Abstract
Immediate early genes have previously demonstrated a rapid increase in gene expression after various behavioral paradigms. The main focus of this article is to identify a molecular marker of circuit activation after manual whisker stimulation or exploration of a novel environment. To this end, we investigated the dynamics of ARC transcription in adult male rats during whisker somatosensation throughout the whisker barrel circuit. At various time points, tissue was biopsied from the ventral posterior medial nucleus (VPM) of the thalamus, primary somatosensory barrel field (S1BF) cortex and hippocampus for quantification using real-time PCR and western blot. Our results show that there were no significant differences in ARC gene or protein expression in the VPM after both types of stimulation. However, manual whisker stimulation resulted in increased ARC gene expression at 15, 30, 60 and 300 minutes in the S1BF, and 15 minutes in the hippocampus (p<0.05). Also, exploration of a novel environment resulted in increased ARC mRNA expression at 15 and 30 minutes in the S1BF and at 15 minutes in the hippocampus (p<0.05). The type of stimulation (manual versus exploration of a novel environment) influenced the magnitude of ARC gene expression in the S1BF (p<0.05). These data are the first to demonstrate that ARC is a specific, quantifiable and input dependent molecular marker of circuit activation which can serve to quantify the impact of brain injury and subsequent rehabilitation on whisker sensation.
Keywords: ARC, whisker stimulation, S1BF, VPM, hippocampus
Introduction
Immediate-early genes (IEGs) are defined as genes that have the fastest possible transcriptional activation in response to neural stimulation ranging from acoustic and visual stimulation, sleep deprivation, fear, and other learning paradigms [1–5]. IEGs have been effective at detecting activation in neural circuits associated with specific behaviors [6]. In order to evaluate the impact of circuit damage and subsequent recovery, it would be useful to have a IEG as a molecular marker of circuit activation that is specific, measurable and input-dependent. Therefore, the purpose of this study is to evaluate ARC as this molecular marker of circuit activation after manual whisker stimulation or exploration of a novel environment in the whisker barrel circuit.
The activity-regulated cytoskeleton-associated (ARC; also known as Arg3.1) IEG was first identified as a plasticity gene [7], which is rapidly activated and transcribed in response to circuit activity. In fact, one epoch of behavioral activity rapidly induces the transcription of ARC mRNA [8]. Different behavioral and learning tasks have been reported to induce transcription of ARC mRNA [9]. In a study by Ivanova et al., (2011), 10 minutes of sound exposure in mice induced the greatest amount of expression of ARC mRNA in cortical layers 3–6 of the auditory cortex at 30 minutes post-stimulation [10]. Spatial learning and memory processes also modified the expression of ARC in the frontal cortex and hippocampus, respectively [6, 11]. Additionally, a single, brief exploratory experience was sufficient to drive an initial (5 minutes) and a temporally delayed (8–24 hour) wave of hippocampal ARC protein expression [12]. These data support behavior-dependent ARC gene activation in functionally relevant neural circuits, thus implying a potential role for ARC expression after circuit activation as a means to identify circuit disruption.
Rodents explore an environment with their whiskers, through the sense of touch. They whisk their vibrissae back and forth, brushing them against the floor, objects, walls, etc. to perceive their environment [13–15]. Whisker deflection drives mechanoreceptors on sensory neurons in the trigeminal ganglia. Sensory neurons project axons that then synapse in the barrelettes of the principal sensory trigeminal nucleus (pr5), which then synapse within neuronal clusters, barreloids, in the ventral posterior medial (VPM) thalamic nucleus. Then, VPM neurons project topographically to barrel fields in layer IV of the primary somatosensory cortex (S1BF) [16]. The whisker barrel circuit is a highly organized and well understood pathway that offers the opportunity to study modalities of intact circuit activation and synaptic transmission in an experimental model that can be assessed over time. Understanding of in vivo neural circuits in rodents directly translates to neural circuit function in humans, thus providing an opportunity to characterize neurological deficits and test treatment paradigms in existing experimental models.
Although there is abundant work in regards to circuit activation and plasticity modeled in the whisker circuit, the time course for ARC and its use as a molecular marker of whisker barrel circuit stimulation has not been elucidated. With this premise, this study proposes to investigate the normal time course of ARC gene and protein expression in the whisker barrel circuit using two methods of whisker somatosensory stimulation: manual whisker stimulation and voluntary exploration of a novel environment. Our data show that after manual whisker stimulation and exploration of a novel environment, there were no significant differences in ARC gene or protein expression in the VPM. However, both tasks revealed the induction of ARC gene and protein expression in S1BF and gene expression in hippocampus. This is the first demonstration of a time course for ARC upregulation in response to whisker somatosensation. These results support the potential for ARC as a molecular biomarker of circuit activation in an intact in vivo neural circuit, which may serve as a functional paradigm to quantify the impact of disease, drugs treatments, behavioral experiences and subsequent repair of compromised circuits.
Methods
Animals and whisker somatosensation procedures
Adult male Sprague-Dawley rats (300–350g; Harlan, Indianapolis, IN) were used in the present study. Animals were housed in a 12 hr light/dark cycle and allowed access to food and water ad libitum. All experimental procedures followed the guidelines established in the Guide for the Care and Use of Laboratory Animals (U.S. Department of Health and Human Services) and were approved by the Institutional Animal Care and Use Committee at St. Joseph’s Hospital and Medical Center (Phoenix, AZ). Sixty-four rats were divided into three groups; naïve, manual whisker stimulation and exploration of novel environment. Naïve animals did not receive stimulation. Naïve animals were removed from their cage and prepared for perfusion and dissection (n=6). Rats included in the manual whisker stimulation group were introduced to the behavior box (57.1 × 39.4 × 15.2 cm) lined with an absorbent pad and allowed to habituate for 5 minutes. Then, the rats’ whiskers were manually stimulated on both mystacial pads with a wooden applicator stick for three consecutive 5 minute periods, following our published protocol for the whisker nuisance task (15 minutes total; n=3–6/time point) [17]. After manual whisker stimulation, animals were returned to their home cage and prepared for perfusion and dissection at eight time points following manual whisker stimulation (15, 30, 60, 90, 120, 180, 240, 300 minutes). The novel environment group was introduced to the same behavioral box lined with an absorbent pad and allowed to habituate for 5 minutes. For the next 15 minutes they were free to explore their environment, voluntarily using their whiskers, but did not have their whiskers stimulated manually (n=3/time point). At the end of the 15 minutes, rats were returned to their home cages until they were prepared for perfusion and dissection at 15, 30, 60 and 120 minutes following the end of the exploration period.
