ABSTRACT
Termination of Saccharomyces cerevisiae RNA polymerase II (Pol II) transcripts occurs through two alternative pathways. Termination of mRNAs is coupled to cleavage and polyadenylation while noncoding transcripts are terminated through the Nrd1-Nab3-Sen1 (NNS) pathway in a process that is linked to RNA degradation by the nuclear exosome. Some mRNA transcripts are also attenuated through premature termination directed by the NNS complex. In this paper we present the results of nuclear depletion of the NNS component Nab3. As expected, many noncoding RNAs fail to terminate properly. In addition, we observe that nitrogen catabolite-repressed genes are upregulated by Nab3 depletion.
KEYWORDS: nitrogen metabolism, noncoding RNA, termination, transcription
INTRODUCTION
Saccharomyces cerevisiae RNA polymerase II (Pol II) synthesizes both mRNAs and noncoding RNAs (ncRNAs), including snRNAs, snoRNAs, and a large number of RNAs with unknown functions (1, 2). The latter class includes cryptic unstable transcripts (CUTs) and stable uncharacterized transcripts (SUTs) (3, 4). Both coding and noncoding transcripts originate from promoters located in nucleosome-free regions in a process that requires both general transcription factors and gene-specific factors (5–7), but termination of these different classes of Pol II transcripts takes place through two different processes. Stable transcripts like mRNA and SUTs terminate through a process linked to cleavage and polyadenylation while snoRNAs and CUTs terminate through the Nrd1-Nab3-Sen1 (NNS) pathway (8–14).
The NNS complex contains two RNA-binding proteins, Nrd1 and Nab3, which recognize specific sequences in nascent transcripts and direct termination in a process that requires the RNA helicase Sen1 (8, 9, 15–20). NNS interacts with Pol II in part through binding of Nrd1 to phosphorylated Ser5 on the C-terminal domain (CTD) (21–23), a pattern of CTD phosphorylation most prominent in the early stages of the transcription cycle, limiting NNS termination to promoter-proximal transcripts (24–29). The NNS complex also interacts with the TRAMP (Trf4/Trf5-Air1/Air2-Mtr4 polyadenylation) complex to couple termination to processing by the nuclear exosome (30–33).
While the two yeast Pol II termination pathways generally operate on distinct sets of transcripts, there are some genes that can use either termination pathway (11). In some cases NNS acts downstream of genes as a fail-safe termination mechanism to ensure that transcripts that fail to terminate through the cleavage/polyadenylation mechanism do not read through into downstream genes (34, 35). In other cases NNS functions upstream to terminate transcripts before the poly(A) site is reached. For example, Nrd1 autoregulates its own transcription by binding, along with Nab3, to sites in the 5′ end of its nascent pre-mRNA (9, 36). The result of this binding leads to premature termination of the majority of Nrd1 transcripts close to the 5′ end. Nrd1 transcripts that escape the NNS pathway go on to terminate through the cleavage/polyadenylation pathway, producing a mature mRNA. A similar form of regulation occurs with a number of genes encoding enzymes involved in nucleotide synthesis (37–39). In these cases, however, the use of alternative start sites leads to pre-mRNAs that have or do not have the Nrd1 and Nab3 binding sites in their 5′ untranslated regions (UTRs) and therefore either terminate prematurely through NNS or elongate to produce a mature mRNA.
We along with others have previously used the anchor-away strategy (40) to create conditional mutants of Nrd1 that lead to nuclear depletion in the presence of rapamycin (7, 41). This approach has led to the identification of NNS termination sites and has identified a limited set of mRNAs that are controlled through NNS. In this paper, we present the results of Nab3 nuclear depletion through the anchor-away method. We observe the expected readthrough of known NNS terminators downstream of ncRNAs and an increase in transcription of several mRNAs known to be regulated by NNS. In addition to these known targets, we show that some genes regulated by nitrogen catabolite repression (NCR) are regulated by NNS.
RESULTS
Nab3 anchor-away experiments.
To examine the effect on transcription of depleting Nab3 from the nucleus, we C-terminally tagged Nab3 with the FRB (FKBP12-rapamycin binding) domain in an anchor-away strain that contains an HTB (6His-TEV-biotin, where TEV is tobacco etch virus) tag on the RNA polymerase subunit Rpb2 (15, 40, 41). FRB-tagged Nab3 protein is normally expressed and renders this strain sensitive to rapamycin (see Fig. S1A and B in the supplemental material), indicating that Nab3 performs an essential nuclear function. We noticed that the NAB3-FRB strain grows slightly more slowly than a control strain even in the absence of rapamycin, indicating that the C-terminal tag is a hypomorphic mutation (Fig. S1C). We performed photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) on the NAB3-FRB RPB2-HTB strain to map changes in the position of actively transcribing Pol II in response to Nab3 depletion. Cross-linking took place 30 min after the addition of rapamycin when cells were still growing (Fig. S1D). To control for effects of the tag, we carried out PAR-CLIP on the FRB-tagged strain with or without rapamycin and a control strain that does not contain an FRB tag but does include the downstream HIS3 marker that was used to introduce the tag. We also conducted PAR-CLIP on an NAB3-HTB strain to map Nab3 binding sites on nascent RNA transcripts (15, 41, 42). PAR-CLIP data sets were obtained from two biological replicates for each of these conditions. The R values for these data sets were greater than 0.98, and thus we combined replicates for the read maps.
Regulation of snoRNAs and CUTs.
