Abstract
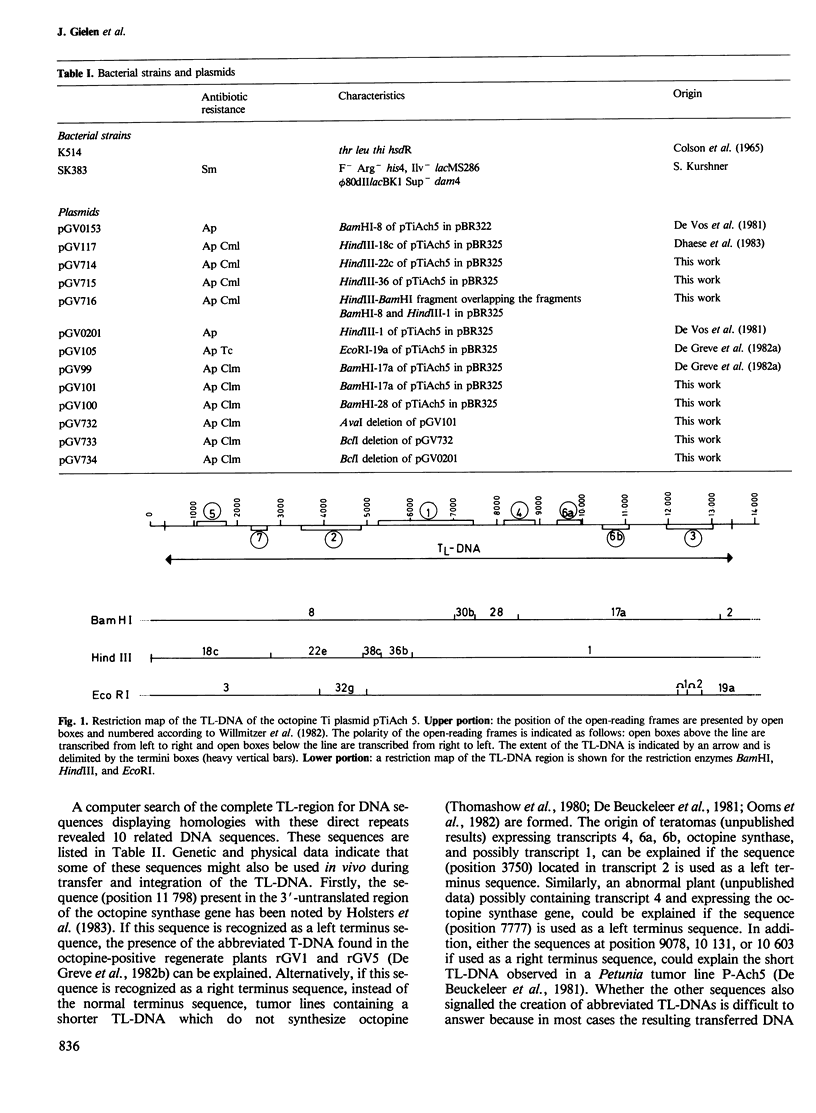
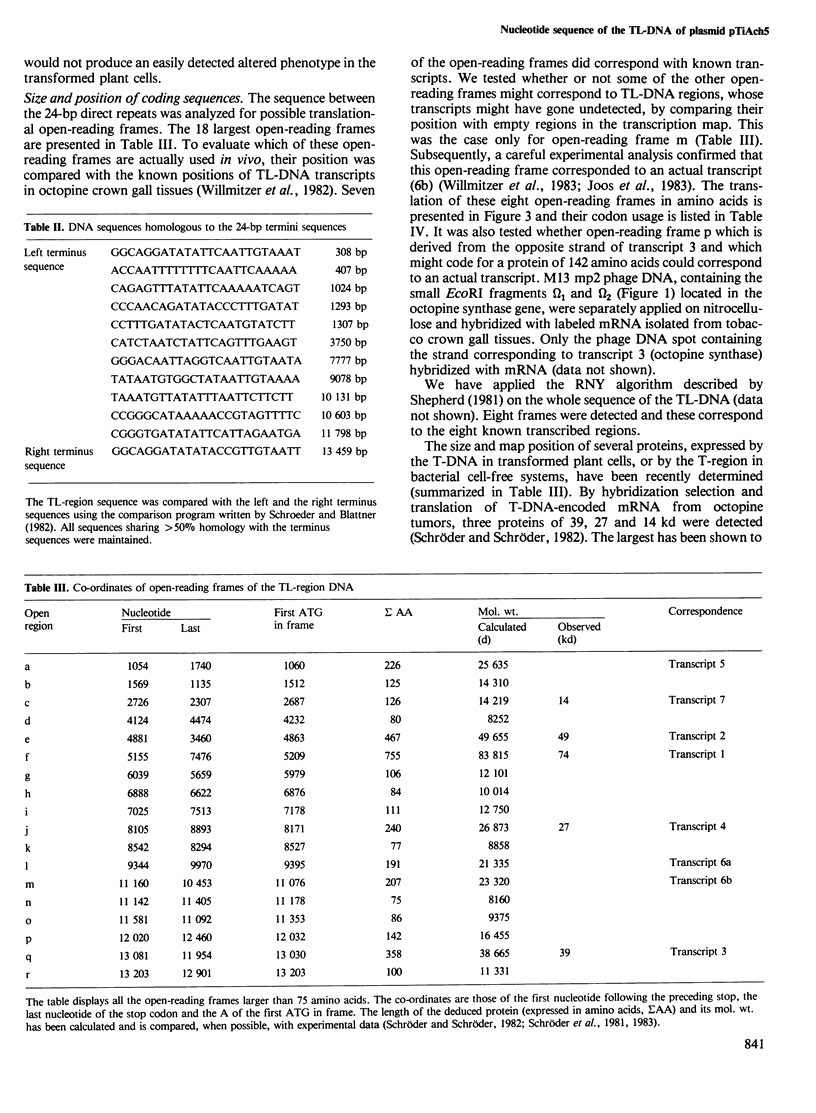
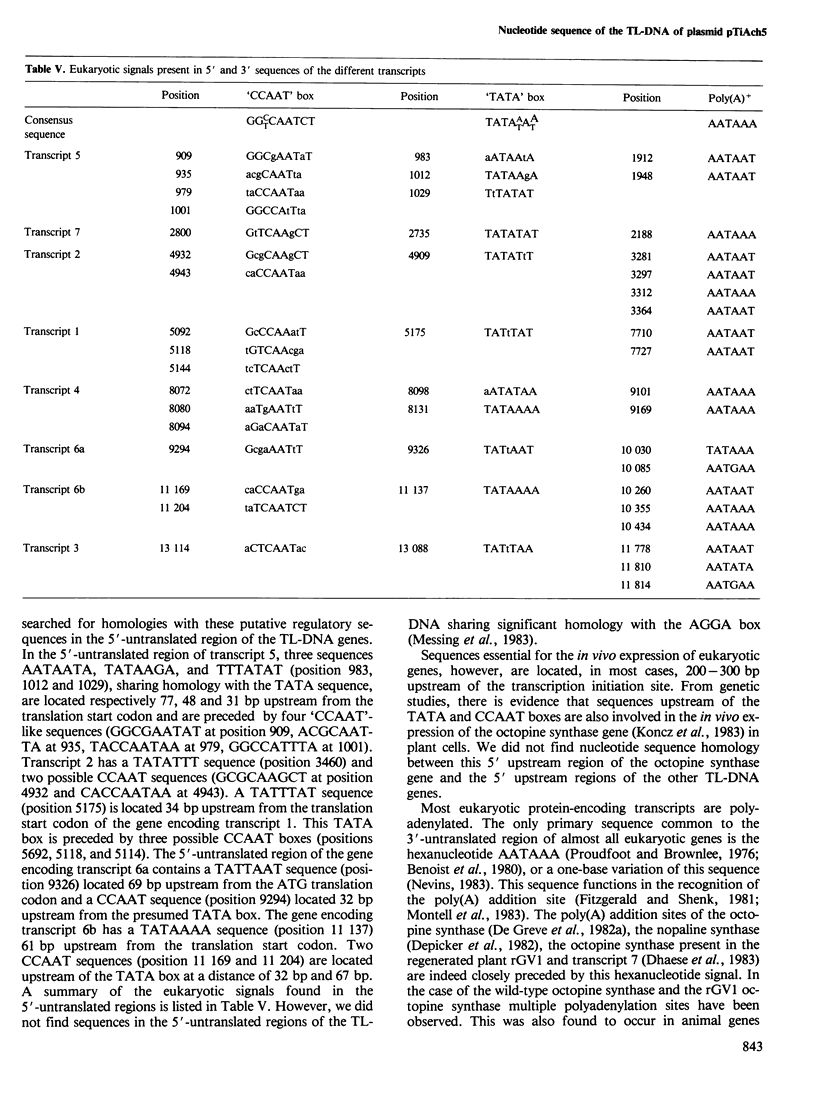
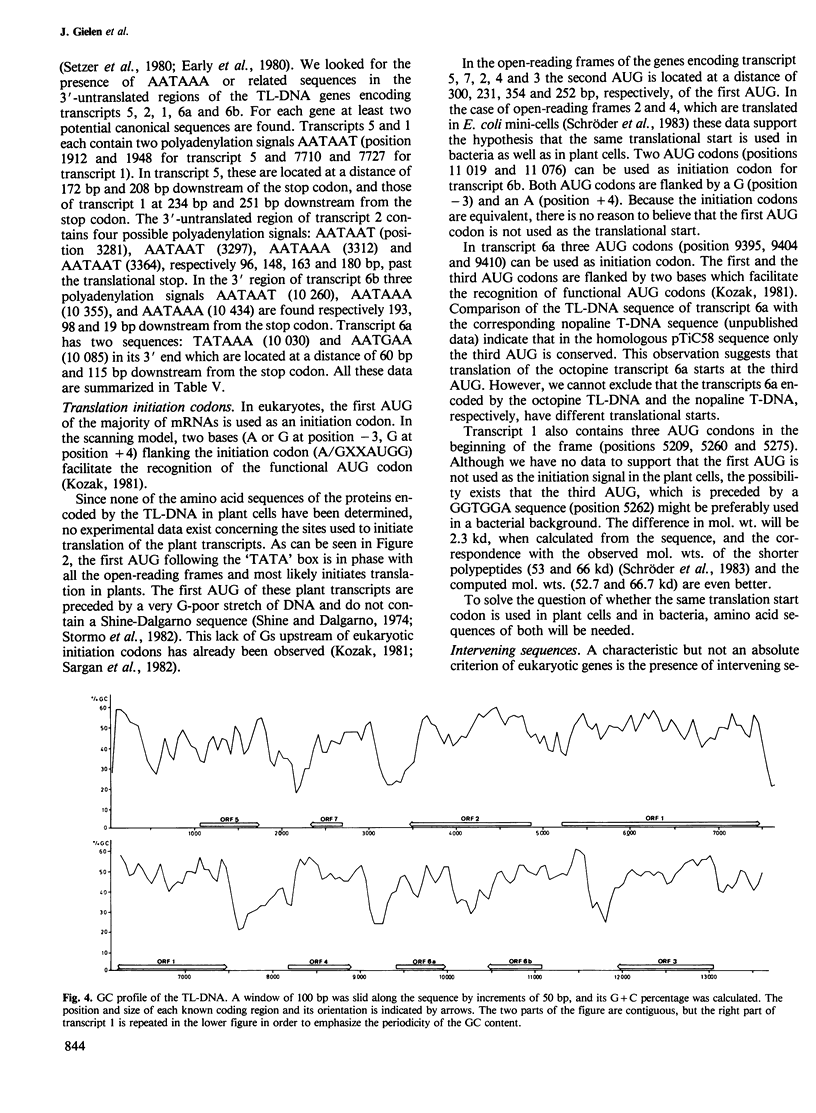
We have determined the complete primary structure (13 637 bp) of the TL-region of Agrobacterium tumefaciens octopine plasmid pTiAch5 . This sequence comprises two small direct repeats which flank the TL-region at each extremity and are involved in the transfer and/or integration of this DNA segment in plants. TL-DNA specifies eight open-reading frames corresponding to experimentally identified transcripts in crown gall tumor tissue. The eight coding regions are not interrupted by intervening sequences and are separated from each other by AT-rich regions. Potential transcriptional control signals upstream of the 5' and 3' ends of all the transcribed regions resemble typical eukaryotic signals: (i) transcriptional initiation signals ('TATA' or Goldberg- Hogness box) are present upstream to the presumed translational start codons; (ii) ' CCAAT ' sequences are present upstream of the proposed 'TATA' box; (iii) polyadenylation signals are present in the 3'-untranslated regions. Furthermore, no Shine-Dalgarno sequences are present upstream of the presumed translational start codons.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betlach M., Hershfield V., Chow L., Brown W., Goodman H., Boyer H. W. A restriction endonuclease analysis of the bacterial plasmid controlling the ecoRI restriction and modification of DNA. Fed Proc. 1976 Jul;35(9):2037–2043. [PubMed] [Google Scholar]
- Bevan M. W., Chilton M. D. Multiple transcripts of T-DNA detected in nopaline crown gall tumors. J Mol Appl Genet. 1982;1(6):539–546. [PubMed] [Google Scholar]
- Caplan A., Herrera-Estrella L., Inzé D., Van Haute E., Van Montagu M., Schell J., Zambryski P. Introduction of genetic material into plant cells. Science. 1983 Nov 18;222(4625):815–821. doi: 10.1126/science.222.4625.815. [DOI] [PubMed] [Google Scholar]
