Abstract
Objectives
A mutation in the JAK2 gene, V617F, has been identified in several BCR-ABL1 negative myeloproliferative neoplasms (MPN): polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF). Defining the presence or absence of this mutation is an essential part of clinical diagnostic algorithms and patient management. Here, we aimed to evaluate the performance of three PCR-based assays: Amplification Refractory Mutation System (ARMS), High-Resolution Melting analysis (HRM), and Sanger direct sequencing, and compare their results with those obtained by a PCR restriction fragment polymorphism assay (PCR-RFLP).
Design and methods
We used blood samples from 136 patients (PV=20; PMF=20; ET=28, and other MPN suspected cases=68).
Results
Comparable results were observed among the four assays in patients with PV, PMF, and MPN suspected cases. In patients with a diagnosis of ET, the JAK2 V617F mutation was detected in 67.8% of them by the PCR-ARMS and PCR-HRM assay and in 64% of them by the conventional Sanger sequence approach. The PCR-ARMS and PCR-HRM assays were 100% concordant. With these tests, only one of the 20 patients with ET and one of the three patients with clinically suspected MPN gave different results compared with those obtained by the PCR-RFLP.
Conclusions
Our results have demonstrated that the PCR-ARMS and PCR-HRM assays could detect the JAK2 V617F mutation effectively in MPN patients, but PCR-HRM assays are rapid and the most cost-effective procedures.
Keywords: Myeloproliferative, JAK2 V617F, Mutation, Wild type, Screening
1. Introduction
Myeloproliferative neoplasms (MPNs) are BCR-ABL1 negative clonal diseases of hematopoietic stem cells in which there is an increased proliferation of myeloid series with efficient maturation leading to peripheral blood leukocytosis, increased erythrocyte mass, or thrombocytosis. The diseases include chronic myelogenous leukemia (CML), primary myelofibrosis (PMF), polycythemia vera (PV), essential thrombocythemia (ET), hyper eosinophilia, and mast cell disorders. The majority of patients develop the disease slowly, and some MPNs can progress to leukemia. Besides the patient's general clinical symptoms, the treatment depends upon the MPN subtype; therefore, determining the right MPN subtype is important. The previous diagnostic algorithm of MPN was mainly based on their clinical and laboratory measures [1].
This algorithm was critically revised by the World Health Organization (WHO) in 2008 to include molecular and genetic diagnostic criteria such as Janus Kinase 2 (JAK2) c.1849G>T; p.Val617Phe (hereafter referred to as JAK2 V617F); and other histological information [2], [3]. Of note, the mutation of JAK2 is not specific for any single MPN but provides clonal proliferation evidence and thus excludes further consideration of a reactive cause. Available data indicate that 90% of patients with PV and 50–60% of cases of those with PMF or ET have detectable JAK2 V617F mutation [4]. A small proportion of PV cases alternatively carry JAK2 exon 12 mutations [5].
JAK2 is a cytoplasmic tyrosine kinase encoded by a gene located on the short arm of chromosome 9 (9p24.1) and functions as a crucial mediator of signaling for hematopoietic cytokines and hormones, several interleukins, and growth hormones [6]. The V617F mutation is a G>T transversion at nucleotide 1849 of exon 14, leading to a substitution of phenylalanine for valine at codon 617 (V617F), at the negative regulatory domain JAK homology 2 (JH2). This point constitutively activates the production of the JAK2 tyrosine kinase owing to the loss of autoinhibitory control [7]. Overexpression of JAK2 V617F together with the erythropoietin receptor in cells, results in hyperactivation of erythropoietin-induced cell signaling. This gain-of-function mutation of JAK2 probably explains the hypersensitivity of PV progenitor cells to not only include growth factors but also cytokines[8]. The incidence of MPNs is estimated to be approximately 0.5–6.5 per 100,000 people[9].
Detection of a JAK2 V617F mutation is a diagnostically relevant molecular marker in MPN disorders [10]. Due to the diagnostic importance of JAK2 V617F mutation, some protocols have been developed using the molecular approach including restriction fragment length polymorphism (RFLP-PCR) [11], amplification refractory mutation system (ARMS) [12], high-resolution melting (HRM) [13], and direct sequencing [14].