Nuclear depletion of Nab3 results in readthrough transcription of snoRNA and CUTs as has been previously described for Nrd1 depletion (7, 9, 32, 33, 41). Figure 1A shows that snR47 and Nrd1 readthrough transcripts are slightly elevated in the FRB-tagged strain, and this level of readthrough is increased in the presence of rapamycin. This result supports the idea that the C-terminally FRB-tagged Nab3 is slightly defective and could explain the slightly slower growth of this strain.
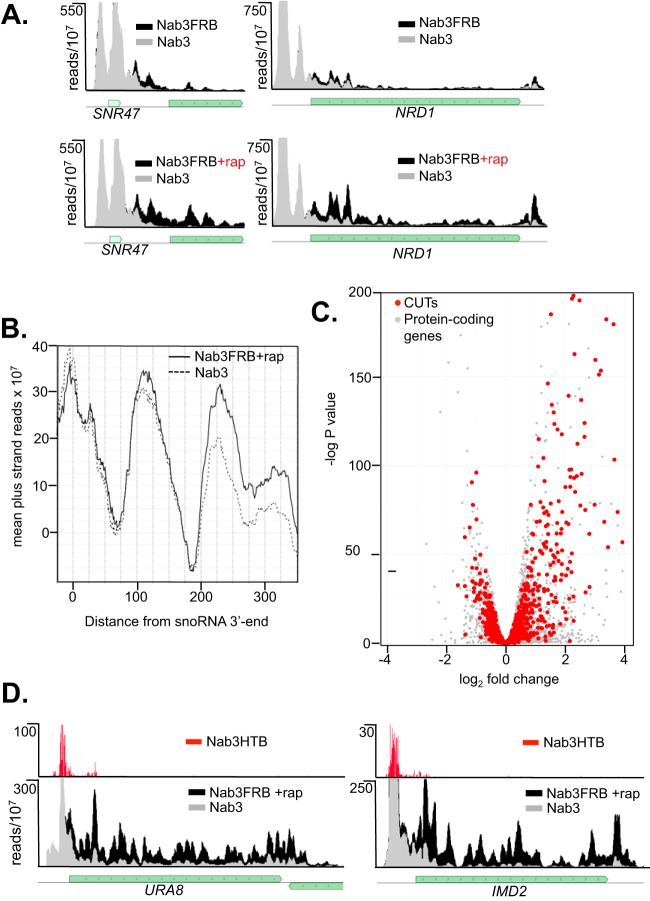
FIG 1.
Readthrough of ncRNA transcripts. (A) Pol II maps of mutant and control NAB3 strains on an snoRNA (SNR47) and the attenuated NRD1 gene. The gray track represents reads from the control strain, while the superimposed black track represents Pol II reads from the NAB3-FRB strain with (+rap) or without nuclear depletion. (B) Anchor plot (67) showing readthrough of snoRNA terminators. (C) Volcano plot showing readthrough of CUTs. (D) Pol II and Nab3 maps of mRNAs regulated by Nab3. rap, rapamycin.
Because the NAB3-FRB strain is a hypomorph, we have compared the Pol II PAR-CLIP results after the anchor-away procedure to those with the untagged NAB3 strain. Figure 1B shows a plot of the averaged sum of Pol II reads downstream of the aligned 3′ end of all snoRNAs. This result indicates that readthrough transcription following Nab3 depletion extends at least 300 bases downstream of snoRNA NNS terminators. Readthrough transcription is also seen with CUTs. Figure 1C is a volcano plot of Pol II reads corresponding to 920 CUTs and extending downstream of the annotated transcript by 450 bases. Comparing Nab3 depletion and wild-type (WT) readthrough data shows that Nab3 depletion results in many CUTs transcribed downstream of their normal termination sites (Fig. 1C, upper right quadrant).
Regulation of protein-coding genes.
In addition to noncoding RNAs, the NNS complex has been shown to bind to a number of mRNAs (7, 15, 19, 35, 43, 44). However, despite the prevalence of Nrd1-Nab3 binding to the 5′ end of mRNAs, there are relatively few genes where regulation by premature termination has been confirmed (7, 15, 35, 41, 43, 45). Nrd1 expression is autoregulated through attenuation and is also sensitive to mutations in Nab3 (9, 36). In our Nab3 anchor-away experiments, we confirm this result (Fig. 1A) and show that several genes involved in nucleotide biosynthesis that have previously been shown to be regulated by Nrd1 (37–39) are also upregulated by Nab3 depletion. Figure 1D shows that IMD2 and URA8 expression is increased when Nab3 is depleted from the nucleus. In both of these cases, PAR-CLIP of Nab3-HTB reveals corresponding Nab3 binding peaks at the 5′ end of the transcript.
NCR.
To look for other protein coding genes that are regulated by Nab3, we plotted the ratio of coding-region reads for the Nab3-depleted data set relative to the WT reads for all yeast genes (Fig. 2A). Only a subset of genes show significantly altered expression, and we focused our attention on those that show the greatest increased expression. Table 1 shows the 48 genes with a greater than 2-fold increase in expression. An example of this class of genes is the GLT1 gene that encodes glutamate synthase (Fig. 2B). Many of the genes that are elevated in response to Nab3 depletion are, like GLT1, regulated by nitrogen catabolite repression (NCR) (Table 1, genes in boldface) (46, 47). This includes genes for a transcription factor that regulates genes involved in nitrogen metabolism (DAL80), an enzyme at the hub of nitrogen metabolism (GLT1), and a number of transporters involved in nitrogen acquisition (GAP1, MEP2, MEP3, and UGA4).
FIG 2.
