels are used, where the boundaries between substructures are placed, and what extent of the hippocampal region is labeled. That paper offered strong motivations for protocol harmonization and took an important first step in that direction, such as the activity of the HSG (http://www.hippocampalsubfields.com/whitepaper).
In addition to the variability between segmentation protocols, a relevant aspect that limits clinical usability of these methods is the paucity of implementations of reliability assessment procedures. Indeed, intra‐ and inter‐rater reliability evaluations are carried out only by a few of the studies reported here.
Repeat‐measurement reliability was assessed in the in vivo study of Wisse et al. (2012) using Intraclass Correlation Coefficient (ICC) and Dice Similarity Comparison (DSC). The reliability measures are usually reported in terms of ICC and DSC scores. The guidelines for the interpretation of ICC agreement measures (whose range is [0, 1]) indicate excellent agreement for ICC ≥ 0.75 (Cicchetti, 1994). The DSC index quantifies the degree of overlap of two segmented regions at the voxel level, accounting for the region size. Its values are in the [0,1] range, and values greater than 80% are considered satisfactory levels of concordance. Wisse et al. concluded that a good‐to‐excellent consistency was achieved between two manual segmentations of one rater for all subfields (ICC varying from 0.74 to 0.98). Furthermore, accuracy measured with DSC was >0.82 for all subfields, with the exception of the smaller subfield CA3 (0.68–0.70).
Inter‐rater reliability was performed by Cho et al. (2010) with two manual tracers. They evaluated the ICC and the sensitivity (Pantel et al., 2000), which indicates the likelihood of agreement between two raters. These estimations were performed for the same protocol applied at both 7T and 1.5T MR images. Paired sample t‐test was used to assess the significance of the differences in reliability and sensitivity between 7T and 1.5T MRI. At 7T, the ICC ranged from 0.93 to 0.97 and sensitivity ranged from 0.85 to 0.91. Moreover, their results clearly demonstrated better boundary detection for hippocampal head, body and tail at 7T than at lower field MRI (P ≤ 0.05), suggesting the usefulness of UHF MR imaging in the accurate diagnosis of diseases related to hippocampal atrophy.
Intra‐ and inter‐rater reliability were also assessed in the study of Yushkevich et al. (2009), regarding ex vivo MRI at 9.4T. The repeatability of the segmentation in terms of Dice overlap by one rater ranged from 0.56 to 0.92. Inter‐rater reliability ranged from 0.47 to 0.87 and was significantly lower for CA1 and CA23 and approximately the same for other subfields. The authors attributed the lower reliability for CA23 to differences in the extension of the label. Lastly, Iglesias et al. (2015) used seven tracers to construct their atlas from ex vivo MRI data. Because the annotation of each scan required about 60 h, they did perform neither repeat measurements nor inter‐rater reliability, but one expert tracer served as quality control for each case, refining the segmentations where necessary. However, the applicability of this atlas, which has been released as part of FreeSurfer v6.0, was demonstrated with experiments on three different publicly available datasets with different types of MRI contrast. The results showed that the atlas and companion segmentation method can discriminate between Alzheimer's disease subjects and elderly controls with 88% accuracy in standard resolution (1mm) T1‐w data, significantly outperforming the atlas in FreeSurfer version 5.3 (86% accuracy) and the classification based on whole hippocampal volume (82% accuracy) (Iglesias et al., 2015).
Regarding semi‐automated methods, Kerchner et al. (2012) found a significant correlation between the output of their protocol and manual measures using statistical Pearson correlation (CA1‐SP: r = 0.74, P ≤ 0.001; CA1‐SRLM: r = 0.75, P ≤ 0.001).
ASHS is the only automated method already applied to UHF MR images. The reliability analysis made by Wisse et al. (2016a) used both DSC and ICC indices to compare manual segmentation of two independent manual tracers and the automated to the corresponding manually segmented regions. About the manual segmentation assessment, the intra‐rater reliability resulted in DSC ranged between 0.66 and 0.87, and ICC ranged between 0.80 and 0.90; on the other hand, the inter‐rater reliability resulted in DSC ranged between 0.57 and 0.82, whereas ICC was very low for the ERC (0.27) but it reached 0.94 for the CA1. ICC and DSC calculated between manual and automated segmentation for each subfield showed that ASHS allowed high accuracy (ICC > 0.74 and DSC > 0.75) for larger subfields, including CA1, DG, and subiculum, and low accuracy for ERC and smaller subfields, including CA2 and CA3. More in detail, the mean generalized DSC across all subfields was 0.80 ± 0.03 in the left hemisphere and 0.79 ± 0.03 in the right hemisphere. The anterior and posterior boundaries of the ERC were an important source of disagreement between the manual and automated segmentation. The accuracy increased by restricting the range of ERC segmentation, suggesting that the ERC segmentation is accurate except at its anterior and posterior segments. In light of these results, the authors concluded that automated segmentation of hippocampal subfields and the ERC at 7T MRI was feasible and that the errors of automated segmentation were comparable or lower than the disagreement between two manual raters applying the same segmentation protocol.