Perfusion and dissection
At a predetermined time, each animal was given a lethal dose of sodium pentobarbital 200 mg/kg (Euthasol®, i.p.). Animals were transcardially perfused with ice-cold phosphate buffered saline (PBS) for 2–3 minutes. The brain was rapidly removed and rinsed with ice-cold PBS. Tissue biopsies (2 mm diameter) taken from the VPM, S1BF and hippocampus were collected from 2 mm thick coronal sections made using a chilled rat brain matrix. Tissue samples from the right hemisphere were, stored in RNAlater® (Invitrogen catalog # AM7020), and kept at −20°C until mRNA was extracted for quantitative PCR. Tissue samples from the left hemisphere were flash frozen and stored in at −80°C until protein was extracted for western blot analysis.
RNA extraction
Total mRNA was extracted from the VPM, S1BF and hippocampus biopsies, previously stored in RNAlater®, using the MagMAX™-96 Total RNA Isolation Kit (Invitrogen catalog; # AM1830). Following the manufacturer’s protocol, biopsies were homogenized in TRI reagent® solution (Invitrogen; # AM9738) and extracted under acidic conditions. Total RNA was further purified using RNA binding beads. The ratio of the absorbance at 260 and 280 nm was used to assess RNA purity and quality (Nanodrop, Thermo-Scientific 2000). All RNA samples were within the established range for pure RNA (1.8–2.1), with an average A260/A280 of 1.99 with a standard deviation of 0.21.
Quantitative PCR
Total RNA was converted to cDNA using the High Capacity RNA-to-cDNA Kit from Life Technologies™ (catalog # 4387406). Each sample of concentrated cDNA was diluted to 5 ng in preparation for quantitative real-time PCR (qPCR) using commercially-available gene expression assays. The Applied Biosystems TaqMan® Gene Expression Assay for ARC (Rn00571208_g1), EgR3 (Rn00567228_m1), Zif268 (Rn00561138_m1) and BDNF (Rn02531967_s1), and were optimized to run under universal thermal cycling conditions, with amplification efficiencies of 100% by the manufacturer. Within each animal, relative gene expression was normalized to the 18S rRNA endogenous control and the expression level in the sham/vehicle group using the 2−ΔΔCT method [18], which relates gene expression to the PCR cycle number at which the fluorescence signals exceed a threshold above baseline. Samples were run in triplicate and 18s was run in duplicate according to manufacturer’s instructions.
Protein extraction
Total protein was extracted from the VPM, S1BF and hippocampus biopsies, previously stored at −80°C. Tissues were homogenized in 250 µl of ice-cold extraction buffer (pH 8.0) containing 0.24 M Tris, 0.74 M NaCl, 100 µl TritonX100 with a protease inhibitor cocktail (complete, Roche Diagnostics; #11836153001). Tissue from hippocampus and S1BF were homogenized with the Precellys®24 machine (Bertin Technologies, Montigny le Bretonneux, France) for 40 seconds. Samples were then centrifuged at 3,000 × g for 15 minutes and the supernatant collected for analysis. The tissue from VPM was homogenized with a sonicator until the solution was completely clear. Samples were then centrifuged at 3,000 × g for 15 minutes and the supernatant collected for analysis. Protein concentrations were determined using the Bicinchoninic acid assay (BCA) using manufactures instructions (Pierce, Rockford, IL).
Western blot
NuPAGE® LDS Sample Buffer (4×) (Life technologies; # NP0007) and NuPAGE® Sample Reducing Agent (10×) (Life technologies; # NP0004) were added to the protein samples and then boiled for 10 minutes at 70 °C. Protein extracts (15 µg) were then electrophoresed in a 4–12% Bis-Tris Midi gel (Life technologies; # WG1403BOX) and transferred to nitrocellulose membranes with iBlot® Transfer Stack (Life Technologies; #IB3010-01) using iBlot® 7-Minute Blotting System (Invitrogen). Blots were blocked in 1× PBS and Odyssey Blocking Buffer (50:50) and incubated with an anti-β-Actin antibody for 12 hours (h) (mouse monoclonal, 1:15000, A2228 SIGMA). After rinsing the blot 3 times with 1X PBS + 1% Tween20 over 30 minutes, blots were incubated with an anti-mouse secondary antibody (1:10000; Licor®) for one hour. Afterwards, blots were washed for 30 minutes and incubated with polyclonal anti-ARC/Arg3.1 antibody for 48h (rabbit polyclonal, 1:500, ab23382, Abcam). Blots are then washed 3 times and incubated with an anti-rabbit secondary antibody (1:20000; Licor®) for one hour. Bands were visualized using the Licor® ODYSSEY® Classic and analyzed by Image Studio™ 4.0 ® software (Lincoln, NE). Densitometry was determined based on band intensity and relative protein expression. Samples were randomized and run in triplicate. ARC protein levels were normalized to β-actin protein levels as a loading control. Replicates for ARC and β-actin were averaged for 1 number per region per each animal and the averaged values were compared over time and to naïve controls.
Statistical analyses
Changes in relative gene and protein expression obtained at each time point were compared over the time course and to naïve animals for both manual whisker stimulation and the novel environment exploration using a one-way ANOVA with Tukey’s post-hoc analysis, unless indicated otherwise. Anatomical regions were analyzed independently. Significance was set at p<0.05. All statistical analyses were performed using Prism 6 (Graph Pad, La Jolla, CA). Data are presented as the mean ± the standard error from the mean (SEM).
Results
ARC is the optimal IEG to study as a molecular marker of circuit activation
A molecular marker of circuit activation should have the capability to indicate circuit activation after stimulation or normal exploration as demonstrated by our two paradigms: manual whisker stimulation and exploration of a novel environment. In addition, the time course of gene expression after circuit activation must consistently result in input-dependent, measurable and statistically significant changes in circuit activation. To this end, we chose three different IEGs and a growth factor: ARC, EgR3, Zif268 and BDNF, which have previously demonstrated an increase in gene expression after various circuit activation paradigms [19–22]. Our data show that ARC had the greatest levels of gene expression after whisker stimulation in the S1BF, with the least amount of variability within animal groups, as demonstrated by smaller error bars in comparison to EgR3, Zif268 and BDNF (Supplementary Figure 1). Based on this information, we focused on characterizing ARC as an indicator of circuit activation.