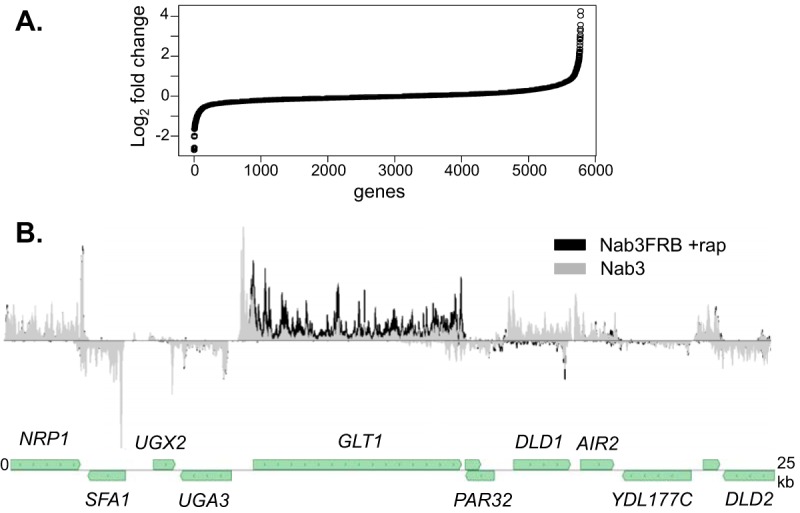
Nab3 depletion changes expression of relatively few genes. (A) Rank order plot of changes in expression of protein coding genes after Nab3 depletion. Genes are ranked from left to right in order of increased expression upon Nab3 depletion. (B) Pol II map showing the increase in expression of GLT1 but no change in adjacent genes.
TABLE 1.
Genes upregulated as a result of Nab3 depletion
| Genea | Fold change in expressionb |
|---|---|
| IMD2 | 6.4 |
| IZH4 | 4.7 |
| DAL80 | 4.6 |
| FRE4 | 4.4 |
| NRD1 | 3.9 |
| URA8 | 3.8 |
| UGA4 | 3.6 |
| HMS1 | 3.4 |
| DAK2 | 3.4 |
| AGP3 | 3.3 |
| YNR066C | 3.3 |
| MEP2 | 3.2 |
| PTR2 | 3.1 |
| SAM2 | 3.0 |
| CRF1 | 2.8 |
| MTO1 | 2.8 |
| BAG7 | 2.7 |
| DUR1,2 | 2.7 |
| RGT1 | 2.7 |
| IMD3 | 2.5 |
| REF2 | 2.5 |
| YDR124W | 2.5 |
| GAP1 | 2.5 |
| DAL1 | 2.5 |
| GLT1 | 2.5 |
| NRK1 | 2.5 |
| OLE1 | 2.4 |
| SUL2 | 2.4 |
| MET6 | 2.3 |
| YCR101C | 2.3 |
| YLR053C | 2.3 |
| MKK2 | 2.2 |
| HEM25 | 2.2 |
| PHD1 | 2.2 |
| TDA4 | 2.2 |
| MEP3 | 2.2 |
| YJL213W | 2.2 |
| TOS2 | 2.1 |
| DSE4 | 2.1 |
| MET3 | 2.1 |
| SPO23 | 2.0 |
| YGK3 | 2.0 |
| OPT2 | 2.0 |
| MET13 | 2.0 |
| FLO5 | 2.0 |
| STE23 | 2.0 |
| URA10 | 2.0 |
| ATG19 | 2.0 |
Genes in boldface are regulated by nitrogen catabolite repression.
Relative to the WT level.
Figure 3A shows the results of a gene set enrichment analysis (GSEA) (48) of the entire set of 90 NCR genes (46). In this analysis genes are ordered from left to right by decreasing enrichment in the anchor-away data set. The enrichment score indicates the degree to which a set of genes is more prevalent at the extremes of the ranked list. The panel to the left shows an enrichment score for Nab3 of 0.6, with the leading edge of NCR genes among the most upregulated genes. The P value for the NCR genes in the Nab3 data is reported by the GSEA algorithm as 0.00, indicating a high degree of significance. The panel to the right is an analysis of NCR genes upregulated in our previous Nrd1 anchor-away experiment (41). Nrd1 depletion yields a lower enrichment score (0.4) and a P value of 0.065. Despite this lower enrichment score, there are some NCR genes among the most upregulated in response to Nrd1 depletion. We have compared the set of genes upregulated in response to Nab3 depletion with our previously published data on genes upregulated in response to Nrd1 depletion (41). As expected, there is significant overlap among these sets of genes (Fig. 3B). About half of genes upregulated by Nab3 depletion are also upregulated after Nrd1 depletion. Among this overlapping set are seven genes regulated by NCR (Table S2).
FIG 3.