The RFLP-PCR exploits variations in homologous DNA sequences and allows rapid detection of JAK2 V617F mutation after the genomic sequences are amplified by PCR. The mutation is distinguished by digestion with a specific restriction enzyme and is identified directly in stained agarose gel. The ARMS technique consists of two complementary PCR reactions. The first reaction contains primers designed to amplify only the wild-type variant while the second reaction contains primers specifically designed to amplify the mutant variant. The HRM assay consists of PCR reaction, followed by a short melting step and subsequent analysis [13], [15], [16]. DNA sequencing is one of the most widely used methods for analysing DNA and has been successfully used to detect any mutation in the sequence being analysed. The DNA sequencing approach is often considered the’‘gold standard’ for DNA sequence analysis; however, this approach is not very sensitive [17]. It has been shown that a mutation must be present in approximately 20% of the sample to be readily detected by direct sequencing [18]. Each of these methods has its own advantages and disadvantages, including sensitivity, ease of performance, and instrumentation constraints.
Results derived from the evaluation of these methods may differ with respect to analytical sensitivity, and specificity [19], [20], [21], [22]. Currently, there is no consensus regarding common analytical aspects that include the type of sample, the clinical relevance of distinguishing heterozygous from homozygous mutations, and the appropriate interpretation and reporting of quantitative results particularly those of low-level mutations [20]. Here, we have evaluated the performance of the PCR-based ARMS, HRM, and Sanger direct sequencing for the detection of JAK2 V617F mutation and compare their results with those obtained by a PCR-RFLP assay.
2. Materials and methods
2.1. Samples
This study was approved by CAPPesq- HCFMUSP under protocol number CAPPesq 9371 for the year 2012. We analyzed blood samples from 136 patients (mean age: 59±17; 50% male) with a clinically suspected diagnosis of MPNs. Patients were classified by the WHO 2008 criteria [3] as one of the following: PV (20 cases), ET (28 cases), or PMF (20 cases). Also, another 68 patients with a clinically suspected diagnosis of MPNs were included. Table 1 demonstrates the main characteristics and the result of the JAK2 V617F mutation analysis by PCR-RFLP in the studied subjects. This methodology was chosen as the gold standard because of its longstanding use.
Table 1.
Clinical and laboratory characteristics of patients included in this study.
| MPNa (n=68) with confirmed diagnosis | MPNa with suspected daignosis (n=68) | |
|---|---|---|
| Age (years) – median (range): | 62.3 (17–87) | 53.6 (15–83) |
| Gender – male/female | 28/40 | 27/41 |
| Hemoglobin (g/dL) – median (range) | 14.0 (7.0–22.0) | 13.5 (6.0–22.0) |
| WBC count (×109 L–1) – median (range) | 9.26 (1.35–66.2) | 8.46 (2.2–58.9) |
| Platelet count (×109 L–1) – median (range) | 527.5 (43.3–1.660) | 179.5 M (14.0–1.110) |
| JAK2 muattion status (%) | 69.1 | 4.4 |
| JAK2 WT | 21 | 65 |
| JAK V617F positive | 47 | 3 |
Myeloproliferative neoplasms.
2.2. DNA extraction and quantification
Genomic DNA was extracted from blood samples using the Blood genomic Prep Mini Spin Kit (GE Healthcare™, UK) following the protocol recommended by the manufacturers. The DNA concentration was determined by spectrophotometry using the Nanodrop ND 1000 (NanoDrop, UNISCIENCE, USA). DNA quality was assessed by the 260:280 and 260:230 spectrophotometric ratios and its integrity by agarose gel electrophoresis. Samples whose 260:280 and 260:230 ratios were <1.8, were excluded. All samples were coded and assayed blindly in duplicate for the JAK2V617F mutation. Code numbers were given to the samples, which were then assayed blindly in duplicate by the investigator. The final result decoding was performed during analysis of data.
