Abstract
A mouth is present in all animals, and comprises an opening from the outside into the oral cavity and the beginnings of the digestive tract to allow eating. This review focuses on the earliest steps in mouth formation. In the first half, we conclude that the mouth arose once during evolution. In all animals, the mouth forms from ectoderm and endoderm. A direct association of oral ectoderm and digestive endoderm is present even in triploblastic animals, and in chordates, this region is known as the extreme anterior domain (EAD). Further support for a single origin of the mouth is a conserved set of genes that form a ‘mouth gene program’ including foxA and otx2. In the second half of this review, we discuss steps involved in vertebrate mouth formation, using the frog Xenopus as a model. The vertebrate mouth derives from oral ectoderm from the anterior neural ridge, pharyngeal endoderm and cranial neural crest (NC). Vertebrates form a mouth by breaking through the body covering in a precise sequence including specification of EAD ectoderm and endoderm as well as NC, formation of a ‘pre‐mouth array,’ basement membrane dissolution, stomodeum formation, and buccopharyngeal membrane perforation. In Xenopus, the EAD is also a craniofacial organizer that guides NC, while reciprocally, the NC signals to the EAD to elicit its morphogenesis into a pre‐mouth array. Human mouth anomalies are prevalent and are affected by genetic and environmental factors, with understanding guided in part by use of animal models. WIREs Dev Biol 2017, 6:e275. doi: 10.1002/wdev.275
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
Multicellular animals need to eat and a mouth is the organ that allows food into the digestive system. It comprises the opening from the outside of the animal, the oral cavity that is connected to the opening and the beginning of the digestive system, the pharynx. Even some single‐celled organisms like Paramecia have a mouth leading into a subcellular intestine.1 Many animals have accessory structures that assist eating and mouth function, and increase complexity of this organ. We hypothesize that the mouth arose once in evolution, and consider two lines of evidence that support this. These include the understanding that the mouth is always built from ectodermal and endodermal lineages. The first multicellular animals with a clear mouth were diploblasts (with ectoderm and endoderm).1, 2, 3 Interestingly, even in triploblastic animals that include mesoderm, the mouth still forms from a region where ectoderm and endoderm directly juxtapose.2, 3 In chordates, we named this region the extreme anterior domain (EAD).4, 5 Another aspect of the ancient origin of the mouth is conservation of gene expression, leading to the proposal of a ‘mouth gene program.’
In initial studies of the Xenopus mouth, we coined the term ‘primary mouth’ to indicate the initial or immature larval mouth, and ‘secondary mouth’ to indicate later elaboration and differentiation of structures to form the mature mouth. Although this nomenclature has been useful to the community, on consideration, we think the general term ‘mouth’ is most useful throughout development. We view mouth development as a continuum, where even at the earliest time after mouth opening, an oral cavity and accessory structures are already forming. This review focuses on the earliest stages of mouth formation, whereby the initial mouth opening forms and the first steps of differentiation are taking place, but before the mouth is mature.
Vertebrates have a complex mouth, derived not only from EAD ectoderm and endoderm that form the oral cavity and pharynx of the digestive tract but also from neural crest (NC) cells that form teeth and jaws. Vertebrates are ‘deuterostomes’ where a mouth breaks through the ectodermal covering and connects to the endodermal digestive tract. In the Xenopus model, development of the vertebrate mouth is associated with reciprocal signaling between the EAD and NC.4, 5 Mouth formation reflects this precision and involves many steps over a long period of development (~2.5 days in Xenopus, 2 weeks in humans). These steps position the mouth‐forming oral ectoderm and digestive endoderm during gastrula and neurula stages and open the mouth as the tadpole is ready to feed. The complexity of mouth formation is one reason for the many human anomalies that include this region.
ORAL EVOLUTION: IS MOUTH DEVELOPMENT CONSERVED?
A mouth is present from the simplest multicellular organisms to humans. The commonality of mouth function is food ingestion, but auxiliary structures may hold, tear or grind food, such as teeth in vertebrates or adult sea urchins and mandibles in insects. In many animals, the mouth has evolved extra functions, including communication and defense, but these are secondary to its role in eating (Figure 1(a) and (b)). The common function of eating could imply that all mouths are homologous structures, or a mouth opening may have arisen multiple times in evolution. To address the question of whether the mouths of all animals derive from an ancient, conserved origin, we discuss embryonic tissue contributions, associated gene expression, and axial position.
Figure 1.
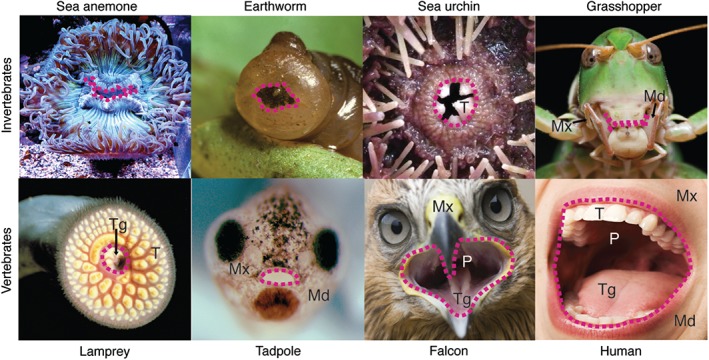
Mouths in adult or larval animals. Frontal views of sea anemone Anthopleura elegantissima, earth worm Lumbricus terrestris, sea urchin Strongylocentrotus purpuratus, grasshopper Anacridium aegyptium, lamprey Petromyzon marinus, tadpole of frog Xenopus laevis, falcon Falco cherrug, and human Homo sapiens. Red dotted line denotes the border of the oral cavity. Md, mandible; Mx, maxilla; P, pharynx; T, teeth; Tg, tongue.
All Mouths are Made from Ectoderm + Endoderm
One of the most persuasive pieces of evidence that the mouth evolved only once is that in all animals the mouth arises from ectodermal and endodermal germ layers.1 In general, the ectoderm is characterized by strong junctions and forms a protective outer covering. The endoderm enters the embryo during gastrulation to form the innermost layer of cells and contributes to the digestive system. The mouth is therefore a joint ectodermal/endodermal structure as it connects an opening in the ectodermal covering to the digestive endoderm.3
This tissue arrangement is obvious in diploblasts where there are only ectodermal and endodermal germ layers.1 Ectodermal, mesodermal, and endodermal germ layers are present in the triploblasts, which include the bilateria—divided into deuterostome and protostome groups. Deuterostomes break a mouth opening through the ectodermal covering. Strikingly, in deuterostomes, the mouth develops as in diploblasts from ectoderm and endoderm, and the mesodermal layer is not involved. Indeed, a unique anterior region of the deuterostome embryo is devoid of mesoderm, so that future oral ectoderm and future foregut (anterior) endoderm are directly juxtaposed2, 3 and go on to form the mouth (Figure 2). In chordates, we named this region the EAD.4 It is unclear whether mesoderm is actively prevented from entering the EAD, perhaps by preferential oral ectoderm/endoderm adhesion, or whether mesoderm is intrinsically unable to migrate to the anterior. The joint ectoderm/endoderm nature of the mouth is consistent with coevolution of the two germ layers to form the mouth opening linked to the digestive system. In this review, we focus primarily on deuterostomes, since mouth development in protostomes is highly variable, and since data including that from the protostome Priapulus caudatus suggests that deuterostomy was the ancestral developmental program in bilaterians.6
Figure 2.