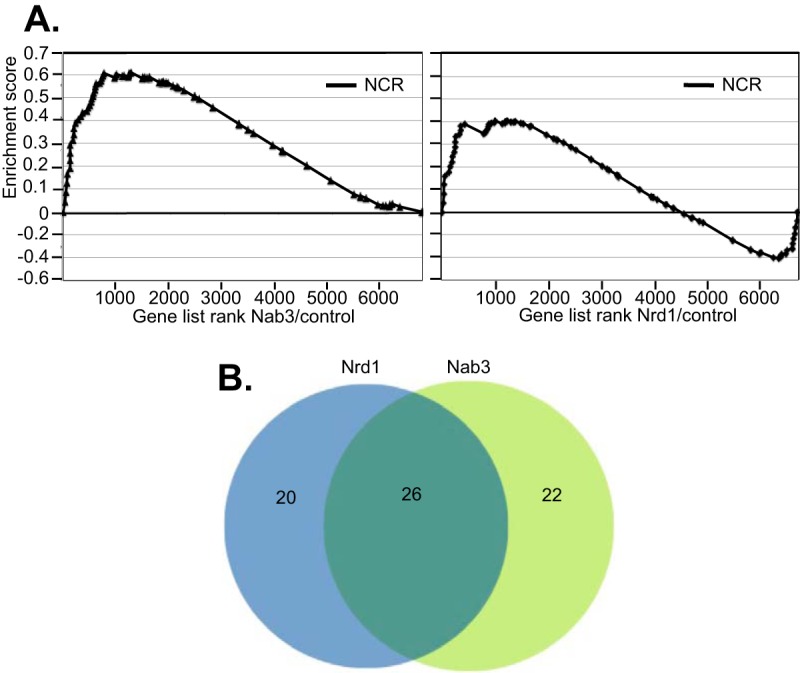
Increased expression of nitrogen catabolite-repressed genes. (A) The left panel is derived from the Nab3 anchor-away experiment, while the right panel is derived from the Nrd1 anchor-away experiment. Both panels show enrichment plots derived from GSEA analysis. In this analysis genes are rank ordered from left to right in order of their ratio of Pol II occupancy. The enrichment score is a running sum of enrichment of the NCR genes. (B) Venn diagram showing overlap of genes upregulated in response to either Nab3 or Nrd1 depletion.
Pol II maps of select NCR-regulated genes (MEP2, DAL80, and GAP1) are shown in Fig. 4. For each of these genes, we observed a peak of Nab3 binding at the 5′ end of the transcript. Nab3 forms a heterodimer with Nrd1 (8, 16), and many ncRNA terminators require both factors for efficient termination (9, 32, 41). One of the genes upregulated in response to Nrd1 depletion is the ammonium permease gene MEP2. Both Nrd1 and Nab3 cross-link to sequences in the 5′ UTR, and upon nuclear depletion the level of transcription is increased across the coding region.
FIG 4.
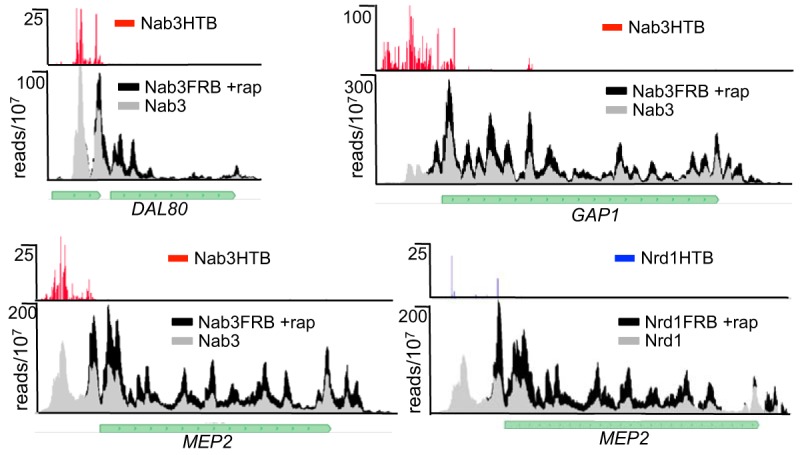
Increased expression of nitrogen catabolite-repressed genes. Pol II and Nab3 binding maps showing examples of Nab3-regulated NCR genes.
Nab3 and Nrd1 motifs.
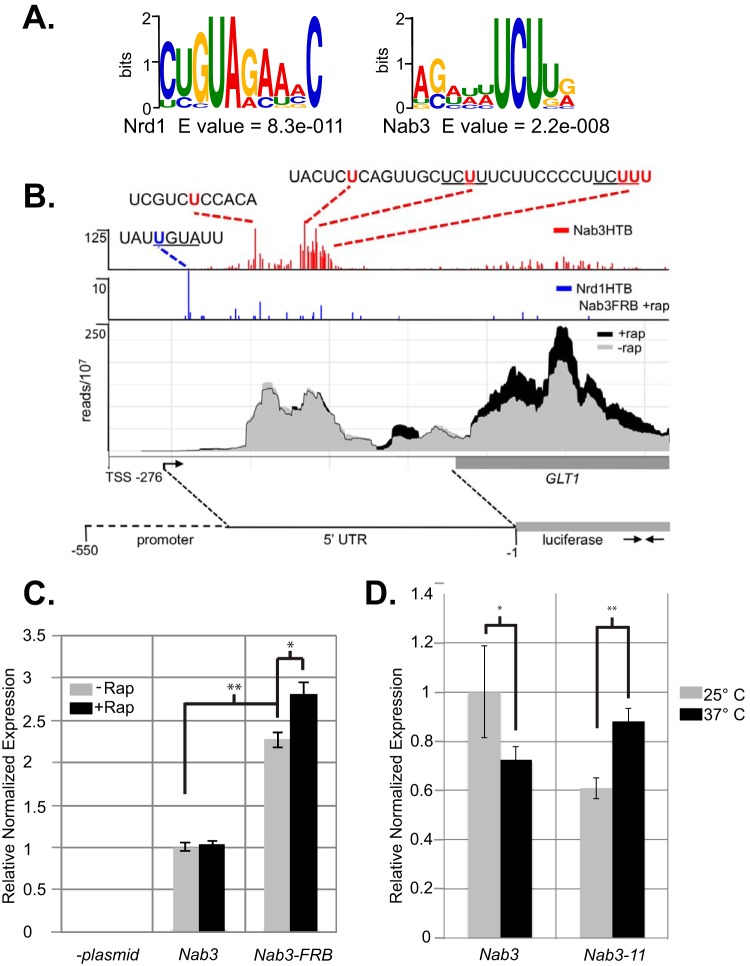
The precise binding sites for Nrd1 and Nab3 can be determined from the presence of T-to-C transitions caused by the 4-thiouracil cross-link (49, 50). Using these T-to-C transitions to find the binding sites, we analyzed the top 40 Nab3 binding peaks among NCR genes and found common motifs corresponding to known Nrd1 and Nab3 binding sites (Table S1) (15–19). Using MEME software (51), we found that the Nrd1 consensus is present in fewer genes but is more statistically significant while the Nab3 consensus is present more often but contains less information (Fig. 5A). Nrd1 cross-links are most often observed at the first U in the UGUA sequence. For Nab3, T-to-C transitions are spread out over a region that often contains several UCU sequences. This pattern is apparent in the cross-linking to the Glt1 5′ UTR shown in Fig. 5B and for MEP3, GAP1, and GAT1 shown in Fig. S2. Multiple cross-linking sites could reflect binding to flexible RNAs by Nab3 or to the presence of multiple Nrd1-Nab3 proteins, as we have previously observed in vitro (16).