2.3. JAK2V617F mutation by restriction fragment length polymorphism (RFLP)
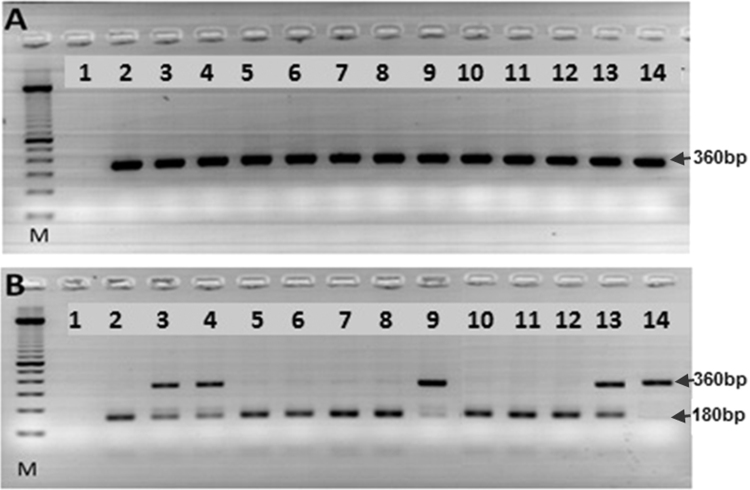
To screen for the presence of the JAK2V617F mutation by restriction digestion, a PCR reaction was prepared to amplify the portion of the JAK2 region that acquires this mutation (360 base pairs) as previously described [23]. The reaction mixture in a final volume of 30 μl was made using 1 U of enzyme Taq polymerase (Invitrogen™, USA) as follows: 100 ng of genomic DNA, 1 μM of primers sense (5′ TGC TGA AAG TAG GAG AAA GTG CAT 3′), 1 μM of antisense primer (5′ TCC TAC AGT GTT TTC AGT TTC AA 3′), 200 μM dNTPs, and 1.5 mM MgCl2. The cycling conditions were as follows: one cycle of 95 °C for two minutes; 35 cycles of 94 °C for 30 s, 56 °C for one minute, 72 °C for one minute, and a final extension at 72 °C for five minutes. All samples were tested in duplicate. The final PCR products were resolved on 3% agarose and visualized with ethidium bromide staining. To ensure no cross-contamination occurred, control experiments with water replacing DNA samples were routinely performed and filter tips were used throughout. The amplified products were then subjected to enzymatic digestion using the restriction enzyme BsaXI (BioLabs™, New England) following the manufacturer's protocol. Following the enzymatic digestion, the 360-bp amplicon remained undigested in mutant JAK2 V617F samples; whereas JAK2 wildtype samples generated a fragment of 180 bp. A human erythroleukemia cell line [HEL], which is known to have more than two mutant alleles per cell [12], was used as a positive control. For negative controls, we used the K562 CML cell line, which does not express JAK2 exon14. Zygosity determination was based on BsaXI restriction digest patterns (Fig. 1).
Fig. 1.
RFLP analysis of JAK2 V617F mutation. (A) PCR amplification of JAK2: Lane M, 100-bp ladder; Lane 1 negative control without template, lanes 3–13, samples from patients, lane 2 normal control (K562 cell line), and lane 14 mutant control (HEL cell line). Arrow indicates the size of the amplified products. (B) BsaXI digestion: Lane M, 100-bp ladder; Lane 1 negative control without template, lanes 2 normal control (K562 cell line), lane 3,4, 9 and 13 samples from patients with a mixed pattern of both mutant and wild-type alleles, and lane 5–7 and 10–12 samples from patients with wildtype allele, and lane 14 is the mutant control (HEL cell line).
2.4. V617F genotyping by amplification refractory mutation system (ARMS)
Patients were genotyped initially by a DNA tetra-primer ARMS assay to specifically amplify the normal and mutant sequences plus a positive control band (DNA fragment spans the site of the JAK2 V617F mutation) in a single reaction as previously reported [24]. Amplifications were performed under standard conditions using AmpliTaq Gold® DNA polymerase (Applied Biosystems, Foster City, CA, USA), annealing temperature 56 °C, and 35 cycles. Amplified products were resolved in an ABI PRISM 3130 and analyzed using the GenScan I D V3.2 (Biosystems, Foster City, CA, EUA).
Results from fragment analysis are interpreted as follows: fragments with 230 bp represent the wild-type allele, and the fragments with 277 bp represent the mutated allele. Patients with an absence of the JAK2 V617F mutation display only a fragment of 230 bp, while individuals with the presence of mutation show a fragment of 277 bp, and patients with mixed populations demonstrate the two fragments. The HEL and K562 CML cell lines were used as positive and negative controls, respectively.