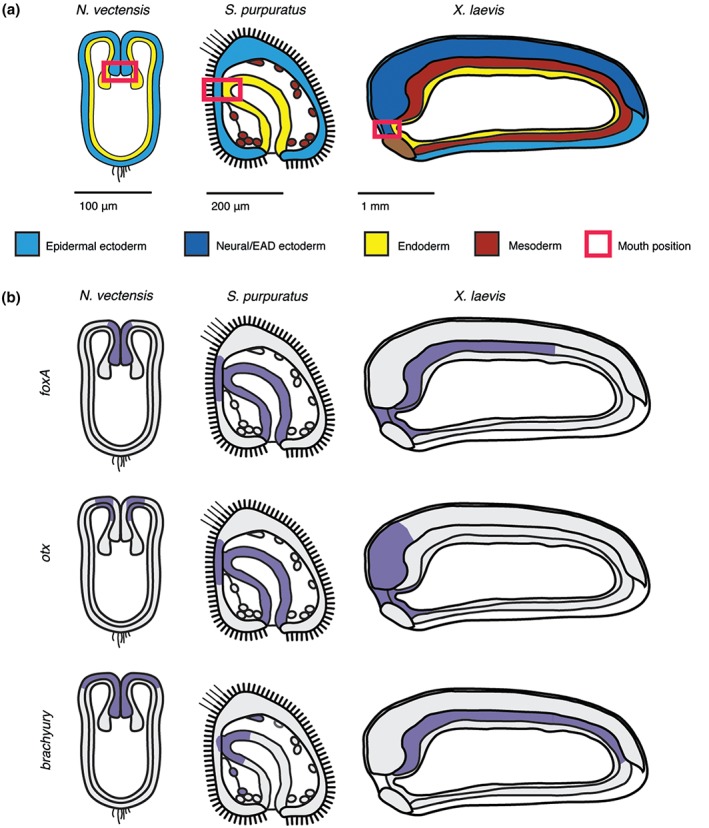
Mouth forms where ectoderm and endoderm are juxtaposed. (a) Position of future mouth relative to germ layers in embryos of three representative animals. Schematics of sagittal sections are shown for the diploblast cnidarian Nematostella vectensis (invertebrate), the triploblasts and deuterostomes sea urchin Strongylocentrotus purpuratus (invertebrate) and frog Xenopus laevis (vertebrate). The red box outlines the mouth‐forming region made up of juxtaposed ectoderm and endoderm. In vertebrates, this region is termed the extreme anterior domain. (b) Ancestral mouth embryonic gene expression domains in N. vectensis, S. purpuratus, and X. laevis (purple). The mouth expression domain of foxA and otx but not brachyury is conserved in vertebrates.
Despite their key association with the mouth, oral ectoderm and digestive endoderm may not be near one another during development. In diploblasts, mouth morphogenesis is fairly straightforward. In the late blastula, the animal pole region is endoderm and the vegetal pole region is ectoderm.7 In Cnidaria, including the sea anemone Nematostella, the blastopore forms at the animal pole, in endoderm (Figure 2). These endodermal cells invaginate and concomitantly, vegetal ectodermal cells move towards the blastopore, which forms the mouth opening.8 Once invagination of the endoderm is complete, ectoderm near the blastopore rolls inward and contributes to the pharynx. In this case, oral ectoderm and digestive endoderm are adjacent to each other throughout gastrulation and move in the same direction towards the blastopore.
Triploblast mouth formation is more complicated as the digestive endoderm must move a considerable distance to meet the oral ectoderm. In deuterostomes, blastula stage embryos have a germ layer position opposite that of diploblasts where ectoderm is located in the animal hemisphere and endoderm in the vegetal hemisphere7 (Figure 2). The blastopore remains associated with endoderm, as in diploblasts, but forms in the vegetal hemisphere. During gastrulation the oral ectoderm remains relatively stationary and the digestive endoderm migrates the entire length of the embryo towards animal pole derived ectoderm, and together these layers form the mouth. A simple example of this morphogenesis is found in sea urchins where the archenteron (endoderm) migrates towards and meets the oral ectoderm.
An additional layer of detail is present in vertebrate embryos where both mouth ectoderm and endoderm are comprised of tissues arising from different locations. In sea urchins and basal deuterostomes that lack a central nervous system or brain, mouth ectoderm arises from the epidermal layers.9 In vertebrates, oral ectoderm derives also from the anterior neural ridge (ANR), the front of the neural plate.5 This dual source of mouth ectoderm is also found in Ciona, an invertebrate chordate, whose mouth is derived from the anterior neuropore.10 Thus in chordates, development of the mouth primordium and anterior neural tube are closely linked. Endoderm making up the vertebrate gut also arises from two locations during gastrulation as the dorsal endoderm migrates anteriorly and ventral endoderm moves posteriorly11 (in the opposite direction). The result of this complementary movement is that foregut endoderm arises from a dorsal source while midgut and hindgut endoderm come primarily from a ventral source. The extraordinary effort that bilateria go to in order to bring pharyngeal endoderm and oral ectoderm together demonstrates the intimate connection between these tissues during mouth formation.
In summary, there is a simple equation for mouth formation: oral ectoderm and digestive endoderm that become associated during gastrulation. Exactly where these tissues derive from in the embryo and how they migrate may have changed over evolutionary time. However, the essence of building a mouth from ectoderm and endoderm is a constant, and we consider this strong evidence for mouth conservation, from diploblasts to triploblasts.
A Conserved Mouth Gene Program
A corollary to the conclusion that oral ectoderm and pharyngeal endoderm are conserved is that similar genes are expressed in these tissues across phyla. Strikingly, three genes, otx, brachyury, and foxA, are expressed throughout the gut endoderm of Cnidaria (Nematostella vectensis), protostomes (Capitella teleta and P. caudatus) and deuterostomes (sea urchins and starfish).6, 12, 13, 14, 15 Otx and foxA are also expressed in mouth ectoderm. The overlapping expression of these three genes in conserved domains across evolution suggests an ancestral gene network regulating mouth and associated gut development16, 17 (Figure 3).
Figure 3.
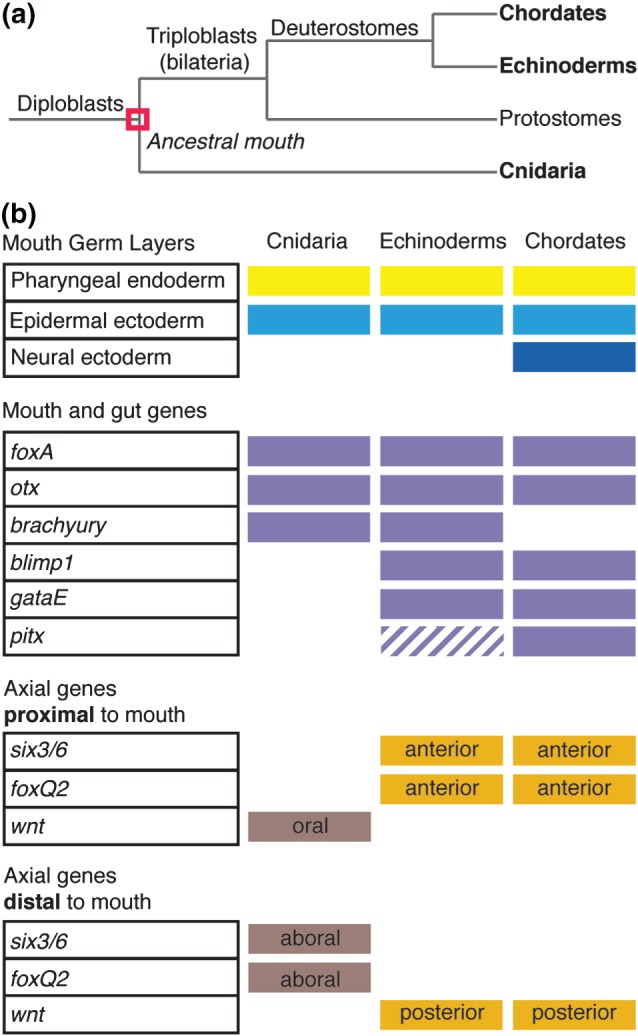
Ancestral mouth was present in the common ancestor of cnidaria and triploblasts. (a) Phylogenetic tree. Chordates, echinoderms, and cnidaria are three distantly related phyla that retain common mouth characteristics, suggesting that the mouth evolved once. (b) Criteria used to evaluate mouth evolution. The mouths of cnidaria, echinoderms, and chordates are all comprised of ectoderm and endoderm and express foxA and otx. There are phylum‐specific characteristics such as neural ectoderm contributing to the chordate mouth and the expression of blimp1, gataE, and pitx in echinoderm and chordates. Analysis of axial positioning genes demonstrates that the cnidarian oral–aboral axis is equivalent to the echinoderm and chordate posterior–anterior axis.
Detailed functional evaluation of these genes has been performed in sea urchins and starfish. otx, brachyury, and foxA are part of the sea urchin endodermal gene regulatory network (GRN), which is one of the most extensively studied GRNs. In general, GRNs comprise ‘kernels’ or evolutionarily inflexible circuits responsible for upstream functions in body patterning, and ‘plug‐ins’ or smaller circuits that have been repeatedly co‐opted for diverse purposes.