- Chilton M. D., Drummond M. H., Merio D. J., Sciaky D., Montoya A. L., Gordon M. P., Nester E. W. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell. 1977 Jun;11(2):263–271. doi: 10.1016/0092-8674(77)90043-5. [DOI] [PubMed] [Google Scholar]
- Colson C., Glover S. W., Symonds N., Stacey K. A. The location of the genes for host-controlled modification and restriction in Escherichia coli K-12. Genetics. 1965 Nov;52(5):1043–1050. doi: 10.1093/genetics/52.5.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beuckeleer M., Lemmers M., De Vos G., Willmitzer L., Van Montagu M., Schell J. Further insight on the transferred-DNA of octopine crown gall. Mol Gen Genet. 1981;183(2):283–288. doi: 10.1007/BF00270630. [DOI] [PubMed] [Google Scholar]
- De Greve H., Dhaese P., Seurinck J., Lemmers M., Van Montagu M., Schell J. Nucleotide sequence and transcript map of the Agrobacterium tumefaciens Ti plasmid-encoded octopine synthase gene. J Mol Appl Genet. 1982;1(6):499–511. [PubMed] [Google Scholar]
- De Vos G., De Beuckeleer M., Van Montagu M., Schell J. Restriction endonuclease mapping of the octopine tumor-inducing plasmid pTiAch5 of Agrobacterium tumefaciens. Plasmid. 1981 Sep;6(2):249–253. doi: 10.1016/0147-619x(81)90070-6. [DOI] [PubMed] [Google Scholar]
- Depicker A., De Wilde M., De Vos G., De Vos R., Van Montagu M., Schell J. Molecular cloning of overlapping segments of the nopaline Ti-plasmid pTiC58 as a means to restriction endonuclease mapping. Plasmid. 1980 Mar;3(2):193–211. doi: 10.1016/0147-619x(80)90109-2. [DOI] [PubMed] [Google Scholar]
- Depicker A., Stachel S., Dhaese P., Zambryski P., Goodman H. M. Nopaline synthase: transcript mapping and DNA sequence. J Mol Appl Genet. 1982;1(6):561–573. [PubMed] [Google Scholar]
- Dhaese P., De Greve H., Gielen J., Seurinck L., Van Montagu M., Schell J. Identification of sequences involved in the polyadenylation of higher plant nuclear transcripts using Agrobacterium T-DNA genes as models. EMBO J. 1983;2(3):419–426. doi: 10.1002/j.1460-2075.1983.tb01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Engler