Time course of ARC gene and protein expression in the VPM after manual whisker stimulation
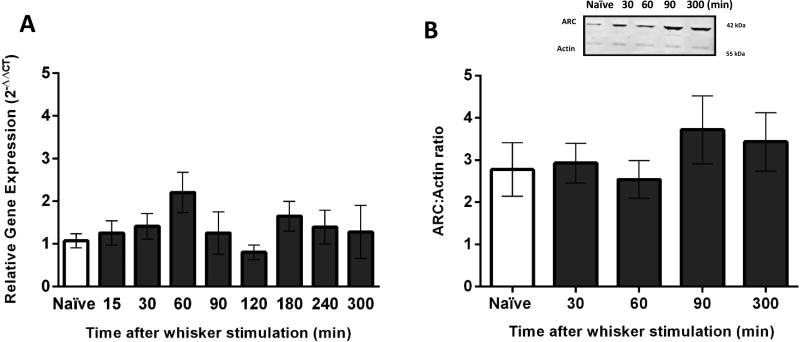
Following manual whisker stimulation, ARC gene expression was examined in VPM tissue at eight different time points: 15, 30, 60, 90, 120, 180, 240 and 300 minutes (n=3–6 rats per group). There were no significant differences in ARC gene expression in comparison to naïve animals at any of eight time points in the VPM (F(8,34)=1.39, p=0.23; Figure 1A).
Figure 1.
ARC gene expression and protein levels in the ventral posterior medial nucleus (VPM) after manual whisker stimulation. (A) The time course of ARC gene expression was assessed in the VPM using quantitative PCR. There was no significant upregulation of ARC gene after manual whisker stimulation in comparison to naïve animals (n=5/time point; naïve=6). (B) The time course of ARC showed no significant increase of ARC protein levels after manual whisker stimulation compared to naïve animals (n=3–5/time point; naïve: n=6). The inset demonstrates a representative western blot of Arc protein from the VPM of naïve and manual whisker stimulation animals, compared to actin. The data are represented as the mean ± standard error of the mean (SEM), where *, p<0.05 in comparison to naïve.
Tissue from the opposite hemisphere was assessed for protein levels using western blot. To demonstrate changes in protein levels, we only chose four time points for western blot based on the time we predicted protein levels to be elevated (30–60 minutes) and resolved (300 minutes). At four time points after manual stimulation (30, 60, 90 and 300 minutes; n=3–6 rats per group) no significant differences in ARC protein expression were detected compared to naïve animals (F(4,26)=0.59, p=0.67; Figure 1B).
Time course of ARC gene and protein expression in the S1BF after manual whisker stimulation
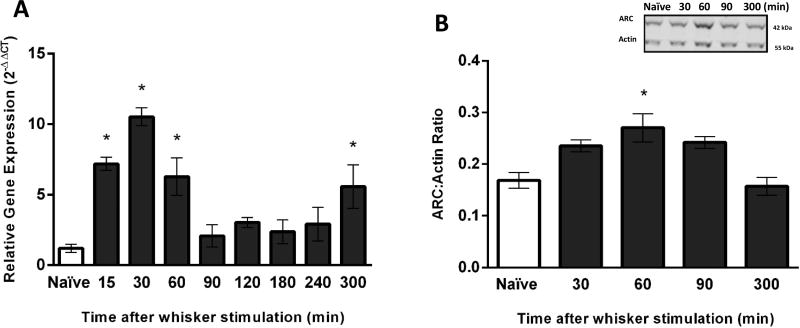
Following manual whisker stimulation, ARC gene expression in the S1BF was assessed at eight time points: 15, 30, 60, 90, 120, 180, 240 and 300 minutes (n=3–6 rats per group). Overall, a one-way ANOVA revealed that whisker stimulation induced a statistically significant change in ARC gene expression over time (F(8,31)=10.57, p<0.0001; Figure 2A). ARC gene expression was elevated to 6-fold, 9-fold and 5-fold at 15, 30 and 60 minutes after whisker stimulation in comparison to naïve, respectively. ARC gene expression was significantly decreased at 120 and 180 minutes after stimulation in comparison to peak expression at 30 minutes. Notably, there was a secondary 4-fold elevation of ARC expression at 300 minutes (5h) relative to naïve animals (Figure 2A).
Figure 2.
ARC gene expression and protein levels in the primary somatosensory barrel field (S1BF) after manual whisker stimulation. (A) The time course of ARC gene expression was assessed in S1BF using quantitative PCR. ARC gene expression was significantly increased at 15, 30, 60 and 300 minutes post-stimulation in comparison to naïve animals (n=5–6/time point). (B) The time course of ARC protein levels in the S1BF were significantly increased at 60 minutes after manual whisker stimulation in comparison to naïve animals (n=3–4/time point). The inset is a representative western blot showing ARC protein expression from the S1BF of (naïve) and manual whisker stimulation animals, compared to actin. The data are presented as the mean ± SEM, where *, p<0.05 in comparison to naïve animals.
Amongst the four time points examined after manual stimulation (30, 60, 90 and 300 minutes; n=3–6 rats per group), ARC protein expression was only significantly elevated at 60 minutes in comparison to naïve animals (F(4,10)=7.97, p=0.004; Figure 2B).
Time course of ARC gene and protein expression in the hippocampus after manual whisker stimulation
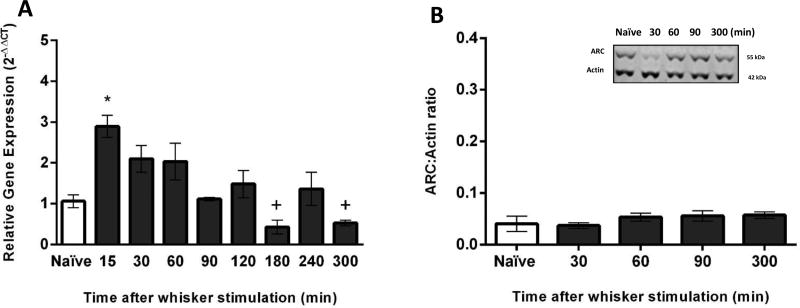
Following manual whisker stimulation, ARC gene expression in the hippocampus was assessed at eight time points: 15, 30, 60, 90, 120, 180, 240 and 300 minutes (n=3–6 rats per group). ARC gene expression at 15 minutes after whisker stimulation was significantly elevated by 1.8-fold compared to naïve animals (F(8,35)=5.92, p<0.0001; Figure 3A). In addition, ARC gene expression was significantly decreased at 180 and 300 minutes after whisker stimulation in comparison to the peak expression at 15 minutes (Figure 3A).