Comparison between GRNs of the distantly related echinoderms sea urchins and starfish, deuterostomes of the Echinoderm phylum, reveals an identical core of transcription factors—otx, brachyury, and foxA, as well as two additional genes—blimp1/krox and gataE. 18, 19 This five‐member kernel regulates the development of digestive (gut) endoderm including that associated with the mouth and each gene is necessary for gut formation. FoxA and otx, independent of their function in endoderm, are also active in oral ectoderm. Transplant experiments in sea urchins demonstrate that ectodermal foxA is required for mouth formation as embryos with ectoderm‐specific foxA loss of function had normal digestive tracts but lacked mouths.17 While functional experiments have not been performed in sea urchins, starfish injected with a dominant negative form of otx formed a truncated archenteron and abnormal mouth ectoderm lacking an invagination corresponding to the mouth.16
Tracing to ancestral diploblasts, is the expression of the five‐membered GRN discovered in Echinoderms also present? As stated earlier, Cnidaria express otx, Brachyury, and foxA in the mouth and gut. However, they lack such specific expression of blimp1 and gataE. 20 Thus otx, brachyury, and foxA likely form an ancestral kernel responsible for mouth and gut formation while blimp1 and gataE appeared in later lineages (Figure 3).
Tracing forward, is the five‐membered GRN an echinoderm‐specific innovation or broadly used by bilateria? Prostomes (C. teleta and P. caudatus) have mouth and/or gut‐specific expression of all five genes.6, 14 This suggests that the echinoderm GRN is used by basal deuterostomes and protostomes as a mouth/gut GRN and that this kernel was present in the deuterostome–protostome ancestor. Conservation of the kernel in chordates is incomplete—perhaps due to gene duplication, different body plans, and added complexity of craniofacial development that involves additional cell types. In vertebrates, foxa and gataE function is largely conserved as both genes are expressed throughout the digestive tract and required for proper development.11 Otx acts early in development as a general anterior patterning gene and later in brain development.21 Blimp1 has a late role in gut development as it controls the transition of the intestinal lining from the neonatal to adult form.22 Of all the members in the conserved mouth gene program, brachyury seems to be most divergent as it is expressed in the mesoderm and regulates convergence and extension of the notochord23 (Figure 3).
An example of chordate‐specific regulation is incorporation of the pitx genes into the ancestral kernel. In chordates, otx activates the pitx class of transcription factors. In Xenopus, pitx1, pitx2, and pitx3 mark the future mouth region (Figure 5). The ascidian Ciona intestinalis expresses pitx2 in the primordial pharynx and anterior neural complex.24 In sea urchins, pitx is expressed in part of the oral ectoderm, however, it is not known whether this expression is required for mouth formation or is associated with left‐right patterning (Figure 3). Functional tests in Xenopus, 3, 25 mouse,26 and human27 demonstrate that pitx1 and pitx2 are required for proper mouth and facial development.
Figure 5.
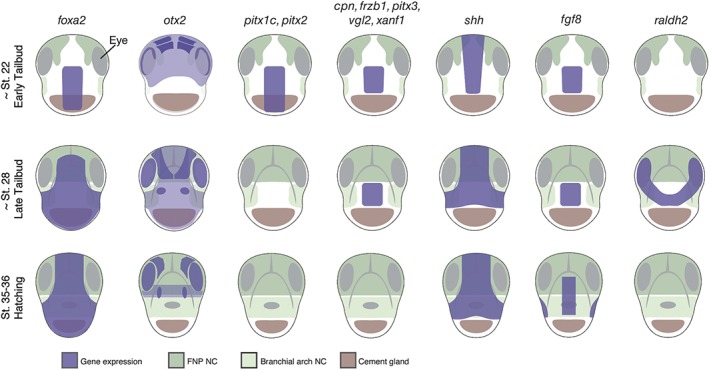
Extreme anterior domain (EAD) gene expression domains in Xenopus. Frontal views of Xenopus tadpole embryos. Selected gene expression domains are shown that include the EAD. These include pitx1c, pitx2, pitx3, vgl2, xanf1, cpn, frzb1, shh, fgf8, and raldh2. See text for details on their function in mouth development.
Overall, many members of the gut gene network first uncovered in sea urchin retain a function in vertebrate mouth and digestive tract development. We propose that these genes form a ‘mouth gene program.’ Although there may be some vertebrate‐specific regulation, the conserved expression of an ancestral kernel comprising otx, brachyury, and foxA in Cnidaria, basal deuterostomes, and protostomes is strong evidence that the mouth arose once during evolution.
Mouth Position Changes Relative to the Body Plan
The final criterion we consider to assess its conservation is whether the mouth is always found in the same position, relative to the primary body axes. More basal groups, such as Cnidaria (hydra, sea anemones), have radially symmetric bodies with an oral/aboral axis where the mouth is located at the oral end. In bilateria, the anteroposterior axis determines the head‐to‐tail set of positions with the mouth almost always located at the anterior end of the animal. Homology between the oral end of Cnidarians and the anterior end of bilaterians would suggest that the mouth arose once at a conserved position along the body axis.
Comparison of Cnidarian and bilaterian body plans is complicated since, unlike bilaterians, where the anterior pole is characterized by a brain or accumulation of nerve cells, Cnidaria lack a centralized nervous system. Additionally, cross species analysis among Cnidarians reveals variable Hox gene expression along the oral–aboral axis.28 Because ‘anterior’ and ‘posterior’ Hox gene expression do not strictly correspond to the oral or aboral pole, the Hox code and its role in axial patterning is likely a bilaterian innovation, and cannot be used as standards of comparison between diploblasts and triploblasts.
Despite these challenges, two pieces of complementary evidence suggest that the anterior, head‐forming region of bilaterians is derived from the aboral domain of the cnidarian–bilaterian ancestor while the posterior region corresponds to the oral pole. First, the transcription factors six3/6 and foxQ2, well‐conserved bilaterian anterior markers, are expressed in the aboral pole of N. vectensis. Functional analysis using morpholino knockdown demonstrates that six3/6 and foxQ2 are required for normal aboral pole development29 (Figure 3). Secondly, Wnt gene expression in Cnidaria is concentrated around the oral pole.30 In bilaterians, Wnt genes are expressed and act in the posterior region during gastrulation, to promote regional identity.31 Functional data in Hydra indicates that Wnts have a similar patterning role as overactivation of Wnt signaling causes ectopic growth of tentacles (an orally associated structure) throughout the body column.32 These data indicate that Wnt signaling is responsible for patterning the oral pole (Figure 3). Together, six3/6 and foxQ2 expression, along with Wnt expression domains, suggest that the oral–aboral axis of diploblasts corresponds to the posterior–anterior axis of triploblasts and bilateria. (Note that the sea urchin/Echinoderm ‘oral/aboral’ axis is not the same as that in diploblasts and is equivalent to the anteroposterior axis.)
These considerations suggest that the ‘mouth gene program’ discussed in the previous section is independent from an ‘anterior’ gene program. Over evolution leading to bilateria, the mouth gene network became associated with the anterior end of the animal, and expression of otx and foxA is close to that of six3/6 and foxQ2 in echinoderms and chordates. In chordates, mouth formation has become tightly associated with the anterior and with brain formation, and mouth ectoderm in the EAD includes and requires neural plate‐derived tissue. Although it does not reflect the ancestral state, this links in a functional way, the mouth of chordates with the anterior of the body.
The Mouth Is a Conserved Structure
In summary, two criteria indicate that the mouth has a common evolutionary origin across animals. First, the mouth always forms from oral ectoderm and digestive endoderm. In triploblasts, mesoderm is never part of the initial mouth. Second, a conserved set of genes that can be considered a mouth gene program can be defined in all animals, including otx and foxA. These considerations indicate that the mouth arose once during evolution and that fundamental aspects of a mouth program have been retained amongst all animals.
STEPS TO FORM A MOUTH: XENOPUS AS A PARADIGM
Of greatest relevance for human health is development of the vertebrate mouth (Figure 1(b)). This derives from a region of juxtaposed ectoderm and endoderm termed the EAD (Figure 2), in conjunction with cells of the cranial NC.2, 4 The EAD forms the mouth opening and the oral cavity, including the oropharynx that is the beginning of the digestive tract, while the NC forms the jaws and teeth. In this section, we review the earliest steps in mouth development, including mouth opening, prior to differentiation of tissues found in the mature mouth.