ASHS has been partially validated in the recent study by Wisse et al. (2016b), where the SRLM surface inferred from ASHS segmentation performed on in vivo 3T images was compared with that obtained from manual ex vivo segmentation (MR acquisition at 9.4 T, see Table 1 for more details). The distance statistics between ASHS‐derived SRLM and ex vivo SRLM was computed using a deformable in vivo to ex vivo alignment. Median surface distance was within a voxel and only 0.1 mm larger than the median distance between manually traced in vivo and ex vivo SRLM surfaces, confirming that ASHS can accurately pick up this anatomical boundary on the in vivo images.
CONCLUSIONS
During last years, the increasing number of patients affected by pathological cognitive deterioration has strengthened the interest of researchers in the investigation on hippocampus. The availability of UHF MRI is changing the target of this research. Differently from past approaches, the possibility to obtain high‐resolution images is increasing knowledge on in vivo radiological anatomy of the hippocampus and mainly of the hippocampal subfields.
In the present review, we provided an overview of the neuroimaging studies performed at UHF MRI with the aim of assessing hippocampal subfield morphology and morphometry. We presented the numerous methods for segmentation and measurement of the hippocampal subregions according to (i) the presence of automatism in the procedure, (ii) the segmented subfields, and (iii) the type of subfield measure. In the latest years, the increasing number of MRI systems that operate at UHF has allowed the quantification of volume and thickness of the inner strata of the hippocampus using images with submillimetric resolution. The features of UHF open new perspectives in the identification of radiological biomarkers for AD at its prodromal clinical stage (i.e., the mild cognitive impairment), and in the improvement of diagnostic accuracy of the disease.
When approaching HF assessment, it is important to remember that pathological cognitive decline is most frequent in the elderly population and it is characterized by a pathological brain atrophy, mainly involving frontotemporal lobes. Nevertheless, a para‐physiological brain atrophy takes place over time also in healthy people, but it is usually diffuse and not confined to frontotemporal regions. Therefore, in order to correctly assess abnormal decline trajectories, the age‐related confounding effects should be accounted for in the analysis, in addition to other confounding variables, such as gender. To account for global effects, the measurements of the whole hippocampus and its subfield are sometimes standardized with respect to an anatomical reference, such as TIV (Wisse et al., 2012), or the ipsilateral hippocampal body length in other cases (Boutet et al., 2014), thus assuming a linear scaling between the sizes of the different structures. Other research groups introduced TIV as covariate in statistical analyses of covariance of hippocampal subfield volumes (Wisse et al., 2014a; Santyr et al., 2016). Despite some researchers normalized the thickness of the CA1 pyramidal cell layer and SRLM to the TIV1/3 (Kerchner et al., 2010), it is not proven that such tiny structures scale linearly with head size. However, the choice of the appropriate anatomical reference quantity and the normalization method are not immediately identifiable.
The main issues limiting the comparison of results among studies are: (i) the absence of guidelines for the choice of target hippocampal subfields; (ii) the lack of standardized technical parameters for acquisition protocols; (iii) the absence of a unique anatomical reference; and (iv) the lack of consistency in the data normalization strategy. The recent work of the Hippocampal Subfield Group (HSG, http://www.hippocampalsubfields.com) aimed to address the second issue, and a white paper for harmonization has been developed (http://www.hippocampalsubfields.com/whitepaper) to define the next steps (for additional details, see Yushkevich et al., 2015a). About the third issue, although Zeineh et al. (2012) developed diffusivity‐based rules for hippocampal subfield manual delineation at 3T, the majority of segmentation protocols refers to anatomical studies. The most applied are the anatomical studies of Duvernoy (1998, 2005), then the works of Insausti and Amaral (2008, 2012), of Mai et al. in 2008, and Amaral and Lavenex in 2007. The interest on high‐resolution anatomical atlases using ex vivo data as a gold standard for hippocampal subfield assessment is increasing. The new automated protocols are based on the ex vivo atlases constructed by Yushkevich et al. (2009, 2015b) and employed in ASHS, and on the segmentations of Iglesias et al. (2015) used in Freesurfer. With respect to the forth issues, a growing body of literature is aiming at finding shared guidelines and reliable methods to correctly account for confounding effects occurring in pathological conditions and also in healthy aging (Edland et al., 2002; Barnes et al., 2010; Voevodskaya et al., 2014).
