Abstract
Sex differences in the development of the normal heart and the prevalence of cardiomyopathies have been reported. The molecular basis of these differences remains unclear. Sex differences in the human heart might be related to patterns of gene expression. Recent studies have shown that sex specific differences in gene expression in tissues including the brain, kidney, skeletal muscle, and liver. Similar data is limited for the heart. Herein we address this issue by analyzing donor and post-mortem adult human heart samples originating from 46 control individuals to study whole-genome gene expression in the human left ventricle. Using data from the genotype tissue expression (GTEx) project, we compared the transcriptome expression profiles of male and female hearts. We found that genes located on sex chromosomes were the most abundant ones among the sexually dimorphic genes. The majority of differentially expressed autosomal genes were those involved in the regulation of inflammation, which has been found to be an important contributor to left ventricular remodeling. Specifically, genes on autosomal chromosomes encoding chemokines with inflammatory functions (e.g. CCL4, CX3CL1, TNFAIP3) and a gene that regulates adhesion of immune cells to the endothelium (e.g., VCAM1) were identified with sex-specific expression levels. This study underlines the relevance of sex as an important modifier of cardiac gene expression. These results have important implications in the understanding of the differences in the physiology of the male and female heart transcriptome and how they may lead to different sex specific difference in human cardiac health and its control.
Introduction
Sex differences in cardiovascular health and disease have been described in multiple clinical observational studies [1]. Previous studies have shown that males and females may vary in the development of the cardiovascular system, resulting in differences in heart size, contractility, and calcium handling [2–4]. Several studies have also demonstrated that sex may play a key role in the susceptibility to various cardiovascular conditions. For example, pulmonary hypertension and heart failure with preserved ejection fraction display a female bias [5, 6], whereas, dilated cardiomyopathy and myocarditis have a male predilection [7, 8]. While many of these clinical differences have been attributed to the presence or absence of sex-specific hormones, namely, estrogen and testosterone [9], a molecular basis for these differences has not been well defined.
Because gene expression is an important determining factor of cellular phenotype, genome-wide transcriptome analysis has been a mainstay of genomics and biomedical research, providing insights into the molecular events underlying human biology and disease. Comparison of the transcription profile between sexes in different tissues such as skin, brain, lung, muscle, and blood has shown widespread differences between males and females. Surprisingly, few studies have described the sex-based differences in both myocardial gene expression in patients without cardiovascular disease. To address this limitation, we analyzed RNA-Seq data obtained from the left ventricle collected from “non-diseased” recently deceased donors in the genotype tissue expression study (GTEX study) [10].
Material and methods
RNA-Seq data set
To study the effect of impact of sex on gene expression in the cardiovascular system, we used the RNA-Seq data set produced by the GTEx consortium (dbGaP accession number: phs000424.v1.p1) that aimed to study human gene expression and regulation in multiple tissues [11]. The entire dataset contains RNA-Seq samples obtained from 544 individuals of both European (≈ 84.3%) and African (≈13.7%) descent and includes samples from various tissues including 190 samples from the left ventricle. We used a subset of data from 46 individuals (29 men and 17 women) who had samples from the left ventricle, who were of European descent, who had no prior documentation of cardiovascular disease, who did not take cardiovascular related medications, and whose cause of death was not related to cardiovascular disease (e.g., stroke, myocardial infarction).
Details on library preparation and sequencing, as well as the data QC pipeline, have been previously described [11]. Briefly, using standard non-strand specific protocol with poly-A selection of mRNA (e.g., the Illumina Tru Seq™ protocol which is a large automated protocol implemented at the Broad Institute), RNA samples meeting QC criteria were sequenced on Illumina HiSeq 2000 instruments. Sequence coverage included a minimum of 50M reads (corresponding to a minimum of 25M 76bp paired-end reads).