While retaining the core‐conserved aspects of mouth development: ectodermal plus endodermal origins and key mouth regulatory genes, vertebrate mouth formation is extremely complex, due to the large number of tissues and cell types present, and the involvement of the NC. Vertebrates are deuterostomes where the mouth breaks through the ectodermal covering to connect the outside with the endodermal digestive tract. Formation of the mouth opening must be carefully coordinated with digestive system development, so that the opening does not form prematurely and become a wound.
The frog Xenopus has proven an outstanding vertebrate model for observation of mouth development, since it undergoes external embryonic development, allowing all stages to be obtained, and since the face is flat owing to the small forebrain. Xenopus mouth development relies on coordinated development of the EAD and NC, in a carefully orchestrated sequence. The earliest steps in mouth formation, including mouth opening appear similar in Xenopus and amniotes,2, 3 although as discussed later in this review, details may differ between species.
Setting Aside the Ectoderm and Endoderm of the EAD
Xenopus mouth development begins at the end of neurulation. At this time, the EAD is defined by future oral ectoderm lying adjacent to pharyngeal endoderm, and genes are expressed that indicate the future mouth4, 5, 33 (Figures 2 and 4). Both ectoderm and endoderm are essential for mouth formation.3 foxa2 and otx2, genes that are part of the conserved mouth gene kernel, are expressed in the developing mouth region (Figure 5).
Figure 4.
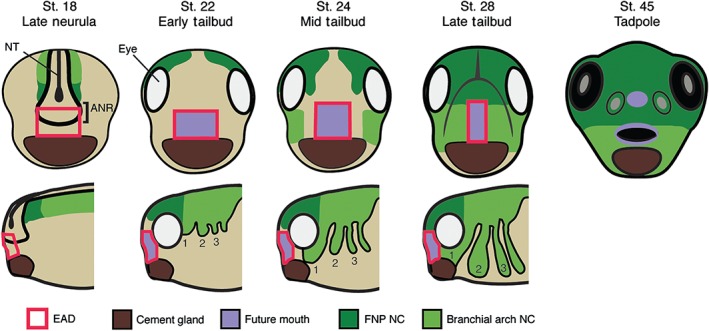
Relationship of the extreme anterior domain (EAD) and neural crest (NC) to the Xenopus mouth. Branchial arch (BA, light green) NC, and frontonasal prominence crest (FNP, dark green) delaminate from the dorsal neural tube (NT) at neurula. The EAD (red outline) is specified including cells arising from the anterior neural ridge (ANR, black outline) and cement gland (CG, brown) tissue. The EAD (purple) will contribute to the lining of the mouth (as well as the nostrils and anterior pituitary). FNP NC migrates anteriorly between the eyes to enter the face while first BA NC migrates bilaterally into the face (late neural‐mid tailbud). Subsequent to NC ingress, EAD ectoderm thins and lengthens to become the ‘pre‐mouth array’ (early‐mid tailbud). NC cells eventually differentiate to form the facial skeleton, including the palate, maxilla, mandible, and connective tissue.
EAD ectoderm derives from the ANR, the anterior boundary of the neural plate.5, 34, 35 At the end of neurulation, a wedge of ectoderm delaminates from the ANR to move ventrally and lie between the epidermal ectoderm and the pharyngeal endoderm. This is EAD ectoderm that will surround the mouth opening and contribute to the oral cavity. Subsequently, a basement membrane forms that separates the developing brain from EAD ectoderm.5 Overlying epidermal ectoderm can be substituted by flank epidermal ectoderm and is therefore not specific for mouth development.2 By early tailbud, EAD ectoderm expresses pitx1, pitx2b, pitx2c, and pitx3 genes, while the underlying pharyngeal endoderm expresses pitx1 and pitx2c 3 (Figure 5). As noted in the Oral Evolution: Is Mouth Development Conserved section, this class of gene is required for Xenopus mouth development.36 An expression microarray screen in our group revealed additional genes expressed in EAD ectoderm and in some cases EAD endoderm, including the Wnt‐inhibitor frzb‐1, kinin–kallikrein pathway factors cpn and kininogen, and the transcription factors vgl2, six1, xanf1, xanf2, and goosecoid 4, 33 (Figure 5). Other signaling factors are also expressed in the EAD or surrounding tissues, including shh, fgf8, and raldh2 37, 38, 39 (Figure 5).
Pharyngeal endoderm is the inner component of the EAD that becomes the epithelial lining of the pharynx.40 It derives from dorsal endoderm that has moved to the anterior of the embryo by the end of gastrulation, and lies anterior to head mesoderm.41, 42 Pharyngeal endoderm specification involves function of the tbx1 transcription factor in Xenopus. 43 Genes expressed in EAD endoderm include kininogen, frzb‐1, and raldh2, 4, 33, 39 and these are required for mouth development (next section) (Figure 5). Pharyngeal endoderm development is also dependent on retinoic acid (RA) signaling.44
Guiding Migratory NC to the Mouth‐Forming Region: The EAD as Organizer
In addition to EAD ectoderm and endoderm, the cranial NC makes a key contribution to mouth formation in Xenopus and other vertebrates, eventually forming the jaws (maxilla and mandible), palate, and upper lip45 (Figures 4 and 6). The NC is a migratory, multipotent population originating from the lateral borders of the neural plate45 that segregates into four streams called branchial arches. First arch crest migrates to the face to lie on either side of the EAD, while the frontonasal prominence crest migrates over the top of the head into the face (Figure 4).45 NC migration is governed by chemotaxis via Sdf1, produced by adjacent regions, together with contact‐inhibition of locomotion (CIL) through N‐cadherin and Wnt/PCP signaling.46 NC cells express complement receptors and secrete complement ligand that promotes cell clustering.47 These dispersion and attraction activities are required for migration of the NC as a group.48 Secreted ephrins and semaphorins promote branchial arch formation.48, 49
Figure 6.
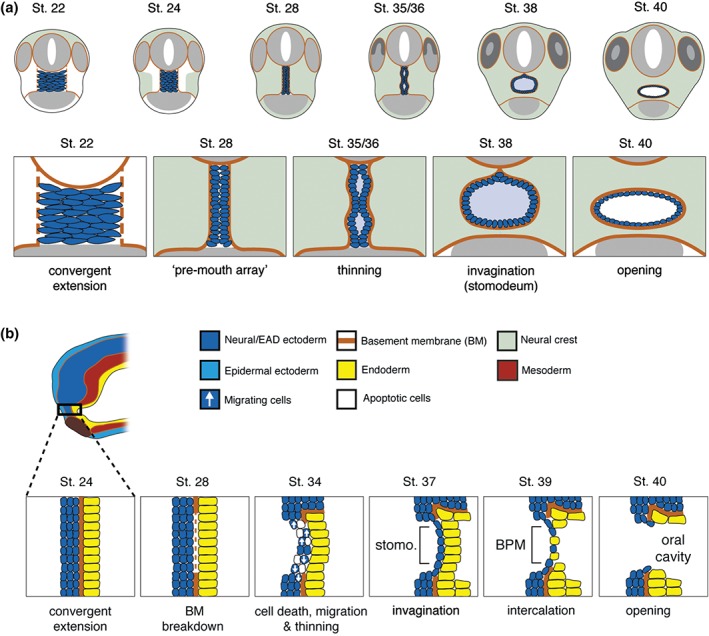
Steps in Xenopus mouth formation. (a) Coronal views of steps to mouth opening. Frontal views of the embryo are shown. The extreme anterior domain (EAD) begins at early tailbud (st. 22) as a wide, short block of cells. By late tailbud (st. 28), the neural crest (NC) migrates to lie on either side of the EAD. Signals from the NC initiate convergent extension in the EAD so that it forms a pre‐mouth array. Apico‐basal polarity is established in the pre‐mouth array, which separates down the midline to form the stomodeum at hatching stages (st. 35/36), that opens into the mouth at tadpole stage (st. 40). NC is in light green. (b) Sagittal views of steps to mouth opening. The EAD from a tailbud embryo showing different germ layers is enlarged in schematics below. Epidermal ectoderm is not shown in enlarged schematics. At late tailbud (st. 28), the pre‐mouth array forms by convergent extension, and the basement membrane (BM) between EAD ectoderm and endoderm disintegrates. The pre‐mouth array opens to form the stomodeal invagination. Stomodeal ectoderm thins concurrent with a burst of apoptosis and migration of ectoderm out of the region at hatching stages (st. 34–37). Intercalation of ectoderm and endoderm produces the buccopharyngeal membrane (BPM), which perforates to open the mouth at tadpole stages (st. 39–40).
