Abstract
Although intensively studied, the high‐resolution crystal structure of the peptide DFNKF, the core‐segment of human calcitonin, has never been described. Here we report how the use of iodination as a strategy to promote crystallisation and facilitate phase determination, allowed us to solve, for the first time, the single‐crystal X‐ray structure of a DFNKF derivative. Computational studies suggest that both the iodinated and the wild‐type peptides populate very similar conformations. Furthermore, the conformer found in the solid‐state structure is one of the most populated in solution, making the crystal structure a reliable model for the peptide in solution. The crystal structure of DFNKF(I) confirms the overall features of the amyloid cross‐β spine and highlights how aromatic–aromatic interactions are important structural factors in the self‐assembly of this peptide. A detailed analysis of such interactions is reported.
Keywords: amyloid beta-peptides, aromatic interactions, crystal structures, iodination, peptides
Introduction
Elucidating the structure and formation mechanism of amyloid fibrils is of paramount importance for current structural biology.1, 2, 3, 4, 5, 6, 7 However, the limited order of fibrils isolated from diseased tissues,8, 9 as well as their great morphological diversity,10, 11 have limited their atomic‐level structural determination. For this reason, a wide array of biophysical tools has been deployed towards reaching this goal, including solid‐state NMR spectroscopy,12, 13, 14, 15, 16 X‐ray diffraction techniques,17, 18, 19, 20, 21, 22 site‐directed spin labelling,23, 24 cryo‐electron microscopy,17, 25, 26 and proline‐scanning mutagenesis, among others.27
An alternative and fruitful strategy towards this same goal is the dissection of amyloidogenic proteins into smaller aggregation‐prone segments28, 29, 30, 31, 32, 33, 34, 35 that may become amenable to X‐ray crystallography allowing their high‐resolution atomic structures to be investigated. In fact, most of these aggregation‐prone segments also form needle‐shaped microcrystals, thus opening the way to structural studies. The crystal structures of these microcrystals have been related to the structures of the fibrils formed by the same peptide or protein.36 This approach has shed important new light on the molecular mechanisms of aggregation into amyloid oligomers and fibrils providing atomic insight into their various polymorphic behaviour.9, 10, 11, 12, 13, 14, 15 For example, these studies allowed the characterization of the “steric zipper”, a motif common to all amyloid structures whereby a self‐complementary interface between a pair of β‐sheets contains tightly interacting side chains,11 as well as the discovery of various oligomeric architectures.28 The pentapeptide DFNKF (H2N‐Asp‐Phe‐Asn‐Lys‐Phe‐COOH, Scheme 1), an amyloidogenic segment derived from the peptide hormone human calcitonin (hCT), is a well‐studied model for amyloid formation.33 In fact, its remarkable ability to form fibrils was already reported in 2002, when Gazit et al. also postulated a possible role of the aromatic residues in the amyloid fibril formation process.34 However, to the best of our knowledge, the high‐resolution crystal structure of this segment has not yet been described. While some authors using solid‐state 13C NMR techniques have reported that DFNKF forms an antiparallel β‐sheet architecture at neutral pH, others have used molecular dynamics simulations and found that the most stable arrangement is the parallel one.12, 37 Here we report for the first time the high‐resolution single crystal X‐ray structure of an iodinated variant of DFNKF, which, we demonstrate by calculations, well represents the wild‐type peptide, thus revealing its virtually unknown packing features as well as the role of the debated aromatic–aromatic interactions involving its phenylalanine residues.
Scheme 1.
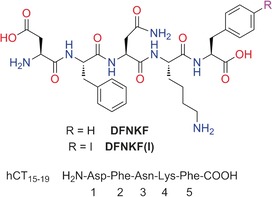
Amino acid sequence and chemical structure of the studied iodinated variant DFNKF(I) of the pentapeptide DFNKF, an amyloidogenic segment derived from the hormone human calcitonin (hCT).
Iodination of peptides and proteins is commonly pursued in order to use the anomalously scattering iodine atom to facilitate single‐wavelength anomalous dispersion38, 39, 40 experiments on in‐house X‐ray sources, thus facilitating structural studies of peptides and proteins. Furthermore, some cases have shown that the introduction of iodinated residues in amino acid sequences may increase their crystallinity.41 Recently, we have demonstrated that iodination at the para‐position of the phenylalanine (Phe) benzene ring in the fifth position of DFNKF does not have much influence on its fibrillating behaviour.42 For this reason, we undertook a detailed computational and crystallographic study on DFNKF(I) with the objective of shedding new light on the controversial nature of the structural organization of this important amyloidogenic peptide (Scheme 1 and section S1 in the Supporting Information).34, 43, 44
Results and Discussion
Conformational analysis of DFNKF segments in solution by MD
In order to assess the possible contribution of iodination to the conformational properties of the DFNKF segment, we initially carried out a molecular dynamics (MD) simulation study on both the wild‐type (WT) peptide and its iodinated variant (Scheme 1). Each peptide was modelled in the extended conformation by using a fully solvated approach at room temperature with the aim of sampling the main conformations that the peptides may adopt in solution. The conformational preferences were extracted by means of a clustering analysis, using the method previously described by Daura et al. (see section S2 in the Supporting Information).45
Structures differing by more than 1 Å in backbone root‐mean‐square deviation (RMSD) along the trajectory were classified as belonging to different clusters (Figure S1). Notably, the three clusters reported in Figure 1 nearly covered 80 % of all structures in the total simulation time for both peptides and, by clustering the backbone conformations in the MD trajectories, no remarkable structural differences were observed in the populations of the WT and iodinated peptides (Figure S2).