A very important issue, which is not always fully addressed in research studies, is the assessment of intra‐ and inter‐rater reliability of segmentation algorithms. Intra‐rater reliability of manual segmentation represents the theoretic upper bound for the agreement of automated with manual segmentation (Wisse et al., 2016a). A lack in reliability assessments of a manual protocol limits both its clinical application and a trustworthy comparison for automated methods. In fact, the benchmark for the semi‐automated and automated segmentation methods should be to introduce an error comparable or lower than the disagreement between two manual raters carrying out the same segmentation. Additionally, semi‐automated and automated segmentation methods could be of limited clinical utility if their reliability and measure reproducibility are not demonstrated. Regarding the hippocampal subfield segmentation, the reported ICC and DSC scores are very variable for different segmented subfields, with unsatisfactory values for small regions (Yushkevich et al., 2009; Wisse et al., 2012).
Despite 7T MRI scanners are spreading in human clinical research, there still are some limitations in the direct application of ex vivo achievements to in vivo sequences. First, the acquisition times of ex vivo studies are much longer than those reasonable for clinical protocols, as evidenced in Table 1. Second, ex vivo MRI exams are free from motion artefacts degrading image quality, such as natural vascular pulsatility and additional movement effects. Even so, the development of new hardware and the improvement of acquisition sequences and post‐processing methods are expected to change for the better the image quality of clinical 7T MRI and to render such ex vivo studies a more reliable reference for future clinical studies.
In spite of the notable number of methods developed at UHF MRI, the majority of protocols is based on manual segmentations, which are extremely time consuming and suffer of intra‐ and inter‐rater variability. Only in the very recent years, three automated segmentation packages have become available: MAGet‐Brain, Freesurfer and ASHS. Nevertheless, the first two methods have not been validated on images acquired at 7T and above. Moreover, the Freesurfer software has shown considerable limitations in its first version 5.3, and, despite its latest version 6.0 overcomes many difficulties, a number of limitations still remain. The necessity of a reliability assessment at UHF for the version 6.0 is also evident, since its applicability has been validated only on MRI data acquired at lower field strengths (up to 3T). By contrast, ASHS was applied on 7T data and a reliability comparison with manual segmentations was performed, showing high accuracy for most subfields. ASHS can be run only when a whole brain T1‐w sequence and an oblique coronal T2‐w sequence oriented along the main axis of the hippocampus have been acquired for each subject. In addition, ASHS allows for the definition and use of user‐defined segmentation protocols, thanks to the ASHS training pipeline. Hence, ASHS presents both the advantages of a fully automated implementation using two MR sequences (T1‐w and T2‐w) and the versatility of being adaptable to different segmentation protocols, if labeled training data are available. The latter feature is particularly interesting as it can allow the comparison among different subfield definitions on the same data and with the same segmentation software, trained on different labeled data. It is highly desirable that in the near future similar segmentation tools include the possibility to handle also additional contrasts, for example SWI, or quantitative images, for example QSM, to fully exploit the added value of UHF MRI.
In conclusion, despite the potential key role of UHF MRI in the study of the selective involvement of the hippocampal subfields in healthy aging, neurodegenerative processes and other pathologies affecting the HF is evident, a stronger effort in neuroimaging research is still necessary to address the following issues: (i) the harmonization of a manual segmentation protocol for the subfields, which is fundamental by itself and constitutes also the reference standard for semi‐automated and automated segmentation tools; (ii) the definition of guidelines for standardized global measures to account for during the analysis, for example total HF volume and TIV; (iii) the development of more versatile and reliable tools for automated segmentation; (iv) inter‐ and intra‐rater reliability assessment of segmentation methods. Once these open issues are fully addressed, the reliability and the reproducibility of the studies will be guaranteed and the inter‐study comparison will be easily allowed. An additional final effort required to the neuroimaging research community is to design more versatile segmentation tools able to accommodate and exploit the complementary information available from different MRI acquisition sequences, whose informative content is particularly enhanced at UHF.