Using facial transplants,4 we discovered that in addition to contributing to the mouth, the EAD is a signaling center that helps direct NC towards the facial midline. At least two signaling pathways act from the EAD. One is the kinin–kallikrein pathway, where the EAD expresses precursors of Kinin ligands and cpn, encoding a Kinin‐processing enzyme. Loss of cpn locally, specifically in the EAD, halts first arch crest migration and leads to failure of mouth formation and an abnormal face.4 Another EAD signal regulates the β‐catenin Wnt pathway, where secreted antagonists frzb‐1 and crescent act within the EAD and also in the developing face, possibly affecting NC.33 EAD pharyngeal endoderm plays a later signaling role in NC development, to induce formation of the cartilaginous skeleton of the mouth and pharynx.44 Thus, EAD endoderm ablation results in an abnormal mouth and pharyngeal skeleton in Xenopus 2 and chick.50
Formation of a Pre‐Mouth Array and the Stomodeum: the NC Signals to EAD Ectoderm
After the NC has come to lie on either side of the EAD, it signals back to the EAD to induce morphogenesis of a ‘pre‐mouth array’5 (Figure 7). This signaling is via the Wnt/PCP pathway where Wnt11 ligand is expressed in the NC and targets the Fzl7 receptor in the EAD. Under control of Wnt/PCP signaling, EAD ectoderm undergoes convergent extension to transition from a wide, short 8 × 8 block of cells (st. 22) to a narrow, tall 20 × 2 cell arrangement we termed the ‘pre‐mouth array’ (st. 28) (Figure 6(a)). Two days later, the pre‐mouth array opens down the middle to form the ‘stomodeum.’ The stomodeum is a highly conserved indentation in bilateria, and indicates the future mouth (tadpole stages, st. 32–40)5 (Figure 6(a) and (b)). The ‘pre‐mouth array’ demonstrates that the stomodeum is organized much earlier than previously understood. Basement membrane breakdown between EAD ectoderm and endoderm (Figure 6(b)) had been considered the first stage of mouth opening, however, the pre‐mouth array precisely sets up the future mouth opening prior to basement membrane breakdown (Figure 6(b)). Our data indicate that mouth development in Xenopus involves reciprocal signaling: from EAD to NC and later from NC to EAD, a sequence that likely coordinates development of tissues and structures leading to proper mouth development (Figures 6 and 7).
Figure 7.
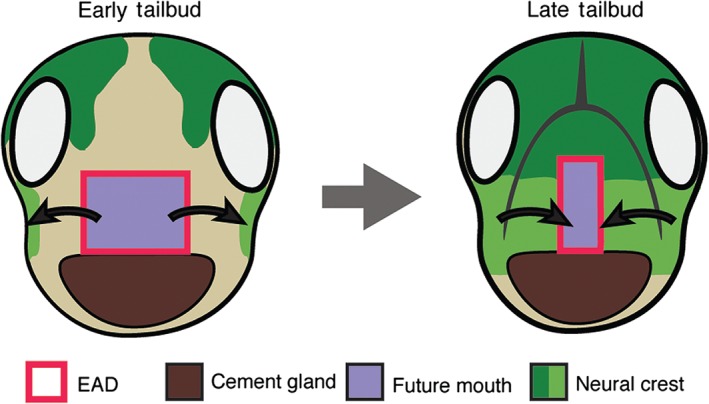
Reciprocal signaling between extreme anterior domain (EAD) organizer and cranial neural crest (NC). The EAD secretes signals, including Kinin peptides, that guide the NC into the face. As they migrate into the face, NC cells secrete factors including Wnt/PCP ligands that stimulate EAD convergent extension to form the ‘pre‐mouth array.’ The pre‐mouth array later opens down the midline to form the stomodeum and edges of the future mouth.
Opening the Mouth: Signals and Steps
Pre‐mouth array formation leads to precisely organized oral ectoderm, juxtaposed to the pharyngeal endoderm. Several additional steps complete mouth formation. During pre‐mouth array formation, the basement membrane separating ectoderm from endoderm is degraded (st. 28) (Figure 6(b)). This is dependent on the β‐catenin Wnt antagonists frzb‐1 and crescent that are expressed in the EAD,33 as well as Hedgehog signaling.51 Subsequently, the pre‐mouth array opens down the midline to form the stomodeum—comprising the borders of the future mouth opening with a central indentation (st. 35–37) (Figure 6(a)). The signal that causes the array to open is unknown, but we speculate that it derives from the underlying endoderm that is maturing into a functional digestive system. Thus, when the pharyngeal endoderm is close to mature, it may signal to the pre‐mouth array ectoderm to elicit its opening. This occurs concomitant with appearance of apical markers on the pre‐mouth array cells that face one another.
EAD ectoderm becomes thinner as cells migrate out of the oral region (st. 32–34) and undergo a burst of apoptosis (st. 34–35).2 The ectoderm and endoderm that form the middle of the stomodeum thin—each becoming a single layer2, 5 (Figure 6(b)). These layers intercalate to form a one or two cell thick ‘buccopharyngeal membrane,’ which perforates to open the mouth (Figure 6(b), st. 40).2 Hedgehog signaling regulates buccopharyngeal membrane perforation51 (st. 39) and recent, elegant data point to c‐Jun N‐terminal kinase (JNK) signaling as a key player in this process, promoting disassembly of adherens junctions via endocytosis.52 Perforation also requires adjacent NC that may provide tension to pull the mouth open.2 Buccopharyngeal membrane perforation is essential, but is more a ‘clean up’ stage, the culmination of processes such as pre‐mouth array formation, which precisely set up the future mouth.
While EAD ectoderm and endoderm are completing mouth opening, NC cells form maxillary and frontonasal ‘prominences’ (cell aggregates) and differentiate into the jaw cartilages, the palate, and the upper lip (st. 37–39).39, 53 Differentiation requires RA signaling from the stomodeum and nasal regions through activation of the homeobox genes lhx8 and msx2. 39 By the time of mouth opening (st. 40), the craniofacial cartilages, connective tissue, jaws, and muscles have begun to differentiate and soon after, the tadpole begins to eat.2
Xenopus as a Model for Mouth Formation in Other Vertebrates
Phases of mouth development appear conserved among anurans and amniotes, indicating that the novel findings made in Xenopus are broadly applicable.3, 54 The juxtaposed oral ectoderm and pharyngeal endoderm comprising the EAD is found in all deuterostomes, and this domain expresses common genes, including pitx genes.3 In vertebrates, the EAD may have slightly different morphologies due to compression of tissue and relative sizes of the ectoderm and endoderm germ layers.36 Comparison across species including mammals, fish, and amphibians demonstrates that these differences in morphologies are associated with two variables—whether yolk is fully internalized into the embryo during gastrulation and how far away the developing head process is located from the yolk. Notably Xenopus laevis has an EAD morphology similar to that of mammals.
We discovered several aspects of mouth formation in Xenopus—including ability of the EAD to act as an organizer,4 dependence of EAD basement membrane degradation on β‐catenin Wnt signaling,33 and pre‐mouth array formation.5 It will be important to test whether these processes are conserved in teleosts and amniotes. Aspects of craniofacial development including NC patterning,55, 56 NC migration,57, 58 and jaw and palate development59, 60, 61have been well studied in other species, and where compared appear similar to that in Xenopus. Mouse and chick have a frontonasal ectodermal zone (FEZ) organizer that is present after NC has arrived in the face. The FEZ lies at the boundary between shh and fgf8 expression in facial ectoderm and controls jaw and cartilage development.62 It is unclear whether Xenopus and zebrafish have a FEZ.