Figure 1.
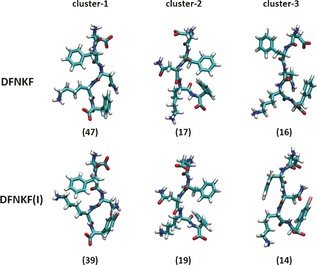
The three most populated structures in solution resulting from MD simulations of both the wild‐type DFNKF and its iodinated variant DFNKF(I). The percentages of the clusters in the MD simulation are shown in parenthesis.
The structures of the three clusters in Figure 1 are very similar with average backbone RMSD below 1 Å (Figure S3). In particular, the representative structure of cluster‐1, common to both DFNKF and DFNKF(I), is rather extended and close to the starting conformation. Conversely, in cluster‐2 both systems adopt a more compact structure with a bent backbone on the Lys4 and with the two Phe rings in close proximity. In cluster‐3, the two peptide segments are again characterized by a rather extended conformation similar to that in cluster‐1.
These initial MD studies demonstrated that the conformational energy landscapes of the two peptides are very similar suggesting that the added iodine atom is not a determining factor in the conformational distribution in solution. Overall, these findings support our initial hypothesis that the iodinated peptide may be used as a good mimic of the WT one.
Single‐crystal X‐ray diffraction analysis of DFNKF(I)
Encouraged by the above MD results, we attempted the crystallization of the iodinated DFNKF. DFNKF(I) was successfully crystallized from a 1:1 mixture of water and acetonitrile by using the sitting‐drop vapour diffusion method (see section S3 in the Supporting Information). Single crystals suitable for X‐ray diffraction analysis were collected after six months and data acquisition was performed using an in‐house X‐ray source (Table S1). The analysis revealed that the asymmetric unit consists of a single molecule of DFNKF(I), in a fully extended conformation similar to those observed in clusters‐1 and ‐3, with four water molecules (Figure S4). Each DFNKF(I) molecule packs by forming three backbone C=O⋅⋅⋅NH hydrogen bonds (HBs) with an adjacent molecule (N2H⋅⋅⋅O1: 3.05(3) Å, 159.8°; N4H⋅⋅⋅O3: 2.91(3) Å, 169.2°; N5H⋅⋅⋅O4: 2.86(4) Å, 164.4°) and one bifurcated HB involving the amide oxygen atom of the asparagine residue with both the N terminus and the backbone nitrogen of the same residue of a neighbouring molecule (N3H⋅⋅⋅O9: 3.01(3) Å, 164.6°; N1H⋅⋅⋅O9: 2.96(2) Å, 150.7°; Figure 2, top).
Figure 2.
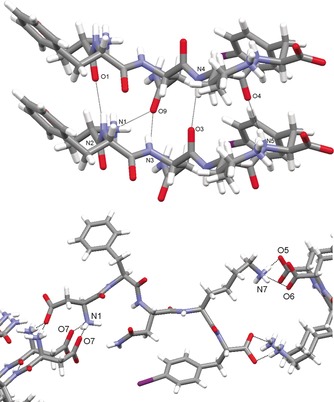
Partial crystal packing views of DFNKF(I) where the hydrogen bonds (dotted black lines) in the β‐sheet (top) and in the lateral self‐assembly are shown (bottom). Colour code: C, grey; O, red; N, light blue; I, purple; H, white.
The lateral self‐assembly of the pentapeptide molecules is driven by electrostatic interactions between the ionisable groups at either end. Specifically, charge‐assisted and bifurcated HBs occur between the N terminus and the aspartic acid side chain (N1H⋅⋅⋅O7: 2.78(3) Å, 173.9°; 2.77(3) Å, 165.9°), and between the C terminus and the lysine side chain of following peptide (N7H⋅⋅⋅O6: 2.81(3) Å, 153.2°; N7H⋅⋅⋅O5: 2.75(3) Å, 159.4°; Figure 2, bottom). As a result of the HB pattern, the DFNKF(I) molecules stack into standard Pauling–Corey parallel β‐sheets as recognized by the fact that each hydrogen‐bonded ring consists of 12 atoms (Figure 2, top) and further confirmed by the backbone torsion angles (Table S2). This finding is in good agreement with theoretical models of DFNKF previously described in the literature,37 while in apparent contrast with the only other example of its experimental structure determined by solid state NMR spectroscopy.12 It is also important to point out that similar to what was shown by previous calculations for DFNKF,46 in the crystal packing of its iodinated derivative the amide groups of the Asn residues stack within a sheet (Asn‐Asn interactions), thus forming the so‐called Asn ladder (Figure 3, top). The stacks of parallel β‐sheets then interact to form the cross β‐structure, which is a key component of the amyloid assembly. The DFNKF(I) β‐strands within each sheet are parallel and exactly in register (Figure 3, top) and, akin to other crystal structures of short amyloidogenic peptides, each pair of sheets is related by a 21 screw axis. The strands in one sheet are antiparallel to those in the next sheet it interacts with, and each sheet is shifted along the screw axis relative to its partner sheet by around 2.4 Å, which is almost half the distance of the overall strand–strand separation of 4.83 Å (Figure 3, bottom).
Figure 3.