REFERENCES
- Amaral A, Lavenex P. 2007. Hippocampal neuroanatomy In: Andersen P, Morris R, Amaral D, Bliss T, O'Keefe J, eds. The Hippocampus Book. New York: Oxford University Press; 37–114. [Google Scholar]
- Apostolova LG, Dinov ID, Dutton RA, Hayashi KM, Toga AW, Cummings JL, Thompson PM. 2006. 3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer's disease. Brain 129:2867–2873. doi:10.1093/brain/awl274. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Zarow C, Biado K, Hurtz S, Boccardi M, Somme J, Honarpisheh H, Blanken AE, Brook J, Tung S, Lo D, Ng D, Alger JR, Vinters H V, Bocchetta M, Duvernoy H, Jack CR, Frisoni GB, EADC‐ADNI Working Group on the Harmonized Protocol for Manual Hippocampal Segmentation. 2015. Relationship between hippocampal atrophy and neuropathology markers: A 7T MRI validation study of the EADC‐ADNI harmonized hippocampal segmentation protocol. Alzheimers Dement 11:139–150. doi:10.1016/j.jalz.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SE, Franz BR, Gur RC, Gur RE, Shapiro RM, Moberg PJ, Trojanowski JQ. 1995. Smaller neuron size in schizophrenia in hippocampal subfields that mediate cortical‐hippocampal interactions. Am J Psychiatry 152:738–748. doi:10.1176/ajp.152.5.738. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. 2005. Unified segmentation. Neuroimage 26:839–851. [DOI] [PubMed] [Google Scholar]
- Barnes J, Ridgway GR, Bartlett J, Henley SMD, Lehmann M, Hobbs N, Clarkson MJ, MacManus DG, Ourselin S, Fox NC. 2010. Head size, age and gender adjustment in MRI studies: A necessary nuisance? Neuroimage 53:1244–1255. doi:10.1016/j.neuroimage.2010.06.025. [DOI] [PubMed] [Google Scholar]
- Boccardi M, Ganzola R, Bocchetta M, Pievani M, Redolfi A, Bartzokis G, Camicioli R, Csernansky JG, de Leon MJ, deToledo‐Morrell L, Killiany RJ, Lehéricy S, Pantel J, Pruessner JC, Soininen H, Watson C, Duchesne S, Jack CR, Frisoni GB. 2011. Survey of protocols for the manual segmentation of the hippocampus: preparatory steps towards a joint EADC‐ADNI harmonized protocol. J Alzheimers Dis 26 (Suppl 3):61–75. doi:10.3233/JAD-2011-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet C, Chupin M, Lehéricy S, Marrakchi‐Kacem L, Epelbaum S, Poupon C, Wiggins C, Vignaud A, Hasboun D, Defontaines B, Hanon O, Dubois B, Sarazin M, Hertz‐Pannier L, Colliot O. 2014. Detection of volume loss in hippocampal layers in Alzheimer's disease using 7 T MRI: A feasibility study. NeuroImage Clin 5:341–348. doi:10.1016/j.nicl.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. 1995. Staging of alzheimer's disease‐related neurofibrillary changes. Neurobiol Aging 16:271–278. doi:10.1016/0197-4580(95)00021-6. [DOI] [PubMed] [Google Scholar]
- Cendes F, Andermann F, Gloor P, Evans A, Jones‐Gotman M, Watson C, Melanson D, Olivier A, Peters T, Lopes‐Cendes I. 1993. MRI volumetric measurement of amygdala and hippocampus in temporal lobe epilepsy. Neurology 43:719–725. [DOI] [PubMed] [Google Scholar]
- Cho Z‐H, Han J‐Y, Hwang S‐I, Kim D, Kim K‐N, Kim N‐B, Kim SJ, Chi J‐G, Park C‐W, Kim Y‐B. 2010. Quantitative analysis of the hippocampus using images obtained from 7.0 T MRI. Neuroimage 49:2134–2140. doi:10.1016/j.neuroimage.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Chupin M, Lehericy S, Colliot O, Marjanska M, Goerke U, Ugurbil K, Moortele P‐F Van de 2009. Three‐dimensional segmentation of the internal structures of the human hippocampus at 7 tesla. Proc 17nd Sci Meet Int Soc Magn Reson Med 49:2009. [Google Scholar]
- Cicchetti DV. 1994. Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychol Assessment 6:284–290. doi:10.1037/1040-3590.6.4.284 [Google Scholar]
- Coras R, Milesi G, Zucca I, Mastropietro A, Scotti A, Figini M, Mühlebner A, Hess A, Graf W, Tringali G, Blümcke I, Villani F, Didato G, Frassoni C, Spreafico R, Garbelli R. 2014. 7T MRI features in control human hippocampus and hippocampal sclerosis: An ex vivo study with histologic correlations. Epilepsia 55:2003–2016. doi:10.1111/epi.12828. [DOI] [PubMed] [Google Scholar]