The process of thinning and perforating the buccopharyngeal membrane appears to be similar in frogs, zebrafish, chick, mouse, and hamster.63, 64, 65, 66 Buccopharyngeal membrane intercalation in all species requires changes in cell adhesion and movement. Electron micrographs in Rana japonica, hamster, and chick show that cellular processes between ectoderm and endoderm germ layers increase the surface area to bring the germ layers immediately adjacent.63, 64, 66 In X. laevis endocytosis of E‐cadherin is required for membrane perforation.67 One difference among species is whether cell death preceeds perforation: this has been observed in X. laevis, Rana japonica, and mouse but not in zebrafish, chick, or hamster.2, 63, 64, 65, 66, 68 When present, cell death begins hours before mouth opening and likely thins cell layers, with other mechanisms utilized for perforation. In X. laevis and R. japonica, EAD ectoderm and endoderm are multiple cell layers thick while in zebrafish and chick this region is thinner and cell death may not be required for tissue thinning. Perforation may be caused by mechanical stress generated by differential growth or movement of tissues surrounding the buccopharyngeal membrane. Consistent with this hypothesis, mouse buccopharyngeal membrane has almost no dividing cells, however, cell division in adjacent areas has not been quantified.65 Inhibition of cell proliferation in X. laevis did not affect mouth opening suggesting that differential cell proliferation is not required for perforation in frog.67 In addition to cell division, movement of surrounding tissues such as the forebrain or facial prominences may generate tension. As embryonic facial morphology of species varies, the specific location and magnitude of forces acting on the buccopharyngeal membrane by surrounding tissue may vary between species.
Human Craniofacial Anomalies Involving the Mouth
Craniofacial anomalies often involve abnormal mouth development, which may go awry frequently due to the many steps involved. These steps may occur very early during mouth formation, and involve the EAD. Later events involving cartilage or bone formation leading to development of the primary or secondary palate may also impact mouth development. Regulation of human mouth development is complex, including genetic and environmental factors.69 Mouth anomalies may occur as part of a ‘syndrome’ if they consistently occur together with phenotypes elsewhere in the face or body,69 or they may specifically only affect the mouth.
Understanding EAD activity in model organisms will lend insight into human craniofacial anomalies, since defects in human EAD signaling may lead to abnormal NC development later manifesting as malformed cartilage and bone. Conversely, abnormal NC signaling to the EAD may lead to abnormal mouth morphology and delayed mouth opening. Symptoms of several syndromes such as Nager syndrome, craniofacial microsomia, and persistent buccopharyngeal membrane may represent outcomes of abnormal EAD function, although these connections are yet unexplored.
Human mouth defects have been associated with genes and signaling pathways identified in vertebrate models, indicating the utility of these for addressing human disorders.70 As details of mouth and other facial features may differ between animals, particularly with regard to palate formation, the model must be chosen carefully. Some of these pathways identified in vertebrates may affect early events, including those surrounding EAD function, while others may impact much later events For example, tbx1 and fgf8 are implicated in human DiGeorge syndrome.71 The Shh pathway is associated with many craniofacial anomalies72 including Pallister–Hall syndrome73 and Grieg cephalopolysyndactly syndrome.74 Disrupting both SHH and β‐catenin WNT signaling promotes facial pathogenesis including that of palate and mouth.37, 75 Genome wide association studies analyzing variation in face morphology finds associated loci harboring candidate genes important for facial development in vertebrate models. Examples include gli3, a member of the Shh pathway, and runx2 a gene that interacts with Shh during bone development, members of the FGF family, endothelin pathway, and semaphorins.76, 77 Genes not obviously involved in signaling such as the nucleolar protein TCOF1 in Treacher Collins Syndrome78 may impact human mouth development.
Mouth development is sensitive to environmental factors including pathogens, teratogens in the form of medicines and other chemicals, especially during the first trimester.69 In general it is unclear what steps in mouth formation these agents impact. Zika virus and cytomegalovirus are both associated with cleft lip and palate.79, 80 Antiseizure medications such as valproate81 and phenytoin,82 as well as RA,83 an anti‐acne medication, are associated with mouth anomalies. Smoking84 and ethanol85 are tightly associated with facial anomalies. Other teratogens affecting the mouth have been defined in animal studies, for example, dioxins86 and dithiocarbamates.87 Maternal health challenges have also been associated with mouth anomalies, including diabetes88 and hyperthyroidism.89 Overall, the landscape of human mouth developmental anomalies is multifactorial, evolving and incomplete.
CONCLUSION
The mouth is a hallmark of multicellular animals and is essential for survival. In the Oral Evolution: Is Mouth Development Conserved section of this review, we drew three key conclusions indicating that the mouth arose once during metazoan evolution. First, in all animals, the mouth is derived from ectoderm and endoderm. Indeed, in deuterostomes a specific region, the EAD, devoid of mesoderm, is fated to form the mouth. Second, we discuss a mouth gene program that coordinates ectodermal and endodermal lineages to form the mouth. A third point is that the chordate mouth has become intimately linked to anterior neural development and includes tissue from this region. In the Steps to form a Mouth: Xenopus as a Paradigm section, we addressed the earliest steps involved in vertebrate mouth formation, using the frog Xenopus as a model. Xenopus also represents the deuterostomes, which open the mouth by breaking through the outer covering of the embryo. A key aspect of productive mouth opening is its coordination with digestive system development. Mouth development has been described in Xenopus in great detail, and comparison with amniotes and teleosts will be important for understanding the universality of processes involved. Human mouth anomalies are associated with environmental factors as well as genes identified directly in affected people and in model systems, indicating the usefulness of these systems for addressing human disorders.
ACKNOWLEDGMENTS
We are grateful for support from the NIDCR (RO1 DE021109 to H. Sive and F30 DE022989 to L. Jacox) and Harvard University (Herschel Smith Graduate Fellowship to L. Jacox).
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
FURTHER READING
- CDC: (http://www.cdc.gov/ncbddd/birthdefects/features/craniofacialdefects.html)
- NIH: Craniofacial Development Resources (http://www.nidcr.nih.gov/Research/ToolsforResearchers/CDR/)
- The Virtual Human Embryo: (http://www.ehd.org/virtual‐human‐embryo/)
- Education section of SDB: (http://www.sdbonline.org/education_resources?STARTROW=1&EDUCATIONRESOURCETYPEID=1)
- Specific pages: (http://www.sdbonline.org/resource?ResourceID=2226)
- Overviews of development by organism: (http://www.sdbonline.org/education_resources?STARTROW=1&EDUCATIONRESOURCETYPEID=4)
- Encyclopedia of Life Sciences: (http://www.els.net/WileyCDA/)
- Specific pages: Dev Bio (http://www.els.net/WileyCDA/ElsTopics/L1‐DVB.html)
REFERENCES
- 1. Reece JB. Campbell Biology: Concepts & Connections. San Francisco, CA: Benjamin Cummings; 2012. [Google Scholar]
- 2. Dickinson AJ, Sive H. Development of the primary mouth in Xenopus laevis . Dev Biol 2006, 295:700–713. [DOI] [PubMed] [Google Scholar]
- 3. Dickinson A, Sive H. Positioning the extreme anterior in Xenopus: cement gland, primary mouth and anterior pituitary. Semin Cell Dev Biol 2007, 18:525–533. [DOI] [PubMed] [Google Scholar]
- 4. Jacox L, Sindelka R, Chen J, Rothman A, Dickinson A, Sive H. The extreme anterior domain is an essential craniofacial organizer acting through Kinin‐kallikrein signaling. Cell Rep 2014, 8:596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jacox L, Chen J, Rothman A, Lathrop‐Marshall H, Sive H. Formation of a “pre‐mouth array” from the extreme anterior domain is directed by neural crest and Wnt/PCP signaling. Cell Rep 2016, 16:1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin‐Duran JM, Janssen R, Wennberg S, Budd GE, Hejnol A. Deuterostomic development in the protostome Priapulus caudatus . Curr Biol 2012, 22:2161–2166. [DOI] [PubMed] [Google Scholar]