2.5. High-resolution melting (HRM) curve analysis
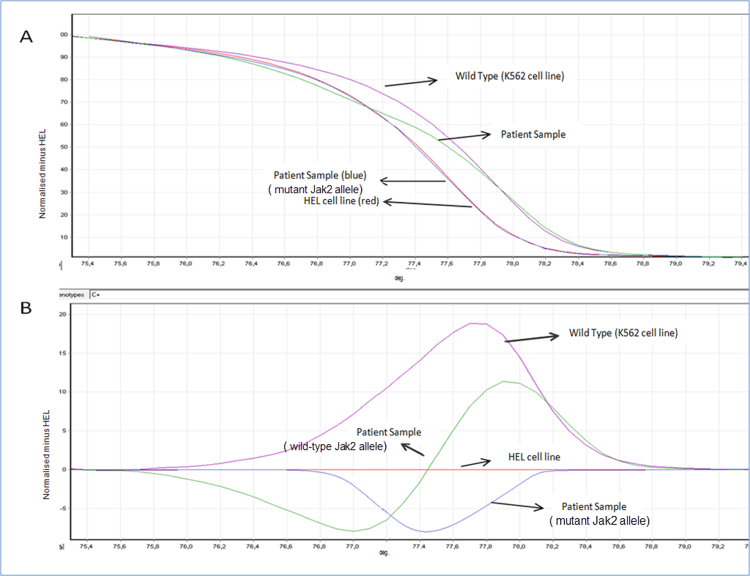
Here, we used the previously published forward primer: 5′ AGC AAG CTT TCT CAC AAG CA 3, reverse primer: 5′ CTG ACA CCT AGC TGT GAT CCT G 3′ (targeting a 155 bp) to detect JAK2 V617F mutation [16]. Wild and mutated JAK2 V617F alleles were amplified with KAPA HRM Fast PCR Kit (Kapa Biosystems). PCR amplification conditions were 50 °C 10 min, 95 °C 30 s, 95 °C 15 s, and 62 °C 1 min, 45 cycles. PCR was performed with ABI 7500 Real-Time PCR system (Applied Biosystems). The HRM PCR reactions were performed and analyzed with the RotorGene real-time analyzer 6000™ (Corbett Life Sciences, Mortlake, Australia). Negative controls and positive controls were used as a reference curve to generate the difference plot. Examples of HRM curves of JAK2 exon 12 mutations are shown in Fig. 2.
Fig. 2.
An example of HRM curves of JAK2 exon 12 mutations. (A) a melting curve profile shows that mutant allele had a melting profile distinct from that of the wildtype allele. (B) Difference plot demonstrates that patient with mutant allele has a characteristic melting curve.
2.6. Direct DNA sequencing
Sanger sequencing was performed on the same amplicons as used for RFLP analysis. The amplified fragments were purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). The purified products were directly sequenced on both strands using the PCR primers and the PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems/Perkin-Elmer, Foster City, CA) in an automated sequencer (ABI 3130, Applied Biosystems). Forward and reverse chromatograms were aligned and compared using the Mutation Surveyor 3.01 software (SoftGenetics, State College, PA).
2.7. Statistical analysis
The percentage of samples correctly identified as positive for the JAK2V617F mutation, the percentage of samples correctly identified as negative, positive predictive, and the negative predictive value for detecting the JAK2V617F mutation were calculated for each of assay. The kappa value (k) was used to determine the level of agreement between each alternative assay and the PCR-RFLP. Negative k indicates that the agreement between tests is less than expected by random chance; a k of 0 indicates that the agreement is the same as chance. K≤0.20=poor agreement, 0.20≤k≤0.40=fair agreement, 0.41≤k≤0.60=moderate agreement, 0.61≤k≤0.80=substantial agreement, k≥0.80=good agreement, and k=1 represents perfect agreement. All statistical analysis was performed using SPSS software v.16.0 (IBM Corp., Armonk, NY, USA).
3. Results
Of the 136 patients evaluated, 20 (14.7%) were diagnosed as PV, 20 (14.7%) as PMF, and 28 (20.6%) as ET according to the 2008 WHO criteria. The remaining 68 (50%) were suspected of having MPNs (Table 1). All samples were subjected to ARMS-PCR methods, HRM-PCR, and direct sequencing, and their results were compared with those obtained by the PCR-RFLP technique. An example of a PCR-RFLP analysis with control is shown in Fig. 1.
Of the 136 samples with a known or suspected diagnosis of an MPN, 50 (36.8%) were positive for the V617F JAK2 mutation by the RFLP assay, and 86 (63.2%) were negative. The proportion of positive patients per disease subtype ranged from 19 (95%) of 20 for PV, eight (40%) of 20 for PMF, 20 (71%) of 28 for ET, and 3 (4%) of 68 patients with a suspected diagnosis of MPNs as shown in Table 2.
Table 2.