Figure 3.
ARC gene expression and protein levels in the hippocampus after manual whisker stimulation. (A) The time course of ARC gene expression was assessed in the hippocampus using quantitative PCR. A significant upregulation of ARC gene expression after manual whisker stimulation was identified at 15 minutes in comparison to naïve animals (n=5–6/time point). In addition, ARC gene expression was significantly decreased at 180 and 300 minutes after whisker stimulation in comparison to the peak expression at 15 minutes (+<0.05). (B) The time course of ARC protein levels in the hippocampus showed no significant increase of ARC after manual whisker stimulation in comparison to naïve animals (n=3–4/time point; naïve: n=6). The representative western blot demonstrates of Arc protein levels from the hippocampus of control (naïve) and manual whisker stimulation animals, compared to actin. The data are presented as the mean ± SEM, where *, p<0.05 in comparison to naïve animals.
ARC protein levels in the hippocampus measured at all four time points (30, 60, 90 and 300) after whisker stimulation (n=3–6 rats per group) revealed no significant differences when compared with the naïve animals (F(4,10)=1.01, p=0.44; Figure 3B).
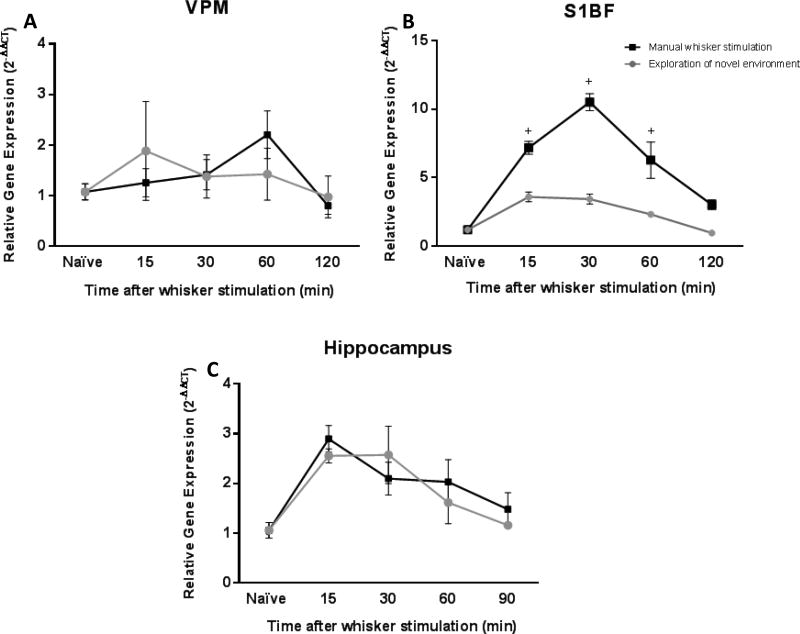
Time course of ARC gene expression after exploring a novel environment
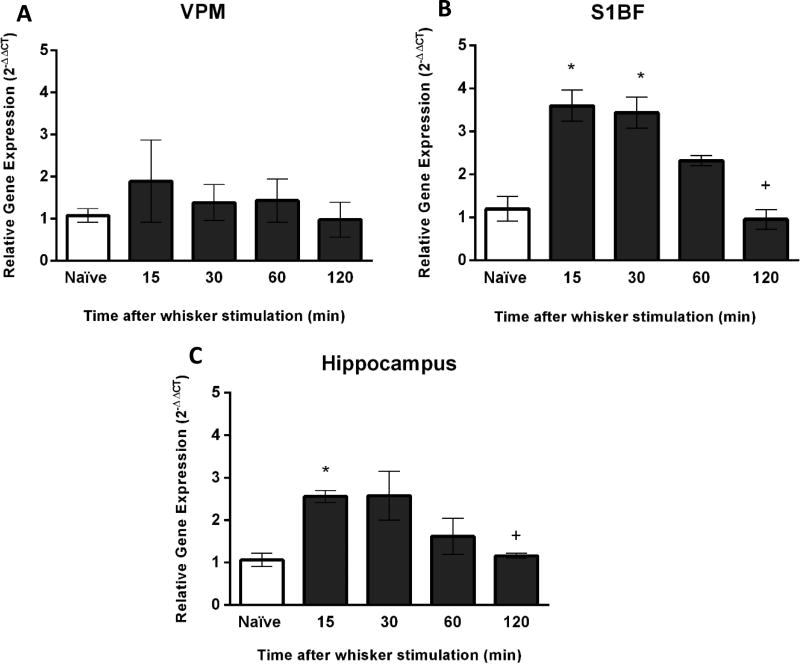
The purpose of these experiments was to quantify ARC gene expression after exploration of a novel environment. Similar to the analysis following manual stimulation, we analyzed the expression of ARC in VPM and S1BF of the whisker circuit and the hippocampus. Consistent to the experiments using manual whisker stimulation, no differences in ARC gene expression in the VPM were detected in comparison to naïve rats after exploring a novel environment (F(4,13)=0.54, p=0.70; Figure 4A). Conversely, in the S1BF at 15 and 30 minutes following exploration of novel environment (n=3 per each group), ARC gene expression was significantly elevated by 2.2 and 2.3-fold, respectively (F(4.13)=15.25, p<0.0001; Figure 4B) in comparison to naïve animals. In the hippocampus, ARC gene expression was significantly increased after exploring the novel environment for 15 minutes (F(4,13)=6.37, p=0.004; Figure 4C). There was a 2-fold increase in comparison to naïve animals for this time point.
Figure 4.
ARC gene expression after exploration of novel environment. (A) The time course of ARC gene expression was assessed using quantitative PCR. In the VPM, there were no changes in ARC gene expression as a function of novel environment exploration (n=3/time point). (B) In the S1BF, a significant upregulation of ARC gene expression was identified at 15 and 30 minutes after exploring a novel environment in comparison to naïve animals (n=3/time point; naïve: n=6). In addition, ARC gene expression was significantly decreased at 120 minutes after exploration of novel environment in comparison to the peak expression at 15 and 30 minutes (+<0.05). (C) In the hippocampus, a significant increase of ARC gene expression was identified at 15 minutes after exploration of novel environment (n=3/time point) In addition, ARC gene expression was significantly decreased at 120 minutes after exploration of novel environment in comparison to the peak expression at 15 minutes (+<0.05). The data are presented as the mean ± SEM, where, * p<0.05 in comparison to naïve animals.