FIG 5.
Nab3 and Nrd1 binding regulates NCR gene expression. (A) Logos corresponding to known Nrd1 and Nab3 binding sites derived from sequences in the top Nab3 binding peaks identified on NCR transcripts. (B) Mapping Nrd1 and Nab3 binding sites in the GLT1 5′ UTR. (C) Reverse transcription-quantitative PCR of luciferase RNA driven by the Glt1 promoter as indicated in panel B. (D) Reverse transcription-quantitative PCR of the endogenous GLT1 gene in NAB3 and nab3-11 strains.
GLT1 regulation.
Mapping Nrd1 and Nab3 binding sites to the 5′ end of an mRNA suggests that the gene is regulated by premature termination but does not rule out regulation by sequences in the coding region or 3′ UTR. To demonstrate that an NCR-regulated gene is regulated through its 5′ sequences, we cloned the Glt1 promoter and 5′ UTR upstream of the luciferase gene. When this plasmid is present in a wild-type strain, we see a low level of reporter mRNA that does not change when rapamycin is added (Fig. 5C). In contrast, in a NAB3-FRB strain we observe an elevated level of expression, and this level increases when Nab3 is depleted from the nucleus. We observed a similar increase in GLT1 expression in a temperature-sensitive nab3-11 strain raised to the nonpermissive temperature (Fig. 5D). This is not as great an increase as seen in the reporter strain, but in the WT strain GLT1 expression decreases at the nonpermissive temperature. Together, these results indicate that GLT1 is regulated by attenuation directed by Nab3 binding to the 5′ UTR.
NAB3-FRB confers resistance to glutamine synthetase inhibition.
The observation that NCR genes are upregulated even though cells are grown in a preferred nitrogen source suggests that Nab3 mutation alters the nitrogen-sensing mechanism. To test the response to nitrogen depletion, we were not able to inhibit the TOR pathway as the anchor-away strain contains the tor1-1 mutation that confers resistance to rapamycin (52). Instead, we used l-methionine sulfoximine (MSX) to inhibit glutamine synthetase and thus deplete cells for glutamine (53, 54). Figure 6 demonstrates that the Nab3-FRB strain is more resistant than the WT to 0.5 mM MSX, presumably due to the partial Nab3 defect caused by the FRB tag. The ability to grow in the presence of MSX is likely due to increased expression of glutamine synthetase and other proteins involved in nitrogen acquisition and metabolism.
FIG 6.
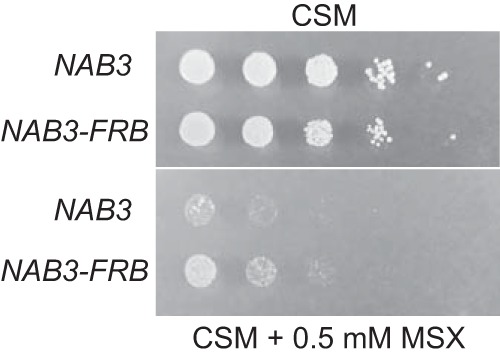
Growth of NAB3-FRB and control strain in the presence or absence of 0.5 mM MSX.
DISCUSSION
Previous work on the yeast NNS pathway has indicated that this alternative Pol II termination pathway is linked to nutrient availability (35, 43, 45, 55). Nrd1 mutants are synthetic lethal, with mutations that activate the Ras-protein kinase A (PKA) signaling pathway, and the pattern of Nrd1 binding changes upon glucose deprivation (42). Recent studies have shown that Nab3 is also important in reshaping the transcriptome in response to glucose deprivation (45, 55). In this paper, we provide additional evidence that Nab3 is important in regulating genes that are required for uptake and catabolism of nonpreferred nitrogen sources, implicating the NNS complex in the regulation of genes in response to either carbon or nitrogen sources.
Saccharomyces cerevisiae prefers ammonia or glutamine as a source of nitrogen but can use a variety of alternative, nonpreferred nitrogen sources (46). Nitrogen availability regulates growth, metabolism, and transcription through a complex interplay of multiple pathways (46, 56, 57). The target of rapamycin complex 1 (TORC1) stimulates expression of growth-related genes in part through the sensing of intracellular amino acid levels (57, 58). The approximately 90 nitrogen catabolite-repressed genes are controlled through a set of four GATA transcription factors that cross-regulate each other in a manner that depends on the source of nitrogen. Although the TORC1 signaling pathway plays a role in GATA factor-mediated NCR-responsive gene transcription (59), other pathways have also been implicated (58, 60–64). In fact, regulation of NCR genes is largely independent of TOR signaling, but details of how different nitrogen sources lead to regulation of NCR genes is not well understood.