- Cosottini M, Frosini D, Pesaresi I, Costagli M, Biagi L, Ceravolo R, Bonuccelli U, Tosetti M. 2014. MR imaging of the substantia nigra at 7 T enables diagnosis of parkinson disease. Radiology 271:831–838. doi:10.1148/radiol.14131448. [DOI] [PubMed] [Google Scholar]
- Costagli M, Donatelli G, Biagi L, Caldarazzo Ienco E, Siciliano G, Tosetti M, Cosottini M. 2016. Magnetic susceptibility in the deep layers of the primary motor cortex in amyotrophic lateral sclerosis. Neuroimage Clin 12:965–969. doi:10.1016/j.nicl.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Flores R, La Joie R, Chételat G. 2015a. Structural imaging of hippocampal subfields in healthy aging and Alzheimer's disease. Neuroscience 309:29–50. doi:10.1016/j.neuroscience.2015.08.033. [DOI] [PubMed] [Google Scholar]
- de Flores R, La Joie R, Landeau B, Perrotin A, Mézenge F, de La Sayette V, Eustache F, Desgranges B, Chételat G. 2015b. Effects of age and Alzheimer's disease on hippocampal subfields. Hum Brain Mapp 36:463–474. doi:10.1002/hbm.22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lanerolle NC, Kim JH, Williamson A, Spencer SS, Zaveri HP, Eid T, Spencer DD. 2003. A retrospective analysis of hippocampal pathology in human temporal lobe epilepsy: Evidence for distinctive patient subcategories. Epilepsia 44:677–687. doi:10.1046/j.1528-1157.2003.32701.x. [DOI] [PubMed] [Google Scholar]
- Dill V, Franco AR, Pinho MS. 2015. Automated methods for hippocampus segmentation: The evolution and a review of the state of the art. Neuroinformatics 13:133–150. doi:10.1007/s12021-014-9243-4. [DOI] [PubMed] [Google Scholar]
- Duvernoy HM. 1998. The Human Hippocampus: Functional Anatomy, Vascularization and Serial Sections with MRI, 2nd ed Berlin, Heidelberg, NY: Springer‐Verlag. [Google Scholar]
- Duvernoy HM. 2005. The Human Hippocampus: Functional Anatomy, Vascularization and Serial Sections with MRI, 3rd ed Berlin, Heidelberg, NY: Springer‐Verlag. [Google Scholar]
- Edland SD, Xu Y, Plevak M, O′Brien P, Tangalos EG, Petersen RC, Jack CR. 2002. Total intracranial volume: Normative values and lack of association with Alzheimer's disease. Neurology 59:272–274. doi:10.1212/WNL.59.2.272. [DOI] [PubMed] [Google Scholar]
- Fischl B. 2012. FreeSurfer. Neuroimage 62:774–781. doi:10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni GB, Ganzola R, Canu E, Rüb U, Pizzini FB, Alessandrini F, Zoccatelli G, Beltramello A, Caltagirone C, Thompson PM. 2008. Mapping local hippocampal changes in Alzheimer's disease and normal ageing with MRI at 3 Tesla. Brain 131:3266–3276. [DOI] [PubMed] [Google Scholar]
- Goncharova II, Dickerson BC, Stoub TR, L. deToledo‐Morrell 1990. MRI of human entorhinal cortex: A reliable protocol for volumetric measurement. Neurobiol Aging 22:737–745. doi:10.1016/S0197-4580(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Henry TR, Chupin M, Lehéricy S, Strupp JP, Sikora MA, Sha ZY, Ugurbil K, Van de Moortele P‐F. 2011. Hippocampal sclerosis in temporal lobe epilepsy: Findings at 7 T. Radiology 261:199–209. doi:10.1148/radiol.11101651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias JE, Augustinack JC, Nguyen K, Player CM, Player A, Wright M, Roy N, Frosch MP, McKee AC, Wald LL, Fischl B, Van Leemput K. 2015. A computational atlas of the hippocampal formation using ex vivo, ultra‐high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage 115:117–137. doi:10.1016/j.neuroimage.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insausti R, Amaral DG. 2008. Entorhinal cortex of the monkey: IV. Topographical and laminar organization of cortical afferents. J Comp Neurol 509:608–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insausti R, Amaral DG. 2012. Hippocampal Formation. In: Atlas of the human brain, Ed 3 (Mai JK, Paxinos G, eds), pp 896–942. London: Elsevier Academic. doi:10.1016/B978‐0‐12‐374236‐0.10024‐0. [Google Scholar]
- Jack CR, Theodore WH, Cook M, McCarthy G. 1995. MRI‐based hippocampal volumetrics: Data acquisition, normal ranges, and optimal protocol. Magn Reson Imaging 13:1057–1064. doi:10.1016/0730-725X(95)02013-J. [DOI] [PubMed] [Google Scholar]