Comparison of JAK2 V617F mutation positive samples as assessed by the PCR-based assays.
| Disorders | n | Positive for JAK2 V617F |
|||
|---|---|---|---|---|---|
| PCR-RFLP | PCR-ARMS | PCR-HRM | Direct Sequencing | ||
| Polycythemia Vera (PV) | 20 | 19 (95%) | 19 (95%) | 19 (95%) | 19 (95%) |
| Primary Mielofibroses (PMF) | 20 | 08 (40%) | 08 (40%) | 08 (40%) | 08 (40%) |
| Essential Thrombocythemia [1] | 28 | 20 (71.4%) | 19 (67.8%) | 19 (67.8%) | 18 (64.3%) |
| Clinically suspected MPNa | 68 | 03 (4.4%) | 02 (2.9%) | 02 (2.9%) | 02 (2.9%) |
Myeloproliferative neoplasms.
The PCR-ARMS and PCR-HRM assays had 100% comparable results (Table 2, Table 3). Both assays had detected the JAK2 V617F mutation in 19 (95%) of the patients with of PV, eight (40%) patients with PMF, 19 (67.8%) in patients with ET, and two patients (2.9%) of those suspected of having MPNs (Table 2). These results were in complete concordance with the results obtained by the PCR-RFLP method in patients with PV and PMF (kappa=1.0). Both the PCR-ARMS and PCR-HRM assays demonstrated an overall sensitivity, specificity, and accuracy of 96%, 100%, and 98.5%, respectively (Table 3). When interpreted in combination, the PCR-ARMS agreed in 48 of the 50 JAK2 V617F PCR-RFLP detectable mutations [96.8% concordances (95% CI 95.6 to 97.7%)]). No sample that was PCR-RFLP negative tested positive by PCR-ARMS or PCR-HRM assay.
Table 3.
Statistical evaluation of PCR-ARMS, PCR-HRM, and direct sequencing to PCR-RFLP.
| PCR-ARMS | PCR-HRM | Direct Sequencing | |
|---|---|---|---|
| Positive predictive value (%) | 100 | 100 | 100 |
| Positive predictive value 95%IC | 92.6–100 | 92.6–100 | 92.4–100 |
| Sensitivity (%) | 96 | 96 | 94 |
| Negative predictive value (%) | 97.7 | 97.7 | 96.6 |
| Negative predictive value 95%IC | 92–99.4 | 92 –99.4 | 90.5–98.8 |
| Specificity (%) | 100 | 100 | 100 |
| Accuracy (%) | 98.5 | 98.5 | 97.8 |
| Concordance (%) | 96.8 | 96.8 | 95.2 |
| Concordance 95%IC | 95.6–97.7 | 95.6–97.7 | 93.3–96.5 |
| Bias correction factor Cb (% accuracy) | 99.9 | 99.9 | 99.9 |
| Weighted Kappa | 0.97 | 0.97 | 0.95 |
| Weighted Kappa 95%IC | 92.4–100 | 92.4–100 | 89.8 to 100 |
The direct sequencing assay was able to detect the JAK2 V617F mutation in 19 (95%), eight (40%), 18 (64.3%) and two (2.9%) in samples of PV, PMF, ET, and MPN respectively. There were two samples from patients with ET disorder that were positive by RFLP and negative by the conventional sequencing method. In comparison with RFLP results, there was an agreement between 47 of 50 samples with detectable JAK2 V617F mutation (Table 2). The overall concordance rate between both tests for detecting the JAK2 V617F mutation was 95.2% (95% CI; 93.3–96.5%). Compared to PCR-RFLP, direct sequencing revealed an overall sensitivity, specificity, and accuracy of 94%, 100%, and 97.8%, respectively (Table 3). Agreement between both tests was 95.2% with a kappa value of 0.95 (95%IC, 89.8 to 100).
4. Discussion
The principal objective for undertaking this study was to evaluate the performance of several JAK2 V617F mutation PCR-based assays. This information was intended to be used in the process of selecting a method with good analytical performance that benefits both laboratories and patients. During the last decades, several PCR-based assays with different analytical sensitivity and specificity parameters have been developed [22]. These assays include genomic direct DNA sequencing [25], RT-PCR-sequencing [11], allele-specific polymerase chain reaction [8], PCR-ARMS [24], PCR-RFLP [11], real-time PCR [26], DNA-melting curve analysis [27], allele blocker PCR [28], and AS-quantitative PCR (AS-PCR) [29].