The results presented here indicate that Nab3 plays a role in nitrogen catabolite repression. Nuclear depletion of Nab3 results in increased expression of many NCR genes, including those for a GATA factor (DAL80), a central nitrogen metabolic enzyme glutamate synthase (GLT1), and several permeases that are specific for different nitrogen sources. We have shown that one of these genes, GLT1, is regulated through its promoter and 5′ UTR. Multiple Nab3 binding sites are occupied in nascent transcripts of many other NCR genes, but whether this binding alone is sufficient to trigger attenuation is not clear. Whether TORC1 signaling is important for NNS regulation was not directly addressed as the anchor-away strain contains the tor1-1 mutation that renders the pathway resistant to rapamycin.
We suggest that Nab3 may be an important component of the regulation of NCR genes. In the presence of ammonia or other rich nitrogen sources, our data demonstrate that Nab3 operates to abrogate expression of GLT1 and many other nitrogen catabolism genes. Given our previous study linking the NNS pathway to the glucose signaling pathway, our current results suggest that NNS may integrate signals from the carbon and nitrogen sensors. By extension, our data suggest that the transcriptional response to carbon and nitrogen source changes involves an Nrd1/Nab3-mediated termination/antitermination regulatory choice that may permit rapid adaptation to environmental change.
Using computational approaches, Chasman and colleagues have discovered interactions among signaling pathways involved in Saccharomyces cerevisiae stress responses (65). One result of these studies was the identification of Pol II CTD phosphorylation as a hub that connects transcriptome changes between growth and stress conditions. Specifically, they showed that CTD Ser5 phosphorylation was essential for Pol II to rapidly relocate in response to stress. The NNS complex role in NCR may represent part of this pathway intersection. Nrd1 preferentially interacts with Ser5-phosphorylated CTD (21, 22). Nrd1 and Nab3 binding to NCR transcripts could normally act together with the appropriate GATA factors to attenuate expression of genes involved in the use of nonpreferred nitrogen sources. Loss of the preferred source would then lead to loss of attenuation, enabling the cell to rapidly reprogram its transcriptome to express genes required for acquisition and metabolism of nonpreferred nitrogen sources. The nature of the signal that impinges on NNS is not known, and whether this involves changes in CTD phosphorylation is also unclear.
MATERIALS AND METHODS
Yeast strains.
Anchor-away and 6His-TEV-biotin (HTB)-tagged strains were constructed as previously described (15, 41). Cells were grown in complete synthetic medium (CSM) containing 2% glucose and 40 mg/liter adenine, in CSM limiting uracil (CSM-Ura) containing 2% glucose, 40 mg/liter adenine, and 60 μM uracil, or in yeast extract-peptone-dextrose (YPD) medium, depending on the experiment. For growth curves, cells were incubated in a 24-well plate using an Infinity 2 (BioTek) instrument. The BioTek was set for orbital shaking (fast; 1-mm amplitude); cells were diluted to an optical density at 600 nm (OD600) of 0.1 per well, and the OD600 was measured every 10 min for 36 h.
Anchor-away and PAR-CLIP experiments.
Cells were grown in 5 ml of YPD medium, seeded into 500 ml of CSM containing 60 μM uracil and 1 μM biotin, and incubated at 30°C until cultures reached an OD600 of 1. Cross-linking was performed as previously described (41). Instead of rapamycin, dimethyl sulfoxide (DMSO) was added as a control sample in the 30-min incubation period before cross-linking. Cross-linked samples with the HTB tag at the C terminus of Rpb1 were processed as previously described (41) with a slight modification to the protocol: the streptavidin beads were resuspended in polynucleotide kinase (PNK) reaction solution (1 U/μl T4 PNK [NEB], 5 mM dithiothreitol [DTT] in PNK buffer), but we excluded the [γ-32P]ATP.
Sequencing and bioinformatics.
Sequencing and demultiplexing were done at the Johns Hopkins University School of Medicine on an Illumina HiSeq instrument. Briefly, raw sequences were trimmed of 3′ adapters as previously described (41) using a wrapper developed by Sarah Wheelan; sequences were condensed to allow no more than a single copy of any exact sequence and aligned to the SacCer3 reference genome (revision R64, February 2011; obtained from the Saccharomyces Genome Database [SGD]) using Bowtie, version 1.1.1, with the following arguments: “-y –best -v 2” to remove sequences that aligned to the tRNA and ribosomal DNA (RDN) regions and “-y –best -v 2 -m 2” for the global alignment. The SAM file format produced from the alignments was then converted to wiggle track format and normalized as previously described (41).
The DESeq package available from Bioconductor was used for differential expression analysis (66). Briefly, the aligned data sets were converted into bam files and fed into the DESeq package pipeline. The default DESeq pipeline was used. DESeq normalized the reads and averaged the replicates to compare expression levels on a per-gene basis. This produced an output file containing fold changes along with their statistical significance. Volcano plots were plotted using these data through R. For Table 1, we removed genes with fewer than 200 reads and genes that are adjacent to known ncRNAs. We used gene set enrichment analysis (GSEA) (48) to further show the differential expression of NCR genes. Read signal was counted exactly as done in the DESeq process to keep these two results comparable and loaded into the GSEA (Broad Institute) Java applet, version 2.2.3, using default settings except that we used the SGD set of yeast genes. GSEA rank orders every gene by a signal-to-noise ratio using multiple replicates and calculates an enrichment score for each set of genes that represents the differential expression of that set of genes.
Quantitative PCR.