- Kerchner G, Bernstein J. 2013. Shared vulnerability of two synaptically‐connected medial temporal lobe areas to age and cognitive decline: A seven tesla magnetic resonance imaging study. J Neurosci 33:16666–16672. doi:10.1523/JNEUROSCI.1915-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchner GA, Hess CP, Hammond‐Rosenbluth KE, Xu D, Rabinovici GD, Kelley DAC, Vigneron DB, Nelson SJ, Miller BL. 2010. Hippocampal CA1 apical neuropil atrophy in mild Alzheimer disease visualized with 7‐T MRI. Neurology 75:1381–1387. doi:10.1212/WNL.0b013e3181f736a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchner GA, Deutsh GK, Zeineh M, Dougherty RF, Saranathan M, Rutt BK. 2012. Hippocampal CA1 apical neuropil atrophy and memory performance in Alzheimer's disease. Neuroimage 63:194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchner GA, Berdnik D, Shen JC, Bernstein JD, Fenesy MC, Deutsch GK, Wyss-Coray T, Rutt BK. 2014. APOE e4 worsens hippocampal CA1 apical neuropil atrophy and episodic memory. Neurology 82:691–697. doi:10.1212/WNL.0000000000000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops AJG, Gerritsen L, van der Graaf Y, Mali WPTM, Geerlings MI. 2010. Basal hypothalamic pituitary adrenal axis activity and hippocampal volumes: The SMART‐medea study. Biol Psychiatry 67:1191–1198. doi:10.1016/j.biopsych.2010.01.025. [DOI] [PubMed] [Google Scholar]
- Konrad C, Ukas T, Nebel C, Arolt V, Toga AW, Narr KL. 2009. Defining the human hippocampus in cerebral magnetic resonance images—An overview of current segmentation protocols. Neuroimage 47:1185–1195. doi:10.1016/j.neuroimage.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraff O, Fischer A, Nagel AM, Mönninghoff C, Ladd ME. 2015. MRI at 7 tesla and above: Demonstrated and potential capabilities. J Magn Reson Imaging 41:13–33. doi:10.1002/jmri.24573. [DOI] [PubMed] [Google Scholar]
- Laakso M, Soininen H, Partanen K, Lehtovirta M, Hallikainen M, Hänninen T, Helkala E‐L, Vainio P, Riekkinen P. 1998. MRI of the hippocampus in Alzheimer's disease: Sensitivity, specificity, and analysis of the incorrectly classified subjects. Neurobiol Aging 19:23–31. doi:10.1016/S0197-4580(98)00006-2. [DOI] [PubMed] [Google Scholar]
- Lucassen P, Heine V, Muller M, van der Beek E, Wiegant V, Ron De Kloet E, Joels M, Fuchs E, Swaab D, Czeh B. 2006. Stress, depression and hippocampal apoptosis. CNS Neurol Disord Drug Targets 5:531–546. doi:10.2174/187152706778559273. [DOI] [PubMed] [Google Scholar]
- Mai, J.K ., Paxinos G., Voss T.. 2008. Atlas of the Human Brain. New York: Academic Press; pp 1–271. [Google Scholar]
- Mueller SG, Stables L, Du AT, Schuff N, Truran D, Cashdollar N, Weiner MW. 2007. Measurement of hippocampal subfields and age‐related changes with high resolution MRI at 4T. Neurobiol Aging 28:719–726. doi:10.1016/j.neurobiolaging.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada T, Matsuzawa H, Igarashi H, Fujii Y, Kwee IL. 2008. In vivo visualization of senile‐plaque‐like pathology in Alzheimer's disease patients by MR microscopy on a 7T system. J Neuroimaging 18:125–129. doi:10.1111/j.1552-6569.2007.00179.x. [DOI] [PubMed] [Google Scholar]
- Pantel J, O'Leary DS, Cretsinger K, Bockholt HJ, Keefe H, Magnotta VA, Andreasen NC. 2000. A new method for the in vivo volumetric measurement of the human hippocampus with high neuroanatomical accuracy. Hippocampus 10:752–758. doi:10.1002/1098-1063(2000)10:6 < 752::AID-HIPO1012 > 3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Parekh MB, Rutt BK, Purcell R, Chen Y, Zeineh MM. 2015. Ultra‐high resolution in‐vivo 7.0T structural imaging of the human hippocampus reveals the endfolial pathway. Neuroimage 112:1–6. doi:10.1016/j.neuroimage.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Jack CR, Xu YC, Waring SC, O′Brien PC, Smith GE, Ivnik RJ, Tangalos EG, Boeve BF, Kokmen E. 2000. Memory and MRI‐based hippocampal volumes in aging and AD. Neurology 54:581–587. Available at: http://www.ncbi.nlm.nih.gov/pubmed/10680786. Accessed October 20, 2016. [DOI] [PubMed] [Google Scholar]