Processing, alignment, and analysis of GTEX sequencing data
A copy of the RNA-Seq raw files was downloaded from NCBI dbGaP repository using Sequence Read Archive Tool Kit (SRA Toolkit v2.5.1). Extracted reads were subsequently aligned to the USC human reference genome assembly hg19 and the human transcriptome sequences (Ensembl v70) using the ultrafast aligner the Spliced Transcripts Alignment to a Reference software (STAR, v. 2.3.0e) with maximum tolerated mismatches set at 10 and the outSAMstrandField intronMotif option enabled [12]. Transcriptome coordinates from mapped reads were converted into genomic coordinates and compared with genome mapped reads to identify reads that map to a unique genome location. This process ensured mapped reads were unique across both the genome and transcriptome, while permitting reads to map to different transcripts of the same gene in the initial transcriptome mapping. Uniquely mapped sequences were inspected using the Integrated Genome Viewer (IGV 1.3.1, Broad Institute) [13]. The number of reads uniquely mapped to each gene were counted using Python-HTSeq count module (version 0.5.3p9) [14]. Differential expression analysis was performed on the counts data using DESeq2 (version 1.1.0, http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html). The DESeq2 package uses a negative binomial model to test for differential expression and a shrinkage estimator for the distribution's variance as detailed previously [15]. Briefly, after read counts were retrieved for each gene, differential gene expression was compared using the DEseq2 package in the following four groups: 1) males and females in the entire cohort, 2) younger males and females <55 years, 3) older males and females ≥ 55 years, and 4) younger (<55 years) and older (≥55 years) within male and female cohorts. Read counts for the male and female were compared to determine the log2-fold change in abundance of each transcript. Raw p-values were then adjusted for multiple testing with the Benjamini-Hochberg procedure. Genes with adjusted p-value of 0.05 or less were termed as differentially expressed genes. The analysis was performed using R 2.15 and R 3.1.3.
Enrichment analysis
To determine molecular networks and canonical signaling pathways that contained the differentially expressed genes, we performed a pathway and network enrichment analysis using the Ingenuity Pathway Analysis (IPA) tool (www.ingenuity.com) by inputting a list of differentially expressed genes between male and female heart tissues. Genes with a 1.5 fold or greater change in expression between the male and female groups were used. The settings for the core analysis were as follows: Ingenuity Knowledge Base; Endogenous Chemicals not included; Direct and Indirect relationships. We determined sex-biased enrichment of several genes annotated by KEGG, Reactome, and BioCarta pathway databases with relevance to cardiovascular health and disease. Fisher's exact test was performed for analysis of canonical signaling pathways. The p-value calculated for a pathway showed the probability of being randomly chosen from all of the curated pathways and was adjusted for multiple hypothesis testing using the Benjamini–Hochberg method. IPA was also used to predict which transcription factors could be responsible for gene expression (upstream regulators option) and whether those transcription factors are activated or inhibited. A corrected p-value of less than 0.05 was used to define significance.
Results
Patient characteristics
The clinical information of the 46 subjects is included in Table 1 and S1 Table. There were no significant differences in the demographic and clinical characteristics between males and females in the entire cohort and in the subset analysis stratified by age (p-value <0.05).
Table 1. Demographic and clinical variables of cohort.