However, these methods have their disadvantages: for example, DNA sequencing has the ability to detect specific DNA-sequence changes but is limited by high cost and low sensitivity[21]. PCR-ARMS, considered one of the most sensitive (0.01%–5%) techniques, requires post-amplification manipulation [15]. The PCR-RFLP is time-consuming and can be complicated by incomplete digestion with the restriction enzyme causing false-positive results [30]. The sensitivity and specificity parameters together with the performance costs are important factors that might impact the utility of these assays and should always be considered when selecting or evaluating these methods. Very few studies have been carried out to evaluate the currently available testing assays for screening the JAK2 point mutation in MPN patients. For instance, Cancovik et al. [19], evaluated the performance of three screening methods (JAK2 Activating Mutation Assay, JAK2 MutaScreen kit, a home-brew melting curve analysis method), and one quantitative method for JAK2 V617F detection in 33 selected samples. The results from the latter study indicate good concordance among all four testing methods when there is a higher concentration of the mutant alleles, while the melting curve assay proved less reliable at lower levels. A comparative study of HRM-PCR and PCR-ARMS assays by Er TK et al. [16] revealed 100% comparable between the tests.
In our study, three different screening techniques were evaluated using a total of 136 patients with confirmed or suspected MPNs and compared their results to those obtained by the PCR-RLFP assay. The PCR-RFLP was used as a reference assay because it was the first method to be described for the detection of the JAK2 V617F mutation and it is a widely used technique.
Our results were consistent with the previously published studies demonstrating perfect concordance among all four testing methods particularly in PV and PMF patients [19], [26]. The discordant results were observed in groups of patients with ET and in patients presenting with clinically suspected MPN. Our results are in agreement with previous studies [16], in which the PCR-HRM analysis of all patients was fully concordant with the ARMS results. With both tests, only one sample of the 20 patients with ET and one of the three patients with clinically suspected MPN gave different results compared with those obtained by the PCR-RFLP. Of note, the JAK2 V617F mutations in both samples were also not detected by direct sequencing. Thus, it is possible that both cases had no mutation and the results obtained by the PCR-RFLP assay were false positives due to incomplete digestion or inferior quality of the DNA template.
PCR-RFLP depends on the restriction enzymes’ success in achieving recognition and cleavage at restriction sites. There is no guarantee that every restriction site will be identified and cleave exactly as predicted. Thus, the results of the present study indicate that both PCR-ARMS and PCR-HRM methods can accurately detect JAK2 V617F mutation. The PCR-ARMS method is based on a multiplex PCR, whereby one intermediate primer will generate a second, smaller fragment in addition to the complete band. The role of the intermediate primer is to introduce bases that are illegitimate to the target sequence to destabilize the annealing process. Thus, in cases with the presence of JAK2 mutation, no annealing with intermediate primer will happen and consequently the side band will not be observed.
The main disadvantage of this method is that it requires post-amplification manipulation. The PCR-HRM method offers some advantages over PCR-ARMS, including ease of performance, speed, and that it does not require any post-PCR processing.
Our evaluation revealed that direct sequencing was a less sensitive method at detecting the JAK2 V617F mutation. The limit for mutation detection by direct sequencing is subjective, and may depend on the experience level of the person interpreting the data. It is also possible that mutations were present below the nominal sensitivity of Sanger sequencing, which can be as high as 20% mutant DNA in the background of the wild type allele [18], [20]. Nowadays, the massive parallel sequencing technologies have been used for standard sequencing applications and have greatly improved the detection of JAK2V617F mutation but they are dependent on the availability of the proprietary equipment [24]. In the future, we might benefit from the incorporation of this technology into routine diagnostic investigations.
5. Conclusions
In conclusion, we have compared the results of four PCR-based assays including PCR-RFLP for detection of JAK2 V617F mutation in blood samples from clinically confirmed and other suspected patients with MPN. Our results have demonstrated that the PCR-ARMS protocol is quick, easy, and can detect the JAK2 V617F mutations effectively in the extracted DNA.
Conflict of interest
None of the authors have any conflict of interest to report.
Acknowledgment
None.