G., Depicker A., Maenhaut R., Villarroel R., Van Montagu M., Schell J. Physical mapping of DNA base sequence homologies between an octopine and a nopaline Ti plasmid of Agrobacterium tumefaciens. J Mol Biol. 1981 Oct 25;152(2):183–208. doi: 10.1016/0022-2836(81)90239-4. [DOI] [PubMed] [Google Scholar]
- Fischer R. L., Goldberg R. B. Structure and flanking regions of soybean seed protein genes. Cell. 1982 Jun;29(2):651–660. doi: 10.1016/0092-8674(82)90181-7. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Garfinkel D. J., Simpson R. B., Ream L. W., White F. F., Gordon M. P., Nester E. W. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981 Nov;27(1 Pt 2):143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- Geraghty D., Peifer M. A., Rubenstein I., Messing J. The primary structure of a plant storage protein: zein. Nucleic Acids Res. 1981 Oct 10;9(19):5163–5174. doi: 10.1093/nar/9.19.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidekamp F., Dirkse W. G., Hille J., van Ormondt H. Nucleotide sequence of the Agrobacterium tumefaciens octopine Ti plasmid-encoded tmr gene. Nucleic Acids Res. 1983 Sep 24;11(18):6211–6223. doi: 10.1093/nar/11.18.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyldig-Nielsen J. J., Jensen E. O., Paludan K., Wiborg O., Garrett R., Jørgensen P., Marcker K. A. The primary structures of two leghemoglobin genes from soybean. Nucleic Acids Res. 1982 Jan 22;10(2):689–701. doi: 10.1093/nar/10.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joos H., Inzé D., Caplan A., Sormann M., Van Montagu M., Schell J. Genetic analysis of T-DNA transcripts in nopaline crown galls. Cell. 1983 Apr;32(4):1057–1067. doi: 10.1016/0092-8674(83)90290-8. [DOI] [PubMed] [Google Scholar]