Since robust responses to gene expression were demonstrated after manual whisker stimulation with small corresponding changes in protein levels, we did not measure protein level in the novel environment group.
Comparison between exploring manual whisker stimulation and novel environment
We compared the impact of the whisker stimulation paradigms on ARC gene expression in each brain region using a 2-way ANOVA with Tukey’s post-hoc analysis. In the VPM, there was no difference in ARC gene expression between manual whisker stimulation and exploring novel environment (F(1,36)=0.00; p=0.99; Figure 5A). However in the S1BF, results revealed that 15, 30 and 60 minutes post-stimulation there were 3, 7 and 4-fold differences, respectively, in ARC gene expression between the stimulation modes (F(1,32)=66.48; p<0.0001, Figure 5B). ARC gene expression levels were higher in animals with manual whisker stimulation in comparison to rats exposed to a novel environment. There was no difference between manual whisker stimulation and exploring novel environment for ARC gene expression in the hippocampus (F(1,36)=0.30; p=0.59, Figure 5C). All together, the comparison of manual whisker stimulation and exploration of a novel environment illustrated a stimulation-dependent increase in ARC gene expression in the S1BF, without impact on the hippocampus, a relay that is more likely activated in response to exploration.
Figure 5.
The level of ARC gene expression in the S1BF can be dependent on the type of whisker stimulation. The line graphs represent the time course of ARC gene expression over time, comparing animals being exposed to manual whisker stimulation (■) with animals using their whiskers to explore a novel environment (●). (A, C) Overall ARC gene expression in the VPM and the hippocampus did not show any differences between manual whisker stimulation animals when compared to exploration of novel environment animals. (B) In the S1BF, ARC gene expression was significantly greater after manual whisker stimulation compared to exploration of novel environment at 15, 30 and 60 min. The data are represented as the mean ± SEM. +, p<0.05 in comparison to exploration of novel environment at the same times post-stimulation by a two-way ANOVA followed by a Bonferoni multiple comparison.
Discussion
Activity-regulated cytoskeleton-associated protein (ARC) is an immediate early gene which is rapidly activated and transcribed in response to circuit activity. In the present study, we investigated the time course of activity-dependent ARC gene and protein expression in the thalamic and cortical relays of the whisker barrel circuit and the hippocampus following two approaches of whisker stimulation. We used both manual whisker stimulation and exploration of novel environment to activate the whisker barrel circuit. Our findings demonstrate that both sensory stimulation paradigms are sufficient to markedly increase ARC gene expression between 15 and 60 minutes and again at 300 minutes following whisker stimulation in the S1BF; with peak gene expression at 30 minutes. These data also indicate that the type of stimulation, in particular, manual whisker stimulation can dramatically influence the magnitude of ARC gene expression in the S1BF. Despite the upregulation of ARC gene expression of 9-fold following manual stimulation at the 30 minutes time point, the concentration of ARC protein in S1BF was modestly increased at 60 minutes, whereas protein levels remained unchanged in the hippocampus and VPM. This is the first study to evaluate ARC as a molecular marker of circuit activation by looking at the time course, specificity (whisker circuit relays vs. the hippocampus), and nature of the whisker stimulation input (manual in comparison to exploration of a novel environment) within two anatomical relays of the whisker barrel circuit, the S1BF and VPM.
In this study we chose to use qPCR and western blot instead of more commonly used in situ hybridization and immunohistochemistry [23] for rapid and comparable gene and protein quantification. Using 2 mm diameter biopsies, we were able to collect VPM, S1BF and hippocampus with discretion. We were able to process one hemisphere for qPCR and the alternate hemisphere for western blot in order to coordinate the time courses for mRNA and protein expression within the same brain/animal. While these methods do not provide the spatial resolution to determine activation in specific cellular layers, they do allow quantitative comparison of the activation levels and thereby identify the optimal time post-stimulation for histological approaches in future studies.
VPM Thalamus
Few studies exist on ARC expression in the thalamus. Our results did not show significant induction of ARC gene or protein levels either by manual whisker stimulation or exploration of novel environment. Most studies investigating the expression of other IEGs in thalamus have focused on visual sensory stimuli [24–26]. Bisler et al, investigated spatial and temporal patterns of c-Fos protein accumulation in the whisker barrel circuit by placing rodents, with all but 2 whiskers shaved, into a novel, enriched environment for 10m, 1h, 6h, 14h, 48h or 5d. Immediately following those times, brains were prepared for immunohistological staining for c-Fos. In the VPM, c-Fos staining was apparent after 1h of enriched environment exposure [27]. Also, thermal stimulation of hind paws of rats over 4 hours revealed the induction of c-fos in posterior thalamic nucleus [28]. Together, Bisler and Bullitt found IEG and protein expression in the thalamus after at least 1 hour of stimulation. In our experiments, we only stimulated for 15 minutes, which may not have been long enough to induce a measurable amount of ARC. It also demonstrates that different early inducible proteins, such as c-Fos, may have substantially different temporal expression; justifying the need to study the parallel time course of gene and protein activation after a controlled bout of behavior over a longer duration.
We suggest that the lack of detectable ARC activation in the VPM may also be in relation to intensity by which the somatosensory stimulation becomes progressively narrower in thalamic neurons. Gilbert & Wiesel describe that the thalamocortical inputs arrive in layer 4 of the cortex and then project to the superficial layers 2 and 3 [29, 30]. The superficial pyramidal neurons project to neurons in layer 5, which in turn project to neurons in layer 6, which project back to the VPM [31]. Even with consideration of trigeminothalamic projection into the VPM, there may be less ARC/IEG activation, making activity dependent changes in ARC more difficult to quantify using qPCR. It is also possible that the 2 mm × 2 mm diameter punch was too large of a biopsy, diluting the increase in gene expression with a larger volume of uninvolved tissue. Future experiments will focus on histological approaches for the VPM, where improved spatial resolution can be obtained.