| All Groups | Young (<55 years old) | Old (≥ 55 years old) | |||||
|---|---|---|---|---|---|---|---|
| Demographic | Male (n = 29) | Female (n = 17) | Male (n = 25) | Female (n = 10) | Male (n = 4) | Female (n = 7) | |
| Age (years) | 42.7±13.3 | 50.5±14.0 | 40.0±11.9 | 42.5±12.8 | 60.7±4.3 | 61.9±4.7 | |
| Caucasian (%) | 96.7 | 82.4 | 96 | 80 | 100 | 85.7 | |
| BMI (kg/m2) | 26.8±4.1 | 26.2±3.9 | 26.±4.4 | 28.1±4.2 | 27.8±2.2 | 23.6±1.1 | |
| Clinical factors (%) | |||||||
| Hypertension | 27 (8/29) | 47 (8/17) | 20(5/25) | 50(5/10) | 75(3/4) | 43(3/7) | |
| Heart disease | 0 | 0 | 0 | 0 | 0 | 0 | |
| COPD/Asthma | 20(6/29) | 6(1/17) | 24(6/25) | 10(1/10) | 0 | 0 | |
| Autoimmune, inflammatory, or infectious disease | 12(3/29) | 0 | 8(2/25) | 0 | 25(1/4) | 0 | |
| Kidney disease | 0 | 6(1/17) | 0 | 0 | 0 | 10(1/7) | |
| Steroid use | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cause of Death (%) | |||||||
| Hemorrhage | 23(7/29) | 29(5/17) | 24(6/25) | 50(5/10) | 25(1/4) | 0 | |
| Blunt injury | 6(2/29) | 12(2/17) | 4(1/25) | 10(1/10) | 25(1/4) | 14(1/7) | |
| SIGSW | 6(2/29) | 0 | 8(2/25) | 0 | 0 | 0 | |
| EtOH abuse | 3(1/29) | 0 | 4(1/25) | 0 | 0 | 0 | |
| Hepatorenal syndrome | 3(1/29) | 0 | 4(1/25) | 0 | 0 | 0 | |
| Shock secondary to burns | 3(1/29) | 0 | 4(1/25) | 0 | 0 | 0 | |
| Trauma | 6(2/29) | 6(1/17) | 8(2/25) | 10(1/10) | 0 | 0 | |
| Smoke inhalation-respiratory disease | 3(1/29) | 0 | 4(1/25) | 0 | 0 | 0 | |
| Hanging | 12(3/29) | 6(1/17) | 8(2/25) | 10(1/10) | 25(1/4) | 0 | |
| Drug intoxication | 12(3/29) | 0 | 8(2/25) | 0 | 25(1/4) | 0 | |
| Trauma | 6(2/29) | 6(1/17) | 8(2/25) | 10(1/10) | 0 | 0 | |
| Asphyxiation | 6(2/29) | 6(1/17) | 8(2/25) | 0 | 0 | 14(1/7) | |
| Seizure | 3(1/29) | 0 | 4(1/25) | 0 | 0 | 0 | |
| Drug overdose | 0 | 18(3/17) | 0 | 10(1/10) | 0 | 29(2/7) | |
| Complications from ALS | 0 | 6(1/17) | 0 | 0 | 0 | 14(1/7) | |
| Liver transplant | 0 | 6(1/17) | 0 | 0 | 0 | 14(1/7) | |
| Not available | 0 | 6(1/17) | 0 | 0 | 0 | 14(1/7) | |
ALS, amyotrophic lateral sclerosis; MVA, motor vehicle accident; SAH, subarachnoid hemorrhage; SIGSW, survivors of self-inflicted gunshot wound.
Differential gene expression based on sex and age
Using a fold change of greater than 1.5 and a cut-off of adjusted p-value (q-value) of 0.05 as defined by the Benjamini–Hochberg procedure, we observed that 178 genes were differentially expressed between the left ventricular tissue obtained from male and female donors. Among genes that were differentially expressed, 124 genes were up regulated in females and 54 genes were up regulated in males (S2 Table, Figs 1 and 2), suggesting a significantly larger number of female- compared to male-biased genes. Similar to previous studies in other tissues obtained from “normal” individuals, the differences in gene expression were relatively modest (e.g., the absolute fold difference for most genes was between 1.5 and 2.0) except for X- and Y-linked genes (Table 2).
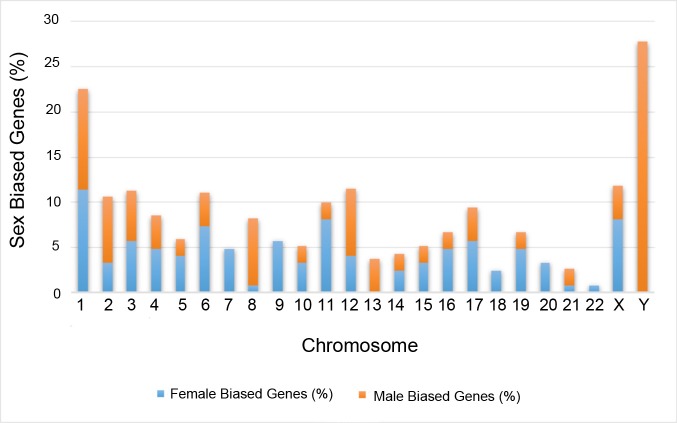
Fig 1. Chromosomal enrichment.