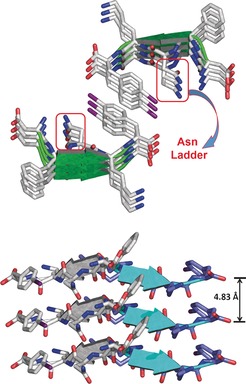
Representation of the DFNKF(I) secondary structure. Top: two facing β‐sheets forming a hydrophobic pocket containing the iodinated aromatic residues. The parallel stacking of the β‐strands leads to the formation of the asparagine ladder. Bottom: three parallel β‐strands in one sheet, which are antiparallel to those in the adjacent sheet. β‐Strands are depicted as cartoons.
There are two different interfaces between the sheets (Figure 4, top). The wet interface is lined with water molecules that completely separate DFNKF(I) molecules, whereby the separation of sheets is around 15.7 Å (Figure S5). In contrast, the dry interface contains no water molecules and therefore the sheets are closer together, being separated by around 10.4 Å (Figure S6). At the wet interface, each polar side chain is hydrated by water molecules that create a HB network involving the carboxylic moieties of adjacent sheets. Conversely, the side chains present at the dry interface (namely Phe and Lys) are securely interdigitated with the same Phe and Lys residues of its interacting partner sheet, thus creating a hydrophobic pocket. All of the above‐described interactions are the basis of the so‐called steric zipper, which is typical of all amyloidogenic peptide structures reported to date.11, 47
Figure 4.
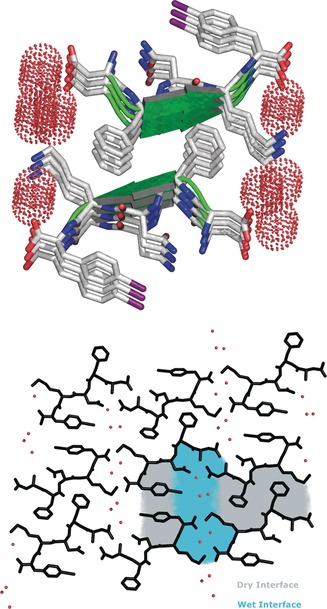
Representation of the steric zipper in DFNKF(I). Top: partial DFNKF(I) crystal packing viewed along the b axis and showing nine sheets. Pairing of the β‐sheets into the steric zippers creates the dry and wet interfaces. The wet interfaces are filled in with water molecules, whereas the closely spaced dry interfaces lack water molecules. Bottom: view along the crystallographic b axis of two facing β‐sheets formed by three parallel β‐strands showing their remarkable shape complementarity, which promotes the formation of the steric zipper. Peptide molecules are shown in black and water molecules are represented by red dots.
According to literature, the steric zipper in the crystal structure of DFNKF(I) can be classified as face‐to‐face–up‐up–class‐1 (Figure 4, bottom).11 This arrangement is common to other amyloidogenic sequences such as GNNQQNY (derived from yeast prion protein Sup35) and VQIVYK (from the Tau protein).11 It has been previously reported that the stacking of the pairs of β‐sheets at the dry interface is the stable structural unit of the cross‐β‐spine and the self‐complementarity of the steric zipper explains how very short peptide sequences are able to form such extended structures like amyloid fibrils.48
Detail of the aromatic–aromatic interactions in DFNKF(I)
Aromatic–aromatic interactions49, 50 (π⋅⋅⋅π) are important stabilizing forces in protein and fibril structures.50, 51, 52 The high‐resolution single‐crystal X‐ray structure of DFNKF(I), thus offers the unique opportunity to probe the aromatic–aromatic interactions between the phenylalanine side chains of this important amyloidogenic segment.53, 54 Specifically, in the parallel β‐sheets, aromatic–aromatic interactions are observed between the rings of Phe2 residues, which adopt a parallel‐displaced arrangement. Stacking parameters are as follows: the centroid–centroid distance55 (R cen) corresponds to the strand–strand separation distance of 4.83 Å (Figure 3, bottom) and the shortest distance between two carbon atoms of the interacting rings R close 56 (R clo_C14–C17) is around 3.4 Å. The offset value, representing the distance between the two centroids projected on a plane defined by the atoms of one ring, is very close to R clo_C14–C17, that is, 3.4 Å, which confirms that the aromatic rings are almost stacked. The same residues are also involved in quite short, thus relatively strong, CH⋅⋅⋅π interactions occurring between the hydrogen atoms of the benzyl CH2 groups and the electron‐rich π‐surface of the aromatic rings; hydrogen atoms (H13A and H13B)‐centroid distance are 2.83 and 3.31 Å, respectively (Figure 5, top). Also, pairs of iodinated rings of the Phe5 residues are observed interacting along the parallel β‐sheets by weak π⋅⋅⋅π interactions occurring in a parallel‐displaced arrangement with a R clo_C28–C32 of 3.29 Å and an offset value of 3.4 Å (Figure 5, bottom).
Figure 5.
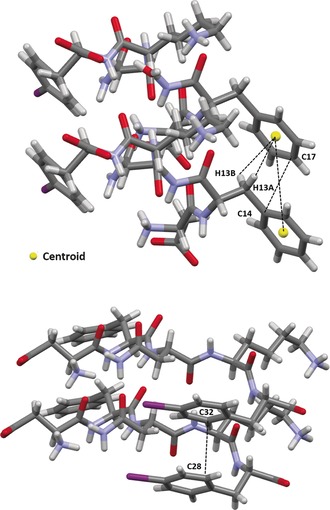
Aromatic–aromatic and CH⋅⋅⋅π interactions involving Phe2 residues (top) and iodinated Phe5 residues (bottom) detected across the parallel β‐sheets of the DFNKF(I) crystal.