S1BF Cortex
In examination of the S1BF, both manual whisker stimulation and voluntary exploration of a novel environment resulted in robust increases in ARC gene expression. There are several studies that demonstrated the expression of other IEGs or related proteins, such as c-Fos, in the S1BF. Manual whisker stimulation over the course of 8 days increased the number of growth factors within layers II–V in somatosensory cortex [32]. Also using immunohistochemistry, Filipkowski et al. (2000) showed two novel approaches to induce whisker stimulation-initiated changes in gene expression of c-Fos in the S1BF: 1) manual whisker stimulation of one side of the rat’s myastacial pad for 20 minutes and 2) placing the rat in a novel wire cage in a dark sound-proof room for 20 minutes. At 2h post-stimulation, immunohistochemical methods detected the greatest levels of c-Fos protein expression in cortical layer IV; with slightly less c-Fos expression observed in layers II/III and V/VI, respectively. These data indicate that whisker stimulation induces robust IEG protein expression in the S1BF. The duration and type of whisker stimulation in Filipkowski’s study shared many similarities with our experimental design. However, in our study, habituation + stimulation lasted 20 minutes, and tissue was processed for protein expression over a time course of 30, 60, 90 and 300 minutes. ARC protein levels peaked at 1h post-stimulation and showed evidence of decline by 90 minutes. Considering these similiarities (whisker stimulation, timing of stimulation) and differences (single time point post-stimulation versus time course) between the studies, the duration of stimulation and the timing of tissue processing are critical elements in study design for assessing intact or pathological circuitry.
Hippocampus
Prior works have documented the gene/protein expression of ARC in the hippocampus when animals are subjected to exploration of a novel environment or learning processes [33]. However, placing the rat in a new environment as a means of stimulating whiskers is rarely used [34], possibly because this type of stimulation is more dependent on the tendency of the animal to explore the new environment and the novelty of elements [35]. Our current data show the peak level of ARC gene expression at 15 minutes post-exposure in the hippocampus, following both manual whisker stimulation and exploration of novel environment. Although our results did not show any significant increase in ARC protein levels in the hippocampus after both behavioral paradigms, data from Ramirez-Amaya et al. (2005) showed that exposure to novel environment will result in significant increase of ARC in hippocampus. In this study, Ramirez-Amaya and colleagues showed that exposure to two sets of novel environment with intervals ranging from 30 minutes to 24h between two sets, will result in expression of ARC at 30 minutes – 2h and 8h – 24h in hippocampal CA and parietal cortex [12]. We suggest that the differences between their and our results could also depend on the different regions of hippocampus (e.g., CA3 and CA1) studied, the duration of the exploration, the novelty of the environment, and the time post-stimulation.
Relationship between voluntary and manual stimulation and gene expression
In manual whisker stimulation experiments, a 5 minute acclimation to the behavior box was followed by 15 minutes of manual whisker stimulation. For the voluntary exploration of the novel environment, rats were placed in the same behavior box, but did not have their whiskers manually stimulated; rather, voluntary exploration resulted in natural whisker stimulation. Both methods of whisker stimulation significantly increased peak ARC gene expression at 30 minutes over a similar time course in S1BF. However, manual whisker stimulation resulted in 3, 7 and 4-fold greater levels of ARC gene expression after 15, 30 and 60 minutes post-stimulation, respectively, in comparison to voluntary exploration of a novel environment. These results confirm that the time course of ARC gene expression is not only repeatable, but input dependent.
While there is an inability to quantify the amount of whisker stimulation that actually occurred (manually or voluntary), one could consider the following scenarios. First, combined exploration of the novel environment while having the whiskers manually stimulated could result in an additive effect. Second, it is also possible that the magnitude of ARC gene expression in the S1BF could depend on the amount of stimulation, whereby less whisker stimulation is achieved during voluntary stimulation in the novel environment. Third, top-down processing involved in voluntary actions could restrict the magnitude of bottom-up sensory signalling [36]. These predictions are supported by two studies. In Filipkowski et al.’s study, two methods for whisker stimulation protocols employed over the same amount of time, produced different-fold changes of c-Fos activation in layer IV, indicating that the method of stimulation may determine the amount of c-Fos activation measured [37]. Another study glued Ni/Fe wire to whiskers and stimulated the whiskers to differing magnitudes using a pulsating magnetic field. They report that the magnitude of c-Fos and zif268 gene expression in the S1BF increased as the intensity of pulsating magnetic field was increased [38]. Regardless, more research is needed to define whether there is stimulus-dependent expression and/or an eventual plateau for ARC gene expression within a given anatomical region.
In our experiments, there was no significant difference between manual whisker stimulation and exploration of a novel environment for ARC gene expression in the hippocampus. Both time courses showed significant, but similar increases in ARC gene expression. Thus higher expression of the ARC gene expression in the hippocampus is primarily the result of spatial exploration of a novel environment and not necessarily the effect of whisker stimulation, confirming the specifity of our stimulation to the whisker barrel circuit. In this situation, top-down processing may restrict the level of circuit activation.
Conclusions
These data are the first to demonstrate that ARC is a specific, quantifiable and an input dependent molecular marker of circuit activation. The time course of ARC gene expression after whisker stimulation in the whisker barrel circuit is repeatable, with the peak of ARC gene expression in the S1BF occurring at 30 minutes. We also showed that different methods of stimulation (manual whisker stimulation versus exploration of novel environment) can influence the magnitude of ARC gene expression in the S1BF, but not in the hippocampus, supporting that ARC gene expression was both input- and whisker circuit-specific. In future studies, ARC, as a molecular marker of circuit activation, could be useful for monitoring the integrity of the thalamocortical circuit under the influence of neurodegeneration, development, disruption (metastasis, brain injury), cortical integration of sensory information, adaptation to an environment, pharmacological intervention or repair.
Supplementary Material
(A) Following manual whisker stimulation, ARC gene expression was examined in S1BF tissue at eight different time points: 15, 30, 60, 90, 120, 180, 240 and 300 minutes. There was a significant increase in gene expression at 15, 30, 60 and 300 minutes after manual whisker stimulation in comparison to naïve animals (F (8, 31) = 10.57, p<0.0001). (B) EgR3 gene expression was examined in S1BF tissue at six different time points: 15, 30, 60, 90, 120 and 300 minutes after manual whisker stimulation. There were no significant differences in EgR3 gene expression in comparison to naïve animals at any time points in the S1BF (F (6, 19)=1.799, p= 0.1530). (C) Zif268 gene expression was examined in S1BF tissue in the same samples used for ARC (A). There were no significant differences in Zif268 gene expression in comparison to naïve animals at any of eight time points in the S1BF (F (8, 33) =1.128, p= 0.3710). (D) BDNF gene expression was examined in S1BF tissue. There was a significant increase at 240 minutes post-stimulation in comparison to naïve animals (F (8,35)=2.322, p= 0.04). The data are presented as the mean ± SEM, where,* p<0.05 in comparison to naïve animals; n=3–6 rats per group.