- Koncz C., De Greve H., André D., Deboeck F., Van Montagu M., Schell J. The opine synthase genes carried by Ti plasmids contain all signals necessary for expression in plants. EMBO J. 1983;2(9):1597–1603. doi: 10.1002/j.1460-2075.1983.tb01630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans J., Deblaere R., Willmitzer L., De Greve H., Hernalsteens J. P., Van Montagu M., Schell J. Genetic Identification of functions of TL-DNA transcripts in octopine crown galls. EMBO J. 1982;1(1):147–152. doi: 10.1002/j.1460-2075.1982.tb01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers M., De Beuckeleer M., Holsters M., Zambryski P., Depicker A., Hernalsteens J. P., Van Montagu M., Schell J. Internal organization, boundaries and integration of Ti-plasmid DNA in nopaline grown gall tumours. J Mol Biol. 1980 Dec 15;144(3):353–376. doi: 10.1016/0022-2836(80)90095-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Nester E. W., Kosuge T. Plasmids specifying plant hyperplasias. Annu Rev Microbiol. 1981;35:531–565. doi: 10.1146/annurev.mi.35.100181.002531. [DOI] [PubMed] [Google Scholar]
- Nevins J. R. The pathway of eukaryotic mRNA formation. Annu Rev Biochem. 1983;52:441–466. doi: 10.1146/annurev.bi.52.070183.002301. [DOI] [PubMed] [Google Scholar]
- Ohmori H., Tomizawa J. I., Maxam A. M. Detection of 5-methylcytosine in DNA sequences. Nucleic Acids Res. 1978 May;5(5):1479–1485. doi: 10.1093/nar/5.5.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms G., Bakker A., Molendijk L., Wullems G. J., Gordon M. P., Nester E. W., Schilperoort R. A. T-DNA organization in homogeneous and heterogeneous octopine-type crown gall tissues of Nicotiana tabacum. Cell. 1982 Sep;30(2):589–597. doi: 10.1016/0092-8674(82)90255-0. [DOI] [PubMed] [Google Scholar]
- Pedersen K., Devereux J., Wilson D. R., Sheldon E., Larkins B. A. Cloning and sequence analysis reveal structural variation among related zein genes in maize. Cell. 1982 Jul;29(3):1015–1026. doi: 10.1016/0092-8674(82)90465-2. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. Eukaryotic promoters? Nature. 1979 May 31;279(5712):376–376. doi: 10.1038/279376a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sargan D. R., Gregory S. P., Butterworth P. H. A possible novel interaction between the 3'-end of 18 S ribosomal RNA and the 5'-leader sequence of many eukaryotic messenger RNAs. FEBS Lett. 1982 Oct 18;147(2):133–136. doi: 10.1016/0014-5793(82)81026-0. [DOI] [PubMed] [Google Scholar]