Total RNA was extracted with acid phenol as previously described (15). Briefly, cells were grown in CSM-Ura with and without rapamycin to an OD of 1, suspended in TES buffer (10 mM Tris [pH 7.5], 10 mM EDTA, 0.5% SDS), and treated with acid phenol for 1 h at 65°C with moderate shaking. Multiple acid phenol washes were used until the phase interface was clean. Chloroform was used to wash any remnants of acid phenol before ethanol precipitation. The total extracted RNA was treated with Turbo DNase-free (AM1907; Ambion), per the most stringent protocol recommended. The resulting DNase-free RNA was then reverse transcribed using a Bio-Rad iScript cDNA synthesis kit. Real-time PCR was performed using iQ SYBR green supermix (Bio-Rad) in a CFX96 real-time PCR detection system (Bio-Rad) using three 20-μl replicates for each sample. Expression of RNA amplified with the luciferase primers (GTGTTCGTGGACGAGGTG and CGATCTTGCCGCCTTTCTTA) or GLT1 primers (CATGTGGTGACTGTAGAAGAGG and GATACGTAGTGCCGTCCATTAG) was normalized to that of ACT1 and graphed relative to that of the WT NAB3 control samples.
Accession number(s).
The accession number for the PAR-CLIP data sets was deposited in the Gene Expression Omnibus database under accession number GSE97345.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Proft (Valencia) for the yeast promoter-luciferase plasmid, Sarah Wheelan (Johns Hopkins) for the trimming computer script, and Brendan Cormack (Johns Hopkins) for thoughtful comments on the manuscript.
This work was supported by grant number RO1GM66108 from the National Institute of General Medical Sciences.
The funders had no role in design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00154-17.
REFERENCES
- 1.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. 2006. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A 103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yassour M, Pfiffner J, Levin JZ, Adiconis X, Gnirke A, Nusbaum C, Thompson DA, Friedman N, Regev A. 2010. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol 11:R87. doi: 10.1186/gb-2010-11-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis CA, Ares M Jr. 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 5.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 7.Schulz D, Schwalb B, Kiesel A, Baejen C, Torkler P, Gagneur J, Soeding J, Cramer P. 2013. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell 155:1075–1087. doi: 10.1016/j.cell.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 8.Conrad NK, Wilson SM, Steinmetz EJ, Patturajan M, Brow DA, Swanson MS, Corden JL. 2000. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154:557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 10.Marquardt S, Hazelbaker DZ, Buratowski S. 2011. Distinct RNA degradation pathways and 3′ extensions of yeast non-coding RNA species. Transcription 2:145–154. doi: 10.4161/trns.2.3.16298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porrua O, Libri D. 2015. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat Rev Mol Cell Biol 16:190–202. doi: 10.1038/nrm3943. [DOI] [PubMed] [Google Scholar]
- 12.Mischo HE, Proudfoot NJ. 2013. Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochim Biophys Acta 1829:174–185. doi: 10.1016/j.bbagrm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Proudfoot NJ. 2016. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352:aad9926. doi: 10.1126/science.aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuehner JN, Pearson EL, Moore C. 2011. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12:283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Creamer TJ, Darby MM, Jamonnak N, Schaughency P, Hao H, Wheelan SJ, Corden JL. 2011. Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet 7:e1002329. doi: 10.1371/journal.pgen.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll KL, Ghirlando R, Ames JM, Corden JL. 2007. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA 13:361–373. doi: 10.1261/rna.338407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL. 2004. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol 24:6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porrua O, Hobor F, Boulay J, Kubicek K, D'Aubenton-Carafa Y, Gudipati RK, Stefl R, Libri D. 2012. In vivo SELEX reveals novel sequence and structural determinants of Nrd1-Nab3-Sen1-dependent transcription termination. EMBO J 31:3935–3948. doi: 10.1038/emboj.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wlotzka W, Kudla G, Granneman S, Tollervey D. 2011. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J 30:1790–1803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porrua O, Libri D. 2013. A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat Struct Mol Biol 20:884–891. doi: 10.1038/nsmb.2592. [DOI] [PubMed] [Google Scholar]
- 21.Gudipati RK, Villa T, Boulay J, Libri D. 2008. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol 15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- 22.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. 2008. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol 15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubicek K, Cerna H, Holub P, Pasulka J, Hrossova D, Loehr F, Hofr C, Vanacova S, Stefl R. 2012. Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. Genes Dev 26:1891–1896. doi: 10.1101/gad.192781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corden JL. 2013. RNA polymerase II C-terminal domain: Tethering transcription to transcript and template. Chem Rev 113:8423–8455. doi: 10.1021/cr400158h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eick D, Geyer M. 2013. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev 113:8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 27.Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, Eick D, Ansari AZ. 2010. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol 17:1154–1161. doi: 10.1038/nsmb.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 29.Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. 2010. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol 17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fasken MB, Laribee RN, Corbett AH. 2015. Nab3 facilitates the function of the TRAMP complex in RNA processing via recruitment of Rrp6 independent of Nrd1. PLoS Genet 11:e1005044. doi: 10.1371/journal.pgen.1005044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tudek A, Porrua O, Kabzinski T, Lidschreiber M, Kubicek K, Fortova A, Lacroute F, Vanacova S, Cramer P, Stefl R, Libri D. 2014. Molecular basis for coordinating transcription termination with noncoding RNA degradation. Mol Cell 55:467–481. doi: 10.1016/j.molcel.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. 2006. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the Nrd1-Nab3 pathway in genome surveillance. Mol Cell 23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 33.Arigo JT, Eyler DE, Carroll KL, Corden JL. 2006. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell 23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 34.Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ. 2009. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol Cell 36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webb S, Hector RD, Kudla G, Granneman S. 2014. PAR-CLIP data indicate that Nrd1-Nab3-dependent transcription termination regulates expression of hundreds of protein coding genes in yeast. Genome Biol 15:R8. doi: 10.1186/gb-2014-15-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arigo JT, Carroll KL, Ames JM, Corden JL. 2006. Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell 21:641–651. doi: 10.1016/j.molcel.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Thiebaut M, Colin J, Neil H, Jacquier A, Seraphin B, Lacroute F, Libri D. 2008. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol Cell 31:671–682. doi: 10.1016/j.molcel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 38.Kuehner JN, Brow DA. 2008. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol Cell 31:201–211. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 39.Jenks MH, O'Rourke TW, Reines D. 2008. Properties of an intergenic terminator and start site switch that regulate IMD2 transcription in yeast. Mol Cell Biol 28:3883–3893. doi: 10.1128/MCB.00380-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haruki H, Nishikawa J, Laemmli UK. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 41.Schaughency P, Merran J, Corden JL. 2014. Genome-wide mapping of yeast RNA polymerase II termination. PLoS Genet 10:e1004632. doi: 10.1371/journal.pgen.1004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jamonnak N, Creamer TJ, Darby MM, Schaughency P, Wheelan SJ, Corden JL. 2011. Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA 17:2011–2025. doi: 10.1261/rna.2840711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darby MM, Serebreni L, Pan X, Boeke JD, Corden JL. 2012. The Saccharomyces cerevisiae Nrd1-Nab3 transcription termination pathway acts in opposition to Ras signaling and mediates response to nutrient depletion. Mol Cell Biol 32:1762–1775. doi: 10.1128/MCB.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol 6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bresson S, Tuck A, Staneva D, Tollervey D. 2017. Nuclear RNA decay pathways aid rapid remodeling of gene expression in yeast. Mol Cell 65:787–800. doi: 10.1016/j.molcel.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Godard P, Urrestarazu A, Vissers S, Kontos K, Bontempi G, van Helden J, Andre B. 2007. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol Cell Biol 27:3065–3086. doi: 10.1128/MCB.01084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. 2014. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 38:254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. 2010. PAR-CliP—a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp 41:2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heitman J, Movva NR, Hall MN. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 53.Manning JM, Moore S, Rowe WB, Meister A. 1969. Identification of l-methionine S-sulfoximine as the diastereoisomer of l-methionine SR-sulfoximine that inhibits glutamine synthetase. Biochemistry 8:2681–2685. doi: 10.1021/bi00834a066. [DOI] [PubMed] [Google Scholar]
- 54.Crespo JL, Powers T, Fowler B, Hall MN. 2002. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci U S A 99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Nues R, Schweikert G, de Leau E, Selega A, Langford A, Franklin R, Iosub I, Wadsworth P, Sanguinetti G, Granneman S. 2017. Kinetic CRAC uncovers a role for Nab3 in determining gene expression profiles during stress. Nat Commun 8:12. doi: 10.1038/s41467-017-00025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ljungdahl PO, Daignan-Fornier B. 2012. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 190:885–929. doi: 10.1534/genetics.111.133306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broach JR. 2012. Nutritional control of growth and development in yeast. Genetics 192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tate JJ, Cooper TG. 2013. Five conditions commonly used to down-regulate TOR complex 1 generate different physiological situations exhibiting distinct requirements and outcomes. J Biol Chem 288:27243–27262. doi: 10.1074/jbc.M113.484386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beck T, Hall MN. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 60.Tate JJ, Feller A, Dubois E, Cooper TG. 2006. Saccharomyces cerevisiae Sit4 phosphatase is active irrespective of the nitrogen source provided, and Gln3 phosphorylation levels become nitrogen source-responsive in a sit4-deleted strain. J Biol Chem 281:37980–37992. doi: 10.1074/jbc.M606973200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tate JJ, Georis I, Dubois E, Cooper TG. 2010. Distinct phosphatase requirements and GATA factor responses to nitrogen catabolite repression and rapamycin treatment in Saccharomyces cerevisiae. J Biol Chem 285:17880–17895. doi: 10.1074/jbc.M109.085712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tate JJ, Rai R, Cooper TG. 2005. Methionine sulfoximine treatment and carbon starvation elicit Snf1-independent phosphorylation of the transcription activator Gln3 in Saccharomyces cerevisiae. J Biol Chem 280:27195–27204. doi: 10.1074/jbc.M504052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Georis I, Tate JJ, Cooper TG, Dubois E. 2011. Nitrogen-responsive regulation of GATA protein family activators Gln3 and Gat1 occurs by two distinct pathways, one inhibited by rapamycin and the other by methionine sulfoximine. J Biol Chem 286:44897–44912. doi: 10.1074/jbc.M111.290577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feller A, Georis I, Tate JJ, Cooper TG, Dubois E. 2013. Alterations in the Ure2 αCap domain elicit different GATA factor responses to rapamycin treatment and nitrogen limitation. J Biol Chem 288:1841–1855. doi: 10.1074/jbc.M112.385054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chasman D, Ho YH, Berry DB, Nemec CM, MacGilvray ME, Hose J, Merrill AE, Lee MV, Will JL, Coon JJ, Ansari AZ, Craven M, Gasch AP. 2014. Pathway connectivity and signaling coordination in the yeast stress-activated signaling network. Mol Syst Biol 10:759. doi: 10.15252/msb.20145120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Homann OR, Johnson AD. 2010. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol 8:49. doi: 10.1186/1741-7007-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.