EgR3, Zif268 and BDNF demonstrate a higher variance from the mean (as demonstrated by the error bars) compared to ARC. For BDNF, there is a significant increase at 240 min in comparison to naïve. However, ARC shows a significant differences at both early (15–30 minutes) and late (300 minutes) time points after whisker stimulation. The lower variance of ARC gene expression between animals in the same group and the early increase in ARC gene expression makes it a more ideal candidate for rapid assessment of circuit-directed activity after whisker stimulation.
Highlights.
These experiments investigated ARC as a molecular marker of circuit activation
Whisker stimulation revealed a primary (30 minute) and secondary (300 minute) peak of ARC gene expression in the S1BF
The time course of ARC gene expression is repeatable between paradigms and the magnitude of ARC mRNA transcription is input dependent
Manual whisker stimulation-induced ARC gene expression is specific to the S1BF, not the hippocampus.
Acknowledgments
The authors wish to thank Maria Cristina Morganti-Kossmann, PhD and Taylor Colburn for valuable feedback on the manuscript, as well as Jenna Ziebell, PhD and Daniel Griffiths for their technical contribution with western blots. Research reported in this manuscript was supported, in part, by National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award numbers NIH R03 NS-077098 and NIH R01 NS-065052, and Phoenix Children’s Hospital Mission Support Funds.
References
- 1.Avey MT, Phillmore LS, MacDougall-Shackleton SA. Immediate early gene expression following exposure to acoustic and visual components of courtship in zebra finches. Behavioural brain research. 2005;165:247–53. doi: 10.1016/j.bbr.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Bailey DJ, Wade J. FOS and ZENK responses in 45-day-old zebra finches vary with auditory stimulus and brain region, but not sex. Behavioural brain research. 2005;162:108–15. doi: 10.1016/j.bbr.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 3.Rosen JB, Adamec RE, Thompson BL. Expression of egr-1 (zif268) mRNA in select fear-related brain regions following exposure to a predator. Behavioural brain research. 2005;162:279–88. doi: 10.1016/j.bbr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang JY, et al. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nature neuroscience. 2011;14:848–56. doi: 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Semba K, Pastorius J, Wilkinson M, Rusak B. Sleep deprivation-induced c-fos and junB expression in the rat brain: effects of duration and timing. Behavioural brain research. 2001;120:75–86. doi: 10.1016/s0166-4328(00)00362-4. [DOI] [PubMed] [Google Scholar]
- 6.Vazdarjanova A, Ramirez-Amaya V, Insel N, Plummer TK, Rosi S, Chowdhury S, et al. Spatial exploration induces ARC, a plasticity-related immediate-early gene, only in calcium/calmodulin-dependent protein kinase II-positive principal excitatory and inhibitory neurons of the rat forebrain. The Journal of comparative neurology. 2006;498:317–29. doi: 10.1002/cne.21003. [DOI] [PubMed] [Google Scholar]
- 7.Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, et al. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–45. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 8.Korb E, Finkbeiner S. Arc in synaptic plasticity: from gene to behavior. Trends in neurosciences. 2011;34:591–8. doi: 10.1016/j.tins.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzingounis AV, Nicoll RA. Arc/Arg3.1: linking gene expression to synaptic plasticity and memory. Neuron. 2006;52:403–7. doi: 10.1016/j.neuron.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 10.Ivanova TN, Matthews A, Gross C, Mappus RC, Gollnick C, Swanson A, et al. Arc/Arg3.1 mRNA expression reveals a subcellular trace of prior sound exposure in adult primary auditory cortex. Neuroscience. 2011;181:117–26. doi: 10.1016/j.neuroscience.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzowski JF, Setlow B, Wagner EK, McGaugh JL. Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, and c-fos, and zif268. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:5089–98. doi: 10.1523/JNEUROSCI.21-14-05089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramirez-Amaya V, Vazdarjanova A, Mikhael D, Rosi S, Worley PF, Barnes CA. Spatial exploration-induced Arc mRNA and protein expression: evidence for selective, network-specific reactivation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:1761–8. doi: 10.1523/JNEUROSCI.4342-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brecht M, Preilowski B, Merzenich MM. Functional architecture of the mystacial vibrissae. Behavioural brain research. 1997;84:81–97. doi: 10.1016/s0166-4328(97)83328-1. [DOI] [PubMed] [Google Scholar]
- 14.Carvell GE, Simons DJ. Biometric analyses of vibrissal tactile discrimination in the rat. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1990;10:2638–48. doi: 10.1523/JNEUROSCI.10-08-02638.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutson KA, Masterton RB. The sensory contribution of a single vibrissa's cortical barrel. Journal of neurophysiology. 1986;56:1196–223. doi: 10.1152/jn.1986.56.4.1196. [DOI] [PubMed] [Google Scholar]
- 16.Woolsey TA, Rovainen CM, Cox SB, Henegar MH, Liang GE, Liu D, et al. Neuronal units linked to microvascular modules in cerebral cortex: response elements for imaging the brain. Cerebral cortex. 1996;6:647–60. doi: 10.1093/cercor/6.5.647. [DOI] [PubMed] [Google Scholar]
- 17.McNamara KC, Lisembee AM, Lifshitz J. The whisker nuisance task identifies a late-onset, persistent sensory sensitivity in diffuse brain-injured rats. Journal of neurotrauma. 2010;27:695–706. doi: 10.1089/neu.2009.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Cunha C, Angelucci A, D'Antoni A, Dobrossy MD, Dunnett SB, Berardi N, et al. Brain-derived neurotrophic factor (BDNF) overexpression in the forebrain results in learning and memory impairments. Neurobiology of disease. 2009;33:358–68. doi: 10.1016/j.nbd.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Gine E, Echeverry-Alzate V, Lopez-Moreno JA, Lopez-Jimenez A, Torres-Romero D, Perez-Castillo A, et al. Developmentally-induced hypothyroidism alters the expression of Egr-1 and Arc genes and the sensitivity to cannabinoid agonists in the hippocampus. Possible implications for memory and learning. Molecular and cellular endocrinology. 2013;365:119–28. doi: 10.1016/j.mce.