References
- 1.Jaffe E.S., Harris N.L., Stein H., Vardiman J.W. Vol. 185. IARC Press; Lyon: 2001. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues; p. 352. [Google Scholar]
- 2.Swerdlow S.H., Campo E., Harris N.L., Jaffe E.S., Pileri S.A., Stein H. IARC Press; Lyon: 2008. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. [Google Scholar]
- 3.Vardiman J.W., Thiele J., Arber D.A., Brunning R.D., Borowitz M.J., Porwit A. Vol. 114. 2009. The 2008 Revision of the World Health Organization (WHO) Classification of Myeloid Neoplasms and Acute leukemia: Rationale and Important Changes; pp. 937–951. (Blood). [DOI] [PubMed] [Google Scholar]
- 4.Tefferi A., Lasho T.L., Gilliland G. JAK2 mutations in myeloproliferative disorders. N Engl. J. Med. 2005;353:1416–1417. doi: 10.1056/NEJMc051878. [DOI] [PubMed] [Google Scholar]
- 5.Scott L.M. The JAK2 exon 12 mutations: a comprehensive review. Am. J. Hematol. 2011;86:668–676. doi: 10.1002/ajh.22063. [DOI] [PubMed] [Google Scholar]
- 6.Gadina M., Hilton D., Johnston J.A., Morinobu A., Lighvani A., Zhou Y.J. Signaling by type I and II cytokine receptors: ten years after. Curr. Opin. Immunol. 2001;13:363–373. doi: 10.1016/s0952-7915(00)00228-4. [DOI] [PubMed] [Google Scholar]
- 7.Lindauer K., Loerting T., Liedl K.R., Kroemer R.T. Prediction of the structure of human Janus kinase 2 (JAK2) comprising the two carboxy-terminal domains reveals a mechanism for autoregulation. Protein Eng. 2001;14:27–37. doi: 10.1093/protein/14.1.27. [DOI] [PubMed] [Google Scholar]
- 8.Zhao R., Xing S., Li Z., Fu X., Li Q., Krantz S.B. Identification of an acquired JAK2 mutation in polycythemia vera. J. Biol. Chem. 2005;280:22788–22792. doi: 10.1074/jbc.C500138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson P., Kutti J., Andreasson B., Safai-Kutti S., Vilen L., Wedel H. Trends in the incidence of chronic Philadelphia chromosome negative (Ph-) myeloproliferative disorders in the city of Goteborg, Sweden, during 1983-99. J. Intern Med. 2004;256:161–165. doi: 10.1111/j.1365-2796.2004.01357.x. [DOI] [PubMed] [Google Scholar]
- 10.Campbell P.J., Scott L.M., Buck G., Wheatley K., East C.L., Marsden J.T. Definition of subtypes of essential thrombocythaemia and relation to polycythaemia vera based on JAK2 V617F mutation status: a prospective study. Lancet. 2005;366:1945–1953. doi: 10.1016/S0140-6736(05)67785-9. [DOI] [PubMed] [Google Scholar]
- 11.Baxter E.J., Scott L.M., Campbell P.J., East C., Fourouclas N., Swanton S. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 12.Jelinek J., Oki Y., Gharibyan V., Bueso-Ramos C., Prchal J.T., Verstovsek S. JAK2 mutation 1849G>T is rare in acute leukemias but can be found in CMML, Philadelphia chromosome-negative CML, and megakaryocytic leukemia. Blood. 2005;106:3370–3373. doi: 10.1182/blood-2005-05-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rapado I., Grande S., Albizua E., Ayala R., Hernandez J.A., Gallardo M. High resolution melting analysis for JAK2 Exon 14 and Exon 12 mutations: a diagnostic tool for myeloproliferative neoplasms. J. Mol. Diagn. 2009;11:155–161. doi: 10.2353/jmoldx.2009.080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tefferi A., Lasho T.L., Schwager S.M., Steensma D.P., Mesa R.A., Li C.Y. The JAK2(V617F) tyrosine kinase mutation in myelofibrosis with myeloid metaplasia: lineage specificity and clinical correlates. Br. J. Haematol. 2005;131:320–328. doi: 10.1111/j.1365-2141.2005.05776.x. [DOI] [PubMed] [Google Scholar]