Furthermore, aromatic–aromatic interactions take place between neighbouring β‐sheets. Two adjacent iodinated phenyl moieties of different strands adopt a T‐shape arrangement (edge‐to‐face) with R cen of 4.8 Å and R clo_C28–C29 of 3.56 Å. This motif is further stabilised by the presence of two weak hydrogen bonds involving the iodine atom of the interacting iodinated ring. The HB donors are positioned on two different DFNKF(I) molecules, one hydrogen atom is located at the meta position of the same aromatic ring involved in the T‐shape motif (H28⋅⋅⋅I distance 3.4 Å), while the other hydrogen atom belong to the benzyl CH2 unit of the translated molecule (H26B⋅⋅⋅I distance 3.5 Å; Figure 6, top). This T‐shape arrangement of the iodinated rings promotes the formation of an additional hydrophobic pocket where no water molecules are detected (Figure 3, top, and Figure 4, bottom). As a result of this antiparallel edge‐to‐face organization of the aromatic rings, the corresponding iodine atoms point radially towards the wet interface without displaying any further intermolecular contacts.
Figure 6.
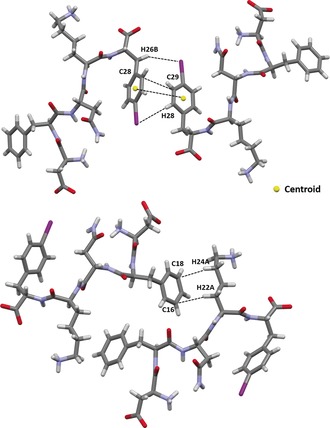
Top: aromatic–aromatic interactions and hydrogen–iodine contacts promote the T‐shape arrangement (edge‐to‐face) between two adjacent iodinated Phe5 residues. Bottom: CH⋅⋅⋅π interactions occurring between the aromatic surface of the Phe2 residue and the methylene hydrogen atoms on the lysine stabilize the steric zipper in the DFNKF(I) crystal structure.
Finally, the steric zipper is stabilized by intermolecular contacts involving the aromatic rings. In fact, the relatively acidic methylene hydrogen atoms on the lysine residues interact through CH⋅⋅⋅π interactions with the electron‐rich π‐surface of the aromatic ring of the Phe2 ring (distances for C16⋅⋅⋅H22A and C18⋅⋅⋅H24A are 2.84 and 2.87 Å, respectively; Figure 6, bottom).
Noncovalent interaction (NCI) analysis on the DFNKF(I) crystal
It is quite clear from above that noncovalent interactions involving aromatic residues cooperate with hydrogen bonds to stabilize the parallel β‐sheet arrangement and influence the overall crystal packing of the DFNKF(I) segment. In order to better visualize these interactions, we performed noncovalent interaction (NCI) analysis57, 58 on the DFNKF(I) crystal by exploiting electron distributions resulting from density functional theory (DFT) calculations at the B3LYP/DZP Level.59, 60, 61
In particular, we studied sub‐packing of the crystal showing various interacting dimers (see section S4 in the Supporting Information): 1) a dimer of the unit cell (without water molecules); 2) a peptide dimer along the crystallographic a axis; 3) a peptide dimer along the crystallographic b axis (β‐sheet); 4) a dimer of the steric zipper (Figure S7). Furthermore, with the aim of evaluating the influence played by the iodine atom on the noncovalent interaction patterns, the corresponding four non‐iodinated systems, bearing hydrogen atoms at correctly optimised positions, were also investigated.
Alongside the strong hydrogen bonds that enable the construction of the β‐sheets, the NCI analysis confirms the presence of numerous interactions involving the phenyl rings of the Phe2 and Phe5 residues, for both simulated systems, showing quite extended interaction isosurfaces at the aromatic moieties. Interestingly, the calculations show an intramolecular interaction between the iodine atom and the NH2 moiety of the Asn residue (Figure 7 a and c, and Figure S9). This contact contributes to stabilize the “U‐shape” conformation adopted by the Asn3‐Lys4‐Phe(I)5 sub‐segment favouring the orientation of the aromatic ring towards the formation of π⋅⋅⋅π interactions along the crystallographic a axis (Figure S10). As a result, the NCI isosurfaces between the phenyl rings of the Phe5 in the T‐shape arrangement are more extended in the iodinated segment than in the WT one (Figures 7 a and b). The reason for this difference is that the iodine atoms partially share their electron density with nearby hydrogen atoms further promoting the stabilization of the packing. The electron‐density studies also show the presence of NCI isosurfaces between the acidic methylene hydrogen atoms on the lysine residues and the π‐surface of the Phe2 ring in the steric zipper highlighting the importance of CH⋅⋅⋅π contacts in the construction of the dry interfaces (Figure 7 c and Figure S12). Overall, the NCI analysis allows visualization of the extended network of stabilizing interactions involving the aromatic rings and shows that the iodine atom actively contributes to the formation of weak intra‐ and intermolecular contacts that strengthen the aromatic–aromatic pairing. In this view, the iodine atom on Phe5 may play the role of potentially limiting the conformational freedom of the peptide chains in the initial steps of aggregation, thus favouring ordered packing of the chains and, as observed in our case, the crystallization of the DFNKF segment.
Figure 7.