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Giuliani D, Ottani A, Minutoli L, Stefano VD, Galantucci M, Bitto A, et al. Functional recovery after delayed treatment of ischemic stroke with melanocortins is associated with overexpression of the activity-dependent gene Zif268. Brain, behavior, and immunity. 2009;23:844–50. doi: 10.1016/j.bbi.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Yun SH, Keblesh J, Trommer BL, Xiong H, Radulovic J, et al. Egr3, a synaptic activity regulated transcription factor that is essential for learning and memory. Molecular and cellular neurosciences. 2007;35:76–88. doi: 10.1016/j.mcn.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messaoudi E, Kanhema T, Soule J, Tiron A, Dagyte G, da Silva B, et al. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:10445–55. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaczmarek L, Chaudhuri A. Sensory regulation of immediate-early gene expression in mammalian visual cortex: implications for functional mapping and neural plasticity. Brain research Brain research reviews. 1997;23:237–56. doi: 10.1016/s0165-0173(97)00005-2. [DOI] [PubMed] [Google Scholar]
- 25.Montero VM. c-fos induction in sensory pathways of rats exploring a novel complex environment: shifts of active thalamic reticular sectors by predominant sensory cues. Neuroscience. 1997;76:1069–81. doi: 10.1016/s0306-4522(96)00417-4. [DOI] [PubMed] [Google Scholar]
- 26.Ons S, Marti O, Armario A. Stress-induced activation of the immediate early gene Arc (activity-regulated cytoskeleton-associated protein) is restricted to telencephalic areas in the rat brain: relationship to c-fos mRNA. Journal of neurochemistry. 2004;89:1111–8. doi: 10.1111/j.1471-4159.2004.02396.x. [DOI] [PubMed] [Google Scholar]
- 27.Bisler S, Schleicher A, Gass P, Stehle JH, Zilles K, Staiger JF. Expression of c-Fos, ICER, Krox-24 and JunB in the whisker-to-barrel pathway of rats: time course of induction upon whisker stimulation by tactile exploration of an enriched environment. Journal of chemical neuroanatomy. 2002;23:187–98. doi: 10.1016/s0891-0618(01)00155-7. [DOI] [PubMed] [Google Scholar]
- 28.Bullitt E. Expression of c-fos-like protein as a marker for neuronal activity following noxious stimulation in the rat. The Journal of comparative neurology. 1990;296:517–30. doi: 10.1002/cne.902960402. [DOI] [PubMed] [Google Scholar]
- 29.Chu YF, Yen CT, Lee LJ. Neonatal whisker clipping alters behavior, neuronal structure and neural activity in adult rats. Behavioural brain research. 2013;238:124–33. doi: 10.1016/j.bbr.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 30.Meyer HS, Wimmer VC, Hemberger M, Bruno RM, de Kock CP, Frick A, et al. Cell type-specific thalamic innervation in a column of rat vibrissal cortex. Cerebral cortex. 2010;20:2287–303. doi: 10.1093/cercor/bhq069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Killackey HP, Sherman SM. Corticothalamic projections from the rat primary somatosensory cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:7381–4. doi: 10.1523/JNEUROSCI.23-19-07381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallett H, Churchill L, Taishi P, De A, Krueger JM. Whisker stimulation increases expression of nerve growth factor- and interleukin-1beta-immunoreactivity in the rat somatosensory cortex. Brain research. 2010;1333:48–56. doi: 10.1016/j.brainres.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–44. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 34.Welker E, Rao SB, Dorfl J, Melzer P, van der Loos H. Plasticity in the barrel cortex of the adult mouse: effects of chronic stimulation upon deoxyglucose uptake in the behaving animal. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1992;12:153–70. doi: 10.1523/JNEUROSCI.12-01-00153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anokhin KV, Mileusnic R, Shamakina IY, Rose SP. Effects of early experience on c-fos gene expression in the chick forebrain. Brain research. 1991;544:101–7. doi: 10.1016/0006-8993(91)90890-8. [DOI] [PubMed] [Google Scholar]
- 36.Zembrzycki A, Chou SJ, Ashery-Padan R, Stoykova A, O'Leary DD. Sensory cortex limits cortical maps and drives top-down plasticity in thalamocortical circuits. Nature neuroscience. 2013;16:1060–7. doi: 10.1038/nn.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filipkowski RK, Rydz M, Berdel B, Morys J, Kaczmarek L. Tactile experience induces c-fos expression in rat barrel cortex. Learning & memory. 2000;7:116–22. doi: 10.1101/lm.7.2.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melzer P, Steiner H. Stimulus-dependent expression of immediate-early genes in rat somatosensory cortex. The Journal of comparative neurology. 1997;380:145–53. doi: 10.1002/(sici)1096-9861(19970331)380:1<145::aid-cne11>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Following manual whisker stimulation, ARC gene expression was examined in S1BF tissue at eight different time points: 15, 30, 60, 90, 120, 180, 240 and 300 minutes. There was a significant increase in gene expression at 15, 30, 60 and 300 minutes after manual whisker stimulation in comparison to naïve animals (F (8, 31) = 10.57, p<0.0001). (B) EgR3 gene expression was examined in S1BF tissue at six different time points: 15, 30, 60, 90, 120 and 300 minutes after manual whisker stimulation. There were no significant differences in EgR3 gene expression in comparison to naïve animals at any time points in the S1BF (F (6, 19)=1.799, p= 0.1530). (C) Zif268 gene expression was examined in S1BF tissue in the same samples used for ARC (A). There were no significant differences in Zif268 gene expression in comparison to naïve animals at any of eight time points in the S1BF (F (8, 33) =1.128, p= 0.3710). (D) BDNF gene expression was examined in S1BF tissue. There was a significant increase at 240 minutes post-stimulation in comparison to naïve animals (F (8,35)=2.322, p= 0.04). The data are presented as the mean ± SEM, where,* p<0.05 in comparison to naïve animals; n=3–6 rats per group.
EgR3, Zif268 and BDNF demonstrate a higher variance from the mean (as demonstrated by the error bars) compared to ARC. For BDNF, there is a significant increase at 240 min in comparison to naïve. However, ARC shows a significant differences at both early (15–30 minutes) and late (300 minutes) time points after whisker stimulation. The lower variance of ARC gene expression between animals in the same group and the early increase in ARC gene expression makes it a more ideal candidate for rapid assessment of circuit-directed activity after whisker stimulation.