- 15.Lay M., Mariappan R., Gotlib J., Dietz L., Sebastian S., Schrijver I. Detection of the JAK2 V617F mutation by LightCycler PCR and probe dissociation analysis. J. Mol. Diagn. 2006;8:330–334. doi: 10.2353/jmoldx.2006.050130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Er T.K., Lin S.F., Chang J.G., Hsieh L.L., Lin S.K., Wang L.H. Detection of the JAK2 V617F missense mutation by high resolution melting analysis and its validation. Clin. Chim. Acta. 2009;408:39–44. doi: 10.1016/j.cca.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Bakker E. Is the DNA sequence the gold standard in genetic testing? Quality of molecular genetic tests assessed. Clin. Chem. 2006;52:557–558. doi: 10.1373/clinchem.2005.066068. [DOI] [PubMed] [Google Scholar]
- 18.Li J., Wang L., Mamon H., Kulke M.H., Berbeco R., Makrigiorgos G.M., Replacing P.C.R. with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat. Med. 2008;14:579–584. doi: 10.1038/nm1708. [DOI] [PubMed] [Google Scholar]
- 19.Cankovic M., Whiteley L., Hawley R.C., Zarbo R.J., Chitale D. Clinical performance of JAK2 V617F mutation detection assays in a molecular diagnostics laboratory: evaluation of screening and quantitation methods. Am. J. Clin. Pathol. 2009;132:713–721. doi: 10.1309/AJCPFHUQZ9AGUEKA. [DOI] [PubMed] [Google Scholar]
- 20.Gong J.Z., Cook J.R., Greiner T.C., Hedvat C., Hill C.E., Lim M.S. Laboratory practice guidelines for detecting and reporting JAK2 and MPL mutations in myeloproliferative neoplasms: a report of the Association for Molecular Pathology. J. Mol. Diagn. 2013;15:733–744. doi: 10.1016/j.jmoldx.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Frantz C., Sekora D.M., Henley D.C., Huang C.K., Pan Q., Quigley N.B. Comparative evaluation of three JAK2V617F mutation detection methods. Am. J. Clin. Pathol. 2007;128:865–874. doi: 10.1309/LW7Q3739RBRMBXXP. [DOI] [PubMed] [Google Scholar]
- 22.Greiner T.C. Diagnostic assays for the JAK2 V617F mutation in chronic myeloproliferative disorders. Am. J. Clin. Pathol. 2006;125:651–653. doi: 10.1309/NXXT-GRCX-D0TM-A3C2. [DOI] [PubMed] [Google Scholar]
- 23.Steensma D.P., Dewald G.W., Lasho T.L., Powell H.L., McClure R.F., Levine R.L. The JAK2 V617F activating tyrosine kinase mutation is an infrequent event in both “atypical” myeloproliferative disorders and myelodysplastic syndromes. Blood. 2005;106:1207–1209. doi: 10.1182/blood-2005-03-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones A.V., Kreil S., Zoi K., Waghorn K., Curtis C., Zhang L. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106:2162–2168. doi: 10.1182/blood-2005-03-1320. [DOI] [PubMed] [Google Scholar]
- 25.Kralovics R., Passamonti F., Buser A.S., Teo S.S., Tiedt R., Passweg J.R. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl. J. Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 26.McClure R., Mai M., Lasho T. Validation of two clinically useful assays for evaluation of JAK2 V617F mutation in chronic myeloproliferative disorders. Leukemia. 2006;20:168–171. doi: 10.1038/sj.leu.2404007. [DOI] [PubMed] [Google Scholar]
- 27.Olsen R.J., Tang Z., Farkas D.H., Bernard D.W., Zu Y., Chang C.C. Detection of the JAK2(V617F) mutation in myeloproliferative disorders by melting curve analysis using the LightCycler system. Arch. Pathol. Lab. Med. 2006;130:997–1003. doi: 10.5858/2006-130-997-DOTJMI. [DOI] [PubMed] [Google Scholar]
- 28.Tan A.Y., Westerman D.A., Dobrovic A. A simple, rapid, and sensitive method for the detection of the JAK2 V617F mutation. Am. J. Clin. Pathol. 2007;127:977–981. doi: 10.1309/1U61JVXTLPPQ7YP1. [DOI] [PubMed] [Google Scholar]
- 29.Lippert E., Boissinot M., Kralovics R., Girodon F., Dobo I., Praloran V. The JAK2-V617F mutation is frequently present at diagnosis in patients with essential thrombocythemia and polycythemia vera. Blood. 2006;108:1865–1867. doi: 10.1182/blood-2006-01-013540. [DOI] [PubMed] [Google Scholar]
- 30.Vannucchi A.M., Pancrazzi A., Bogani C., Antonioli E., Guglielmelli P. A quantitative assay for JAK2(V617F) mutation in myeloproliferative disorders by ARMS-PCR and capillary electrophoresis. Leukemia. 2006;20:1055–1060. doi: 10.1038/sj.leu.2404209. [DOI] [PubMed] [Google Scholar]