- Pipitone J, Park MTM, Winterburn J, Lett TA, Lerch JP, Pruessner JC, Lepage M, Voineskos AN, Chakravarty MM. 2014. Multi‐atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage 101:494–512. doi:10.1016/j.neuroimage.2014.04.054. [DOI] [PubMed] [Google Scholar]
- Pluta J, Yushkevich P, Das S, Wolk D. 2012. In vivo analysis of hippocampal subfield atrophy in mild cognitive impairment via semi‐automatic segmentation of T2‐weighted MRI. J Alzheimers Dis 31:85–99. doi:10.3233/JAD-2012-111931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retico A, Donatelli G, Costagli M, Biagi L, Fantacci ME, Frosini D, Tognoni G, Cosottini M, Tosetti M. 2016. Semiautomatic image processing tool to measure small structures in magnetic resonance images of the brain at 7 Tesla application to hippocampus subfields of patients with mild cognitive impairment. BIOIMAGING 2016—3rd Int Conf Bioimaging, Proceedings; Part 9th Int Jt Conf Biomed Eng Syst Technol BIOSTEC 2016. 2(Biostec):124–128. doi:10.5220/0005818001240128.
- Santyr BG, Goubran M, Lau JC, Kwan BYM, Salehi F, Lee DH, Mirsattari SM, Burneo JG, Steven DA, Parrent AG, de Ribaupierre S, Hammond RR, Peters TM, Khan AR. 2016. Investigation of hippocampal substructures in focal temporal lobe epilepsy with and without hippocampal sclerosis at 7T. J Magn Reson Imaging. 2016 Aug 26. doi:10.1002/jmri.25447. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Simiæ G, Kostoviæ I, Winblad B, Bogdanoviæ N. 1997. Volume and number of neurons of the human hippocampal formation in normal aging and Alzheimer's disease. J Comp Neurol 379:482–494. [DOI] [PubMed] [Google Scholar]
- Small SA, Schobel SA, Buxton RB, Witter MP, Barnes CA. 2011. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat Rev Neurosci 12:585–601. doi:10.1038/nrn3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthana NA, Donix M, Wozny DR, Bazih A, Jones M, Heidemann RM, Trampel R, Ekstrom AD, Scharf M, Knowton B, Turner R, Bookheimer SY. 2015. High‐resolution 7T fMRI of human hippocampal subfields during associative learning. J Cogn Neurosci 27:1194–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theysohn JM, Kraff O, Maderwald S, Schlamann MU, de Greiff A, Forsting M, Ladd SC, Ladd ME, Gizewski ER. 2009. The human hippocampus at 7 T‐in vivo MRI. Hippocampus 19:1–7. doi:10.1002/hipo.20487. [DOI] [PubMed] [Google Scholar]
- Thomas BP, Welch EB, Niederhauser BD, Whetsell WO, Anderson AW, Gore JC, Avison MJ, Creasy JL. 2008. High‐resolution 7T MRI of the human hippocampus in vivo. J Magn Reson Imaging 28:1266–1272. doi:10.1002/jmri.21576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Kolk AG, Hendrikse J, Zwanenburg JJM, Visser F, Luijten PR. 2013. Clinical applications of 7 T MRI in the brain. Eur J Radiol 82:708–718. doi:10.1016/j.ejrad.2011.07.007. [DOI] [PubMed] [Google Scholar]
- van der Zwaag W, Schäfer A, Marques JP, Turner R, Trampel R. 2016. Recent applications of UHF‐MRI in the study of human brain function and structure: A review. NMR Biomed 29:1274–1288. doi:10.1002/nbm.3275. [DOI] [PubMed] [Google Scholar]
- Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinak J, Dickerson BC, Golland P, Fischl B. 2009. Automated segmentation of hippocampal subfields from ultra‐high resolution in vivo MRI. Hippocampus 19:549–557. doi:10.1002/hipo.20615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voevodskaya O, Simmons A, Nordenskjöld R, Kullberg J, Ahlstrüm H, Lind L, Wahlund L‐O, Larsson E‐M, Westman E, Initiative ADN. 2014. The effects of intracranial volume adjustment approaches on multiple regional MRI volumes in healthy aging and Alzheimer's disease. Front Aging Neurosci 6:264. doi:10.3389/fnagi.2014.