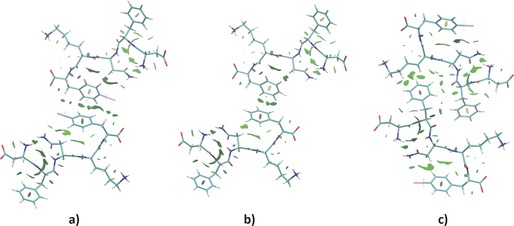
Dimers of DFNKF(I) and DFNKF obtained by NCI calculations where the NCI isosurfaces are shown. A dimer of: a) DFNKF(I), and b) DFNKF formed by the asymmetric unit (without water molecules) and its nearest peptide neighbour along the crystallographic a axis and showing different NCI isosurfaces. The DFNKF(I) molecule displays more extended isosurface contacts around the Phe5 compared to the wild‐type segment. c) The “steric zipper” dimer of the DFNKF(I) shows several NCI isosurface involving acidic methylene hydrogen atoms on the lysine residues and the π‐surface of the Phe2. All the NCI calculations were performed at B3LYP/DZP level. The cut‐offs are set at s=0.5 a.u. and −0.05 a.u.<sign(λ 2)ρ<0.05 a.u., while the NCI colour scale is defined according to the values of sign(λ 2)ρ, with blue indicating strong attractive interactions, green indicating van der Waals interactions and red indicating strong non‐bonded overlaps. For the complete meaning of s, ρ and λ 2, see section S4 in the Supporting Information.
MD conformational analysis of DFNKF(I) molecule in the crystal
Further MD simulations were performed in order to evaluate the effective role of the iodine atom on the conformational flexibility of the peptide. A model for DFNKF(I) was obtained starting from its crystal structure and after simulation at room temperature for a total time of 100 ns. The model of the WT sequence DFNKF crystal was obtained by replacing the iodinated phenylalanine with its hydrogenated counterpart and after simulation under the same conditions (see section S5 in the Supporting Information).
The results show a strong increase in the mobility of the aromatic rings in the DFNKF crystal, as well as higher root mean square fluctuation values for the backbone of Asn3, when compared to the DFNKF(I) crystal simulation. These results nicely mirror those obtained by the NCI analysis where the presence of the isosurface contact between the iodine atom and the Asn3 is highlighted. Removing this contact substantially weakens the crystal packing therefore destabilising it.
Finally, the MD trajectories of the isolated peptides in solution were compared to the crystal backbone conformations to assess their significance under physiological conditions. It has to be noted, however, that the starting structure of the MD simulations was the fully extended one in both peptide systems, which is, therefore, unrelated to the actual crystal conformation. Yet, the time‐dependent RMSD, calculated for the backbone atoms (see sections S2 and S5 in the Supporting Information), demonstrate that a structure close (RMSD ≈0.7 Å) to the crystal conformation was recursively populated during the simulations of both DFNKF and of DFNKF(I). Therefore, the conformation observed in the crystal is representative of one actual conformer in solution.
Based on the cluster decomposition analysis discussed above, the crystal conformation was compared to each of the three most populated clusters (Figure 1). The distribution of RMSD values between the members of each cluster and the crystal structure demonstrated that the third cluster best represents the solution conformer observed in the crystal. This confirms that the crystal structure conformation is significantly populated in solution, thus the structural features observed in the solid state may be used to predict the behaviour of the peptide in solution.
Conclusions
In conclusion, the high resolution crystal structure of the peptide DFNKF, a core‐segment derived from human calcitonin, although intensively studied, has never been described. Herein we have reported the single‐crystal X‐ray structure of an iodinated variant of DFNKF, that is, DFNKF(I). Computational analyses show that this iodinated variant represents the conformational structures of the wild‐type peptide, both in solution and in the solid state. In fact, from the computational point of view, it is suggested that both the iodinated and the wild‐type peptides populate a very similar range of conformations and that the conformer found in the crystal structure was present in solution. This finding highlights that the solid‐state structure is representative of the peptide in solution and thus a good model. Furthermore, our study clearly demonstrates, with solid scientific grounds, why iodination may promote peptide crystallisation, by introducing secondary contacts that stabilize the overall packing as a consequence of the reduced conformational freedom. Iodination, alongside facilitating phase determination, may be developed as a routine strategy to obtain the single‐crystal X‐ray structures of peptide segments otherwise difficult to crystallize.62 Our results shed light on the virtually unknown packing features of DFNKF, which shows the formation of parallel β‐sheets that are laterally stabilized by electrostatic, namely charge‐assisted hydrogen bonds, and hydrophobic interactions. Importantly, the detailed nature of the long‐debated aromatic–aromatic interactions, typically occurring in peptide segments containing phenylalanine63 and tyrosine,64 is described in detail. In particular, parallel‐displaced and edge‐to‐face interactions are observed, along with important CH⋅⋅⋅π interactions involving the side‐chain methylene hydrogens of Phe. Interfering with these interactions involving aromatic residues may provide clues for future design of fibrillation inhibitor and disruptors. Efforts in this direction are currently underway in our laboratory and will be reported elsewhere.
Experimental Section
Computational analysis molecular dynamics (MD): monomers in solution
GROMACS 4.6.3 was used for the MD simulations.65 DFNKF and DFNKF(I) peptides were modelled in extended conformations and inserted in a fully solvated cubic box with a volume of 81 nm3, containing 2647 TIP4P water molecules.66 The systems were subjected to energy minimization by using steepest descent algorithm for 5000 steps. A single trajectory was calculated, with a total simulation time of 1.1 microsecond. The first 500 ps were discarded to account for equilibration and frames were saved every 2 ps. The calculation of the electrostatic forces utilized the PME implementation of the Ewald summation method. The LINCS67 algorithm was used to constrain all bond lengths, while SETTLE was used for the water molecules.68 See the Supporting Information for more detail.