- 7. Martindale MQ, Hejnol A. A developmental perspective: changes in the position of the blastopore during bilaterian evolution. Dev Cell 2009, 17:162–174. [DOI] [PubMed] [Google Scholar]
- 8. Magie CR, Daly M, Martindale MQ. Gastrulation in the cnidarian Nematostella vectensis occurs via invagination not ingression. Dev Biol 2007, 305:483–497. [DOI] [PubMed] [Google Scholar]
- 9. Yoshikawa S. Oral/aboral ectoderm differentiation of the sea urchin embryo depends on a planar or secretory signal from the vegetal hemisphere. Dev Growth Differ 1997, 39:319–327. [DOI] [PubMed] [Google Scholar]
- 10. Veeman MT, Newman‐Smith E, El‐Nachef D, Smith WC. The ascidian mouth opening is derived from the anterior neuropore: reassessing the mouth/neural tube relationship in chordate evolution. Dev Biol 2010, 344:138–149. [DOI] [PubMed] [Google Scholar]
- 11. Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol 2009, 25:221–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fritzenwanker JH, Saina M, Technau U. Analysis of forkhead and snail expression reveals epithelial‐mesenchymal transitions during embryonic and larval development of Nematostella vectensis . Dev Biol 2004, 275:389–402. [DOI] [PubMed] [Google Scholar]
- 13. Scholz CB, Technau U. The ancestral role of Brachyury: expression of NemBra1 in the basal cnidarian Nematostella vectensis (Anthozoa). Dev Genes Evol 2003, 212:563–570. [DOI] [PubMed] [Google Scholar]
- 14. Boyle MJ, Yamaguchi E, Seaver EC. Molecular conservation of metazoan gut formation: evidence from expression of endomesoderm genes in Capitella teleta (Annelida). Evodevo 2014, 5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peter IS, Davidson EH. A gene regulatory network controlling the embryonic specification of endoderm. Nature 2011, 474:635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hinman VF, Nguyen AT, Davidson EH. Expression and function of a starfish Otx ortholog, AmOtx: a conserved role for Otx proteins in endoderm development that predates divergence of the eleutherozoa. Mech Dev 2003, 120:1165–1176. [DOI] [PubMed] [Google Scholar]
- 17. Oliveri P, Walton KD, Davidson EH, McClay DR. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development 2006, 133:4173–4181. [DOI] [PubMed] [Google Scholar]
- 18. Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc Natl Acad Sci USA 2007, 104:19404–19409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid‐blastula stage. Dev Biol 2010, 340:188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saudemont A, Haillot E, Mekpoh F, Bessodes N, Quirin M, Lapraz F, Duboc V, Rottinger E, Range R, Oisel A, et al. Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet 2010, 6:e1001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boyl PP, Signore M, Annino A, Barbera JP, Acampora D, Simeone A. Otx genes in the development and evolution of the vertebrate brain. Int J Dev Neurosci 2001, 19:353–363. [DOI] [PubMed] [Google Scholar]
- 22. Muncan V, Heijmans J, Krasinski SD, Buller NV, Wildenberg ME, Meisner S, Radonjic M, Stapleton KA, Lamers WH, Biemond I, et al. Blimp1 regulates the transition of neonatal to adult intestinal epithelium. Nat Commun 2011, 2:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamada A, Martindale MQ, Fukui A, Tochinai S. Highly conserved functions of the Brachyury gene on morphogenetic movements: insight from the early‐diverging phylum Ctenophora. Dev Biol 2010, 339:212–222. [DOI] [PubMed] [Google Scholar]
- 24. Boorman CJ, Shimeld SM. Cloning and expression of a Pitx homeobox gene from the lamprey, a jawless vertebrate. Dev Genes Evol 2002, 212:349–353. [DOI] [PubMed] [Google Scholar]
- 25. Khosrowshahian F, Wolanski M, Chang WY, Fujiki K, Jacobs L, Crawford MJ. Lens and retina formation require expression of Pitx3 in Xenopus pre‐lens ectoderm. Dev Dyn 2005, 234:577–589. [DOI] [PubMed] [Google Scholar]
- 26. Lanctot C, Lamolet B, Drouin J. The bicoid‐related homeoprotein Ptx1 defines the most anterior domain of the embryo and differentiates posterior from anterior lateral mesoderm. Development 1997, 124:2807–2817. [DOI] [PubMed] [Google Scholar]
- 27. Amendt BA, Semina EV, Alward WL. Rieger syndrome: a clinical, molecular, and biochemical analysis. Cell Mol Life Sci 2000, 57:1652–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chiori R, Jager M, Denker E, Wincker P, Da Silva C, Le Guyader H, Manuel M, Queinnec E. Are Hox genes ancestrally involved in axial patterning? Evidence from the hydrozoan Clytia hemisphaerica (Cnidaria). PLoS One 2009, 4:e4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sinigaglia C, Busengdal H, Leclere L, Technau U, Rentzsch F. The bilaterian head patterning gene six3/6 controls aboral domain development in a cnidarian. PLoS Biol 2013, 11:e1001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Broun M, Gee L, Reinhardt B, Bode HR. Formation of the head organizer in hydra involves the canonical Wnt pathway. Development 2005, 132:2907–2916. [DOI] [PubMed] [Google Scholar]
- 31. Petersen CP, Reddien PW. Wnt signaling and the polarity of the primary body axis. Cell 2009, 139:1056–1068. [DOI] [PubMed] [Google Scholar]
- 32. Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, Rothbacher U, Holstein TW. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 2000, 407:186–189. [DOI] [PubMed] [Google Scholar]
- 33. Dickinson AJ, Sive HL. The Wnt antagonists Frzb‐1 and Crescent locally regulate basement membrane dissolution in the developing primary mouth. Development 2009, 136:1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Couly GF, Le Douarin NM. Mapping of the early neural primordium in quail‐chick chimeras. I. Developmental relationships between placodes, facial ectoderm, and prosencephalon. Dev Biol 1985, 110:422–439. [DOI] [PubMed] [Google Scholar]
- 35. Eagleson G, Ferreiro B, Harris WA. Fate of the anterior neural ridge and the morphogenesis of the Xenopus forebrain. J Neurobiol 1995, 28:146–158. [DOI] [PubMed] [Google Scholar]
- 36. Soukup V, Horacek I, Cerny R. Development and evolution of the vertebrate primary mouth. J Anat 2013, 222:79–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kurosaka H, Iulianella A, Williams T, Trainor PA. Disrupting hedgehog and WNT signaling interactions promotes cleft lip pathogenesis. J Clin Invest 2014, 124:1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Crump JG, Maves L, Lawson ND, Weinstein BM, Kimmel CB. An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development 2004, 131:5703–5716. [DOI] [PubMed] [Google Scholar]
- 39. Kennedy AE, Dickinson AJ. Median facial clefts in Xenopus laevis: roles of retinoic acid signaling and homeobox genes. Dev Biol 2012, 365:229–240. [DOI] [PubMed] [Google Scholar]
- 40. Barlow LA. Specification of pharyngeal endoderm is dependent on early signals from axial mesoderm. Development 2001, 128:4573–4583. [DOI] [PubMed] [Google Scholar]
- 41. Keller RE. Vital dye mapping of the gastrula and neurula of Xenopus laevis. II. Prospective areas and morphogenetic movements of the deep layer. Dev Biol 1976, 51:118–137. [DOI] [PubMed] [Google Scholar]
- 42. Keller RE. Vital dye mapping of the gastrula and neurula of Xenopus laevis. I. Prospective areas and morphogenetic movements of the superficial layer. Dev Biol 1975, 42:222–241. [DOI] [PubMed] [Google Scholar]
- 43. Koop D, Chen J, Theodosiou M, Carvalho JE, Alvarez S, de Lera AR, Holland LZ, Schubert M. Roles of retinoic acid and Tbx1/10 in pharyngeal segmentation: amphioxus and the ancestral chordate condition. Evodevo 2014, 5:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Graham A, Okabe M, Quinlan R. The role of the endoderm in the development and evolution of the pharyngeal arches. J Anat 2005, 207:479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development 2010, 137:2605–2621. [DOI] [PubMed] [Google Scholar]
- 46. Carmona‐Fontaine C, Matthews HK, Kuriyama S, Moreno M, Dunn GA, Parsons M, Stern CD, Mayor R. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature 2008, 456:957–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carmona‐Fontaine C, Theveneau E, Tzekou A, Tada M, Woods M, Page KM, Parsons M, Lambris JD, Mayor R. Complement fragment C3a controls mutual cell attraction during collective cell migration. Dev Cell 2011, 21:1026–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mayor R, Theveneau E. The neural crest. Development 2013, 140:2247–2251. [DOI] [PubMed] [Google Scholar]