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, Jack CR, Jagust W, Liu E, Morris JC, Petersen RC, Saykin AJ, Schmidt ME, Shaw L, Siuciak JA, Soares H, Toga AW, Trojanowski JQ. 2012. The Alzheimer's disease neuroimaging initiative: A review of papers published since its inception. Alzheimer's Dement 8:S1–S68. doi:10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MJ, Kawas CH, Stewart WF, Rudow GL, Troncoso JC. 2004. Hippocampal neurons in pre‐clinical Alzheimer's disease. Neurobiol Aging 25:1205–1212. doi:10.1016/j.neurobiolaging.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Wisse LEM, Gerritsen L, Zwanenburg JJM, Kuijf HJ, Luijten PR, Biessels GJ, Geerlings MI. 2012. Subfields of the hippocampal formation at 7T MRI: In vivo volumetric assessment. Neuroimage 61:1043–1049. doi:10.1016/j.neuroimage.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Wisse LEM, Biessels GJ, Heringa SM, Kuijf HJ, Koek DL, Luijten PR, Geerlings MI. 2014a. Hippocampal subfield volumes at 7T in early Alzheimer's disease and normal aging. Neurobiol Aging 35:2039–2045. doi:10.1016/j.neurobiolaging.2014.02.021. [DOI] [PubMed] [Google Scholar]
- Wisse LEM, Biessels GJ, Geerlings MI. 2014b. A critical appraisal of the hippocampal subfield segmentation package in freesurfer. Front Aging Neurosci 6:261. doi:10.3389/fnagi.2014.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse LEM, Kuijf HJ, Honingh AM, Wang H, Pluta JB, Das SR, Wolk DA, Zwanenburg JJM, Yushkevich PA, Geerlings MI. 2016a. Automated hippocampal subfield segmentation at 7T MRI. Am J Neuroradiol 37:1050–1057. doi:10.3174/ajnr.A4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse LEM, Adler DH, Ittyerah R, Pluta JB, Robinson JL, Schuck T, Trojanowski JQ, Grossman M, Detre JA, Elliott MA, Toledo JB, Liu W, Pickup S, Das SR, Wolk DA, Yushkevich PA. 2016b. Comparison of in vivo and ex vivo MRI of the human hippocampal formation in the same subjects. Cereb Cortex. 2016 Sep 24. doi:10.1093/cercor/bhw299. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. 2006. User‐guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 31:1116–1128. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Avants BB, Pluta J, Das S, Minkoff D, Mechanic‐Hamilton D, Glynn S, Pickup S, Liu W, Gee JC, Grossman M, Detre JA. 2009. A high‐resolution computational atlas of the human hippocampus from postmortem magnetic resonance imaging at 9.4 T. Neuroimage 44:385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Wang H, Pluta J, Das SR, Craige C, Avants BB, Weiner MW, Mueller S. 2010. Nearly automatic segmentation of hippocampal subfields in in vivo focal T2‐weighted MRI. Neuroimage 53:1208–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Amaral RSC, Augustinack JC, Bender AR, Bernstein JD, Boccardi M, Bocchetta M, Burggren AC, Carr VA, Chakravarty MM, Chételat G, Daugherty AM, Davachi L, Ding SL, Ekstrom A, Geerlings MI, Hassan A, Huang Y, Iglesias JE, La Joie R, Kerchner GA, LaRocque KF, Libby LA, Malykhin N, Mueller SG, Olsen RK, Palombo DJ, Parekh MB, Pluta JB, Preston AR, Pruessner JC, Ranganath C, Raz N, Schlichting ML, Schoemaker D, Singh S, Stark CEL, Suthana N, Tompary A, Turowski MM, Van Leemput K, Wagner AD, Wang L, Winterburn JL, Wisse LEM, Yassa MA, Zeineh MM. 2015a. Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: Towards a harmonized segmentation protocol. Neuroimage 111:526–541. doi:10.1016/j.neuroimage.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Pluta J, Wang H, Ding SL, Xie L, Gertje E, Mancuso L, Kliot D, Wolk SR, Wolk DA. 2015b. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum Brain Map 36:258–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineh MM, Holdsworth S, Skare S, Atlas SW, Bammer R. 2012. Ultra‐high resolution diffusion tensor imaging of the microscopic pathways of the medial temporal lobe. Neuroimage 62:2065–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineh MM, Chen Y, Kitzler HH, Hammond R, Vogel H, Rutt BK. 2015. Activated iron‐containing microglia in the human hippocampus identified by magnetic resonance imaging in Alzheimer disease. Neurobiol Aging 36:2483–2500. doi:10.1016/j.neurobiolaging.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