Peptide crystallization
DFNK(para‐iodophenylalanine, DNFKF(I)) crystals were grown by using the sitting‐drop vapour diffusion method. The peptide was dissolved in a 1:1 mixture of deionized water (18.2 MΩ cm) and acetonitrile at room temperature at 1 mg mL−1. A drop of 20 μL was deposited and put in vapour diffusion with a reservoir containing only deionized water (18.2 MΩ cm). Single crystals were grown over a six‐month period.
X‐ray crystallography data acquisition
Crystals of DFNK(para‐iodophenylalanine) were extremely thin, curved yellow sheets or laminae. X‐ray diffraction data were collected from DFNK(para‐iodophenylalanine) crystals with a Bruker APEX‐II diffractometer equipped with microfocus and CCD detector by using MoKα radiation (λ=0.71073 Å) and with a collection of 600“/frame. The crystal was cryo‐cooled (103 K) for data collection (Bruker KRYOFLEX). The structure was solved by SIR200269 and refinements were carried out by full‐matrix least‐squares on F 2 using the SHELXL70 program. See the Supporting Information for more detail.
Noncovalent interaction (NCI) analysis on the DFNKF(I) crystal
Noncovalent interaction (NCI) analyses57, 58 were performed on model systems extracted from the crystal structure of the DFNKF(I) peptide (Figure S7) and the missing hydrogen atoms of the water molecules were added by using the rules present in the hydrogen Gromacs database.71 Afterwards, exploiting the Gaussian09 quantum chemistry package,72 for each system we carried out DFT calculations using the B3LYP functional73 and the DZP basis‐set.60, 61 These quantum mechanical computations provided the electron distributions that were employed in the NCI calculations performed by means of the software NCIPLOT‐3.0.58 See the Supporting Information for more detail.
Computational analysis of the crystal model and molecular dynamics
The model of the crystal was created by using the procedure described by Case et al.74 The asymmetric units (ASU) described in Table S2 in the Supporting Information were replicated to obtain a “supercell” of 3×5×3 using the program Chimera (UCSF Chimera: a visualization system for exploratory research and analysis).75 The obtained “supercell” contains 90 peptide fragments and 360 water molecules. The supercell was used to define the central tetrahedral simulation box, of size (55.917, 24.160, 65.589 Å), which was subjected to periodic boundary conditions (PBCs). Using this setup, the virtual PBC images of the system were in contact with the main box, mimicking an infinite crystal. See the Supporting Information for more detail.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
The European Research Council is gratefully acknowledged for the Starting Grant ERC‐2012‐StG_20111012 FOLDHALO (Grant Agreement Number 307108) to P.M.
A. Bertolani, A. Pizzi, L. Pirrie, L. Gazzera, G. Morra, M. Meli, G. Colombo, A. Genoni, G. Cavallo, G. Terraneo, P. Metrangolo, Chem. Eur. J. 2017, 23, 2051.
References
- 1. Selkoe D. J., Physiol. Rev. 2001, 81, 741–767. [DOI] [PubMed] [Google Scholar]
- 2. Cukalevski R., Yang X., Meisl G., Weininger U., Bernfur K., Frohm B., Knowles T. P. J., Linse S., Chem. Sci. 2015, 6, 4215–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dobson C. M., Nature 2003, 426, 884–889. [DOI] [PubMed] [Google Scholar]
- 4. Frenkel-Pinter M., Richman M., Belostozky A., Abu-Mokh A., Gazit E., Rahimipour S., Segal D., Chem. Eur. J. 2016, 22, 5945–5952. [DOI] [PubMed] [Google Scholar]
- 5. Palmal S., Ranjan Maity A., Kumar Singh B., Basu S., Jana N. R., Jana N. R., Chem. Eur. J. 2014, 20, 6184–6191. [DOI] [PubMed] [Google Scholar]
- 6. Greenwald J., Riek R., Structure 2010, 18, 1244–1260. [DOI] [PubMed] [Google Scholar]
- 7. Peacock H., Luo J., Yamashita T., Luccarelli J., Thompson S., Hamilton A. D., Chem. Sci. 2016, 7, 6435–6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Selkoe D. J., Nature 2003, 426, 900–904. [DOI] [PubMed] [Google Scholar]
- 9. Chiti F., Dobson C. M., Annu. Rev. Biochem. 2006, 75, 333–366. [DOI] [PubMed] [Google Scholar]
- 10. Kreplak L., Aebi U., Adv. Protein Chem. 2006, 73, 217–233. [DOI] [PubMed] [Google Scholar]
- 11. Sawaya M. R., Sambashivan S., Nelson R., Ivanova M. I., Sievers S. A., Apostol M. I., Thompson M. J., Balbirnie M., Wiltzius J. J. W., McFarlane H. T., Madsen A. Ø., Riekel C., Eisenberg D., Nature 2007, 447, 453–457. [DOI] [PubMed] [Google Scholar]
- 12. Naito A., Kamihira M., Inoue R., Saito H., Magn. Reson. Chem. 2004, 42, 247–257. [DOI] [PubMed] [Google Scholar]