- 49. Yu HH, Moens CB. Semaphorin signaling guides cranial neural crest cell migration in zebrafish. Dev Biol 2005, 280:373–385. [DOI] [PubMed] [Google Scholar]
- 50. Benouaiche L, Gitton Y, Vincent C, Couly G, Levi G. Sonic hedgehog signalling from foregut endoderm patterns the avian nasal capsule. Development 2008, 135:2221–2225. [DOI] [PubMed] [Google Scholar]
- 51. Tabler JM, Bolger TG, Wallingford J, Liu KJ. Hedgehog activity controls opening of the primary mouth. Dev Biol 2014, 396:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Houssin NS, Bharathan NK, Turner SD, Dickinson AJG. The role of JNK during buccopharyngeal membrane perforation, the last step of embryonic mouth formation. Dev Dyn 2017, 246:100–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Szabo‐Rogers HL, Smithers LE, Yakob W, Liu KJ. New directions in craniofacial morphogenesis. Dev Biol 2010, 341:84–94. [DOI] [PubMed] [Google Scholar]
- 54. Young NM, Hu D, Lainoff AJ, Smith FJ, Diaz R, Tucker AS, Trainor PA, Schneider RA, Hallgrimsson B, Marcucio RS. Embryonic bauplans and the developmental origins of facial diversity and constraint. Development 2014, 141:1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chai Y, Maxson RE Jr. Recent advances in craniofacial morphogenesis. Dev Dyn 2006, 235:2353–2375. [DOI] [PubMed] [Google Scholar]
- 56. Couly G, Creuzet S, Bennaceur S, Vincent C, Le Douarin NM. Interactions between Hox‐negative cephalic neural crest cells and the foregut endoderm in patterning the facial skeleton in the vertebrate head. Development 2002, 129:1061–1073. [DOI] [PubMed] [Google Scholar]
- 57. Barriga EH, Trainor PA, Bronner M, Mayor R. Animal models for studying neural crest development: is the mouse different? Development 2015, 142:1555–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Theveneau E, Mayor R. Neural crest delamination and migration: from epithelium‐to‐mesenchyme transition to collective cell migration. Dev Biol 2012, 366:34–54. [DOI] [PubMed] [Google Scholar]
- 59. Liu B, Rooker SM, Helms JA. Molecular control of facial morphology. Semin Cell Dev Biol 2010, 21:309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Medeiros DM, Crump JG. New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Dev Biol 2012, 371:121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bush JO, Jiang R. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development 2012, 139:231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hu D, Marcucio RS. Unique organization of the frontonasal ectodermal zone in birds and mammals. Dev Biol 2009, 325:200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Watanabe K, Sasaki F, Takahama H. The ultrastructure of oral (buccopharyngeal) membrane formation and rupture in the anuran embryo. Anat Rec 1984, 210:513–524. [DOI] [PubMed] [Google Scholar]
- 64. Waterman RE, Schoenwolf GC. The ultrastructure of oral (buccopharyngeal) membrane formation and rupture in the chick embryo. Anat Rec 1980, 197:441–470. [DOI] [PubMed] [Google Scholar]
- 65. Poelmann RE, Dubois SV, Hermsen C, Smits‐van Prooije AE, Vermeij‐Keers C. Cell degeneration and mitosis in the buccopharyngeal and branchial membranes in the mouse embryo. Anat Embryol (Berl) 1985, 171:187–192. [DOI] [PubMed] [Google Scholar]
- 66. Waterman RE. Ultrastructure of oral (buccopharyngeal) membrane formation and rupture in the hamster embryo. Dev Biol 1977, 58:219–229. [DOI] [PubMed] [Google Scholar]
- 67. Houssin NS, Bharathan NK, Turner SD, Dickinson AJ. Role of JNK during buccopharyngeal membrane perforation, the last step of embryonic mouth formation. Dev Dyn 2017, 246:100–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Waterman RE, Kao R. Formation of the mouth opening in the zebrafish embryo. Scan Electron Microsc 1982, 3:1249–1257. [Google Scholar]
- 69. Saal HM. Genetic evaluation for craniofacial conditions. Facial Plast Surg Clin North Am 2016, 24:405–425. [DOI] [PubMed] [Google Scholar]
- 70. Van Otterloo E, Williams T, Artinger KB. The old and new face of craniofacial research: how animal models inform human craniofacial genetic and clinical data. Dev Biol 2016, 415:171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Huh SH, Ornitz DM. Beta‐catenin deficiency causes DiGeorge syndrome‐like phenotypes through regulation of Tbx1. Development 2010, 137:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Villavicencio EH, Walterhouse DO, Iannaccone PM. The sonic hedgehog‐patched‐gli pathway in human development and disease. Am J Hum Genet 2000, 67:1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hill P, Wang B, Ruther U. The molecular basis of Pallister Hall associated polydactyly. Hum Mol Genet 2007, 16:2089–2096. [DOI] [PubMed] [Google Scholar]
- 74. Veistinen L, Takatalo M, Tanimoto Y, Kesper DA, Vortkamp A, Rice DP. Loss‐of‐function of Gli3 in mice causes abnormal frontal bone morphology and premature synostosis of the interfrontal suture. Front Physiol 2012, 3:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cobourne MT, Xavier GM, Depew M, Hagan L, Sealby J, Webster Z, Sharpe PT. Sonic hedgehog signalling inhibits palatogenesis and arrests tooth development in a mouse model of the nevoid basal cell carcinoma syndrome. Dev Biol 2009, 331:38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Adhikari K, Fuentes‐Guajardo M, Quinto‐Sanchez M, Mendoza‐Revilla J, Camilo Chacon‐Duque J, Acuna‐Alonzo V, Jaramillo C, Arias W, Lozano RB, Perez GM, et al. A genome‐wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nat Commun 2016, 7:11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang YB, Hu J, Zhang J, Zhou X, Li X, Gu C, Liu T, Xie Y, Liu J, Gu M, et al. Genome‐wide association study identifies multiple susceptibility loci for craniofacial microsomia. Nat Commun 2016, 7:10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci USA 2004, 101:10709–10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Moura da Silva AA, Ganz JS, Sousa PD, Doriqui MJ, Ribeiro MR, Branco MD, Queiroz RC, Pacheco MJ, Vieira da Costa FR, Silva FS, et al. Early growth and neurologic outcomes of infants with probable congenital Zika virus syndrome. Emerg Infect Dis 2016, 22:1953–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Weichert A, Vogt M, Dudenhausen JW, Kalache KD. Evidence in a human fetus of micrognathia and cleft lip as potential effects of early cytomegalovirus infection. Fetal Diagn Ther 2010, 28:225–228. [DOI] [PubMed] [Google Scholar]
- 81. Ornoy A. Valproic acid in pregnancy: how much are we endangering the embryo and fetus? Reprod Toxicol 2009, 28:1–10. [DOI] [PubMed] [Google Scholar]
- 82. Orup HI Jr, Deutsch CK, Holmes LB. Laser light scan analysis of the “anticonvulsant face”. Birth Defects Res A Clin Mol Teratol 2014, 100:905–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Malvasi A, Tinelli A, Buia A, De Luca GF. Possible long‐term teratogenic effect of isotretinoin in pregnancy. Eur Rev Med Pharmacol Sci 2009, 13:393–396. [PubMed] [Google Scholar]
- 84. Shi M, Wehby GL, Murray JC. Review on genetic variants and maternal smoking in the etiology of oral clefts and other birth defects. Birth Defects Res C Embryo Today 2008, 84:16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Murawski NJ, Moore EM, Thomas JD, Riley EP. Advances in diagnosis and treatment of fetal alcohol spectrum disorders: from animal models to human studies. Alcohol Res 2015, 37:97–108. [PMC free article] [PubMed] [Google Scholar]
- 86. Burns FR, Peterson RE, Heideman W. Dioxin disrupts cranial cartilage and dermal bone development in zebrafish larvae. Aquat Toxicol 2015, 164:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. van Boxtel AL, Pieterse B, Cenijn P, Kamstra JH, Brouwer A, van Wieringen W, de Boer J, Legler J. Dithiocarbamates induce craniofacial abnormalities and downregulate sox9a during zebrafish development. Toxicol Sci 2010, 117:209–217. [DOI] [PubMed] [Google Scholar]
- 88. Liu S, Rouleau J, Leon JA, Sauve R, Joseph KS, Ray JG, Canadian Perinatal Surveillance S. Impact of pre‐pregnancy diabetes mellitus on congenital anomalies, Canada, 2002–2012. Health Promot Chronic Dis Prev Can 2015, 35:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Carmichael SL, Ma C, Rasmussen SA, Cunningham ML, Browne ML, Dosiou C, Lammer EJ, Shaw GM. Craniosynostosis and risk factors related to thyroid dysfunction. Am J Med Genet A 2015, 167A:701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]