- Schroeder J. L., Blattner F. R. Formal description of a DNA oriented computer language. Nucleic Acids Res. 1982 Jan 11;10(1):69–84. doi: 10.1093/nar/10.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder G., Klipp W., Hillebrand A., Ehring R., Koncz C., Schröder J. The conserved part of the T-region in Ti-plasmids expresses four proteins in bacteria. EMBO J. 1983;2(3):403–409. doi: 10.1002/j.1460-2075.1983.tb01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder J., Hillebrand A., Klipp W., Pühler A. Expression of plant tumor-specific proteins in minicells of Escherichia coli: a fusion protein of lysopine dehydrogenase with chloramphenicol acetyltransferase. Nucleic Acids Res. 1981 Oct 24;9(20):5187–5202. doi: 10.1093/nar/9.20.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers P. H. Pattern recognition in genetic sequences. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3041–3041. doi: 10.1073/pnas.76.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Nunberg J. H., Schimke R. T. Size heterogeneity in the 3' end of dihydrofolate reductase messenger RNAs in mouse cells. Cell. 1980 Nov;22(2 Pt 2):361–370. doi: 10.1016/0092-8674(80)90346-3. [DOI] [PubMed] [Google Scholar]
- Shah D. M., Hightower R. C., Meagher R. B. Complete nucleotide sequence of a soybean actin gene. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1022–1026. doi: 10.1073/pnas.79.4.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J. C. Method to determine the reading frame of a protein from the purine/pyrimidine genome sequence and its possible evolutionary justification. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1596–1600. doi: 10.1073/pnas.78.3.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. B., O'Hara P. J., Kwok W., Montoya A. L., Lichtenstein C., Gordon M. P., Nester E. W. DNA from the A6S/2 crown gall tumor contains scrambled Ti-plasmid sequences near its junctions with plant DNA. Cell. 1982 Jul;29(3):1005–1014. doi: 10.1016/0092-8674(82)90464-0. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M. F., Nutter R., Montoya A. L., Gordon M. P., Nester E. W. Integration and organization of Ti plasmid sequences in crown gall tumors. Cell. 1980 Mar;19(3):729–739. doi: 10.1016/s0092-8674(80)80049-3. [DOI] [PubMed] [Google Scholar]
- Wieslander L. A simple method to recover intact high molecular weight RNA and DNA after electrophoretic separation in low gelling temperature agarose gels. Anal Biochem. 1979 Oct 1;98(2):305–309. doi: 10.1016/0003-2697(79)90145-3. [DOI] [PubMed] [Google Scholar]
- Willmitzer L., Dhaese P., Schreier P. H., Schmalenbach W., Van Montagu M., Schell J. Size, location and polarity of T-DNA-encoded transcripts in nopaline crown gall tumors; common transcripts in octopine and nopaline tumors. Cell. 1983 Apr;32(4):1045–1056. doi: 10.1016/0092-8674(83)90289-1. [DOI] [PubMed] [Google Scholar]
- Willmitzer L., Schmalenbach W., Schell J. Transcription of T-DNA in octopine and nopaline crown gall tumours is inhibited by low concentrations of alpha-amanitin. Nucleic Acids Res. 1981 Oct 10;9(19):4801–4812. doi: 10.1093/nar/9.19.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmitzer L., Simons G., Schell J. The TL-DNA in octopine crown-gall tumours codes for seven well-defined polyadenylated transcripts. EMBO J. 1982;1(1):139–146. doi: 10.1002/j.1460-2075.1982.tb01137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav N. S., Vanderleyden J., Bennett D. R., Barnes W. M., Chilton M. D. Short direct repeats flank the T-DNA on a nopaline Ti plasmid. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6322–6326. doi: 10.1073/pnas.79.20.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski P., Depicker A., Kruger K., Goodman H. M. Tumor induction by Agrobacterium tumefaciens: analysis of the boundaries of T-DNA. J Mol Appl Genet. 1982;1(4):361–370. [PubMed] [Google Scholar]