- 13. Jaroniec C. P., MacPhee C. E., Astrof N. S., Dobson C. M., Griffin R. G., Proc. Natl. Acad. Sci. USA 2002, 99, 16748–16753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Petkova A. T., Ishii Y., Balbach J. J., Antzutkin O. N., Leapman R. D., Delaglio F., Tycko R., Proc. Natl. Acad. Sci. USA 2002, 99, 16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin N., Chao J. C., Cheng H., Chou F., Chang C., Chen Y., Chang Y., Huang S., Chan J. C. C., Chem. Eur. J. 2010, 16, 5492–5499. [DOI] [PubMed] [Google Scholar]
- 16. Colvin M. T, Silvers R., Zhe Ni Q., Can T. V., Sergeyev I. V., Rosay M., Donovan K. J., Michael B., Wall J. S., Linse S., Griffin R. G., J. Am. Chem. Soc. 2016, 138, 9663–9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Langkilde A. E., Morris K. L., Serpell L. C., Svergun D. I., Vestergaard B., Acta Crystallogr. Sect. A 2015, 71, 882–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sunde M., Blake C., Adv. Protein Chem. 1997, 50, 123–159. [DOI] [PubMed] [Google Scholar]
- 19. Stroud J. C., Liu C., Teng P. K., Eisenberg D., Proc. Natl. Acad. Sci. USA 2012, 109, 7717–7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diaz-Avalos R., Long C., Fontano E., Balbirnie M., Grothe R., Eisenberg D., Caspar D. L. D., J. Mol. Biol. 2003, 330, 1165–1175. [DOI] [PubMed] [Google Scholar]
- 21. Pinto Oliveira C. L., Behrens M. A., Pedersen J. S., Erlacher K., Otzen D., Pedersen J. S., J. Mol. Biol. 2009, 387, 147–161. [DOI] [PubMed] [Google Scholar]
- 22. Vestergaard B., Groenning M., Roessle M., Kastrup J. S., van de Weert M., Flink J. M., Frokjaer S., Gajhede M., Svergun D. I., PLoS Biol. 2007, 5, e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Torok M., Milton S., Kayed R., Wu P., McIntire T., Glabe C. G., Langen R., J. Biol. Chem. 2002, 277, 40810–40815. [DOI] [PubMed] [Google Scholar]
- 24. Serag A. A., Altenbach C., Gingery M., Hubbell W. L., Yeates T. O., Biochemistry 2001, 40, 9089–9096. [DOI] [PubMed] [Google Scholar]
- 25. Jiménez J. L., Guijarro J. I., Orlova E., Zurdo J., Dobson C. M., Sunde M., Saibil H. R., EMBO J. 1999, 18, 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiménez J. L., Tennent G., Pepys M., Saibil H. R., J. Mol. Biol. 2001, 311, 241–247. [DOI] [PubMed] [Google Scholar]
- 27. Williams A. D., Portelius E., Kheterpal I., Guo J. T., Cook K. D., Xu Y., Wetzel R., J. Mol. Biol. 2004, 335, 833–842. [DOI] [PubMed] [Google Scholar]
- 28. Apostol M. I., Perry K., Surewicz W. K., J. Am. Chem. Soc. 2013, 135, 10202–10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kok W. M., Cottam J. M., Ciccotosto G. D., Miles L. A., Karas J. A., Scanlon D. B., Roberts B. R., Parker M. W., Cappai R., Barnham K. J., Hutton C. A., Chem. Sci. 2013, 4, 4449–4454. [Google Scholar]
- 30. Spencer R. K., Li H., Nowick J. S., J. Am. Chem. Soc. 2014, 136, 5595–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Daly S., Kulesza A., Poussigue F., Simon A., Choi C. M., Knight G., Chirot F., MacAleese L., Antoine R., Dugourd P., Chem. Sci. 2015, 6, 5040–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ray S., Das A. K., Drew M. G. B., Banerjee A., Chem. Commun. 2006, 4230–4232. [DOI] [PubMed] [Google Scholar]
- 33. Lakshmanan A., Cheong D. W., Accardo A., Di Fabrizio E., Riekel C., Hauser C. A. E., Proc. Natl. Acad. Sci. USA 2013, 110, 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reches M., Porat Y., Gazit E., J. Biol. Chem. 2002, 277, 35475–35480. [DOI] [PubMed] [Google Scholar]
- 35. Yoo S., Kreutzer A. G., Truex N. L., Nowick J. S., Chem. Sci. 2016, 7, 6946–6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Görbitz C. H., Chem. Commun. 2006, 2332–2334. [DOI] [PubMed] [Google Scholar]
- 37. Zanuy D., Haspel N., Tsai H.-H. G., Ma B., Gunasekaran K., Wolfson H. J., Nussinov R., Phys. Biol. 2004, 1, 89–99. [DOI] [PubMed] [Google Scholar]
- 38. Chen L. Q., Rose J. P., Breslow E., Yang D., Chang W. R., Furey W. F., Sax M., Wang B. C., Proc. Natl. Acad. Sci. USA 1991, 88, 4240–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schneggenburger P. E., Beerlink A., Worbs B., Salditt T., Diederichsen U., ChemPhysChem 2009, 10, 1567–1576. [DOI] [PubMed] [Google Scholar]
- 40. Pham J. D., Chim N., Goulding C. W., Nowick J. S., J. Am. Chem. Soc. 2013, 135, 12460–12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Obrecht D., Altorfer M., Bohdal U., Daly J., Huber W., Labhardt A., Lehmann C., Muller K., Ruffieux R., Schonholzer P., Spiegler C., Zumbrunn C., Biopolymers 1997, 42, 575–626. [Google Scholar]
- 42. Bertolani A., Pirrie L., Houbenov N., Haataja J., Stefan L., Catalano L., Terraneo G., Giancane G., Valli L., Milani R., Ikkala O., Resnati G., Metrangolo P., Nat. Commun. 2015, 6, 7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gavin Tsai H.-H., Zanuy D., Haspel N., Gunasekaran K., Ma B., Tsai C.-J., Nussinov R., Biophys. J. 2004, 87, 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haspel N., Zanuy D., Ma B., Wolfson H., Nussinov R., J. Mol. Biol. 2005, 345, 1213–1227. [DOI] [PubMed] [Google Scholar]
- 45. Daura X., Gademann K., Jaun B., Seebach D., van Gunsteren W. F., Mark A. E., Angew. Chem. Int. Ed. 1999, 38, 236–240; [Google Scholar]; Angew. Chem. 1999, 111, 249–253. [Google Scholar]
- 46. Tsai H.-H. G., Reches M., Tsai C.-J., Gunasekaran K., Gazit E., Nussinov R., Proc. Natl. Acad. Sci. USA 2005, 102, 8174–8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stroud J. C., Acta Crystallogr. Sect. A 2013, 69, 540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nelson R., Sawaya M. R., Balbirnie M., Madsen A. Ø., Riekel C., Grothe R., Eisenberg D., Nat. Cell Biol. 2005, 7, 773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martinez C. R., Iverson B. L., Chem. Sci. 2012, 3, 2191–2201. [Google Scholar]
- 50. Aravinda S., Shamala N., Das C., Sriranjini A., Karle I. L., Balaram P., J. Am. Chem. Soc. 2003, 125, 5308–5315. [DOI] [PubMed] [Google Scholar]
- 51. Burley S. K., Petsko G. A., Science 1985, 229, 23–28. [DOI] [PubMed] [Google Scholar]
- 52. McGaughey G. B., Gagné M., Rappé A. K., J. Biol. Chem. 1998, 273, 15458–15463. [DOI] [PubMed] [Google Scholar]
- 53. Valdes H., Pluhackova K., Hobza P., J. Chem. Theory Comput. 2009, 5, 2248–2256. [DOI] [PubMed] [Google Scholar]
- 54. Yan X., Zhu P., Li J., Chem. Soc. Rev. 2010, 39, 1877–1890. [DOI] [PubMed] [Google Scholar]
- 55. R centroid (R cen): the distance between two centroids, here the distance between the geometric centres of the aromatic rings of nearby phenylalanine residues.
- 56. R close (R clo): the shortest distance between two carbon atoms belonging to different, interacting rings; here the rings are the aromatic phenylalanine residues of nearby peptide strands.
- 57. Johnson E. R., Keinan S., Mori Sánchez P., Contreras García J., Cohen A. J., Yang W., J. Am. Chem. Soc. 2010, 132, 6498–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Contreras-García J., Johnson E. R., Keinan S., Chaudret R., Piquemal J. P., Beratan D. N., Yang W., J. Chem. Theory Comput. 2011, 7, 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Becke A. D., J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar]
- 60. Canal Neto A., Muniz E. P., Centoducatte R., Jorge F. E., J. Mol. Struct. 2005, 718, 219–224. [Google Scholar]
- 61. Barros C. L., de Oliveira P. J. P., Jorge F. E., Canal Neto A., Campos M., Mol. Phys. 2010, 108, 1965–1972. [Google Scholar]
- 62. Collie G. W., Pulka-Ziach K., Guichard G., Chem. Commun. 2016, 52, 1202–1205. [DOI] [PubMed] [Google Scholar]
- 63. Gazit E., Nat. Chem. 2014, 7, 14–15. [DOI] [PubMed] [Google Scholar]
- 64. Rodríguez-Pérez J. C., Hamley I. W., Squires A. M., Phys. Chem. Chem. Phys. 2013, 15, 13940–13950. [DOI] [PubMed] [Google Scholar]
- 65. van der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. C., J. Comput. Chem. 2005, 26, 1701–1718. [DOI] [PubMed] [Google Scholar]
- 66. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., J. Chem. Phys. 1983, 79, 926–935. [Google Scholar]
- 67. Berendsen H. J. C., Grigera J. R., Straatsma T. P., J. Phys. Chem. 1987, 91, 6269–6271. [Google Scholar]
- 68. Miyamoto S., Kollman P. A., J. Comput. Chem. 1992, 13, 952–962. [Google Scholar]
- 69. Burla M. C., Camalli M., Carrozzini B., Cascarano G. L., Giacovazzo C., Polidori G., Spagna R., J. Appl. Crystallogr. 2003, 36, 1103. [Google Scholar]
- 70. Sheldrick G. M., J. Appl. Crystallogr. 2011, 44, 1281–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bekker H., Berendsen H. J. C., Dijkstra E. J., Achterop S., van Drunen R., van der Spoel D., Sijbers A., Keegstra H., Reitsma B., Renardus M. K. R., in Physics Computing 92 (Eds.: R. A. de Groot, J. Nadrchal), World Scientific, Singapore, 1993. [Google Scholar]
- 72.Gaussian 09, Revision D.01, M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery, Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski, D. J. Fox, Gaussian, Inc., Wallingford CT, 2009.
- 73. Stephens P. J., Devlin F. J., Chabalowski C. F., Frisch M. J., J. Phys. Chem. 1994, 98, 11623–11627. [Google Scholar]
- 74. Janowski P. A., Cerutti D. S., Holton J., Case D. A., J. Am. Chem. Soc. 2013, 135, 7938–7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., J. Comput. Chem. 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