For each chromosome, the number of male- and female-biased genes was computed, the table below the graph lists chromosomes that passed the test for enrichment by Fisher exact test with Benjamini correction, P < 0.05.
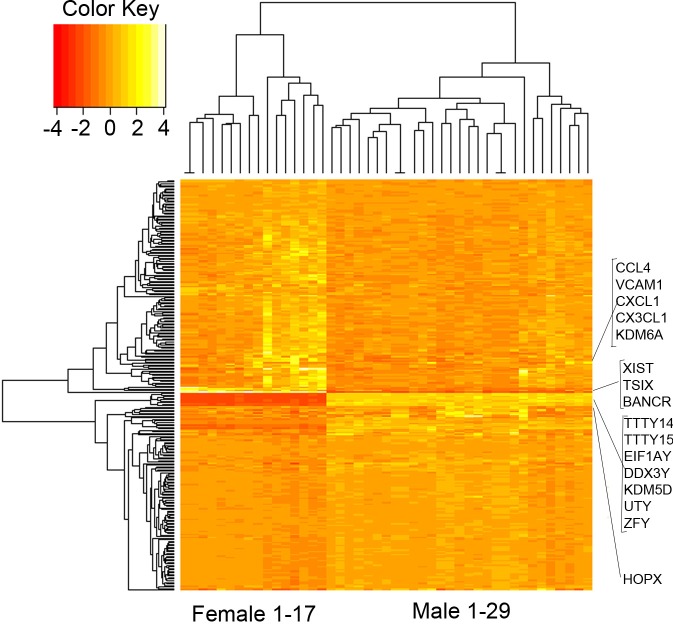
Fig 2. Differentially expressed genes heat map.
Heat map showing differentially expressed genes between male and female LV tissues.
Table 2. Age and sex stratified comparisons of differentially expressed genes, q value <0.05 and fold change > 1.5.
| Sex-Dependent Differences | Absolute Fold Change | Differentially Expressed Genes | Female-Biased Genes | Male-Biased Genes |
|---|---|---|---|---|
| All Samples (n = 46) | 1.5–2.0 | 138 | 105 | 33 |
| 2.0–2.5 | 22 | 17 | 5 | |
| >2.5 | 18 | 2 | 16 | |
| < 55 (n = 32) | 1.5–2.0 | 5 | 4 | 1 |
| 2.0–2.5 | 9 | 7 | 2 | |
| >2.5 | 20 | 4 | 16 | |
| ≥55 (n = 14) | 1.5–2.0 | 9 | 3 | 6 |
| 2.0–2.5 | 6 | 3 | 3 | |
| >2.5 | 22 | 4 | 18 |
Fig 1 shows the distribution of differentially expressed genes in all chromosomes in women and men. The majority of sex-biased genes were autosomal but, as expected, many were located on the sex chromosomes. Xist (X inactive specific transcript) was the sex chromosome linked gene with the highest expression. In addition, eight of these sex chromosome linked genes identified in a previous small microarray study (n = 6) were also differentially expressed in the left ventricular tissue in our cohort (e.g., EIF1AY, RPS4Y1, DDX3Y, JARID1D, USP9Y, ZFY, PRKY, UTY) [16]. Of the autosomal genes not located on a sex chromosome, HOPX, a gene that modulates cardiac development, had the highest expression levels. We then compared the frequencies of significant genes on each chromosome against the total number of genes interrogated on each chromosome. Chromosome 11 was found to have more female-biased differential expression, while chromosomes 8 and Y contained more male-biased genes (Fig 1). To determine the effects of age on gene expression levels, we performed an analysis between the older (≥ 55) and younger cohort (<55). Zero genes met the cutoff for differential expression (q value <0.05 and absolute fold change > 1.5) in the female cohort compared with 2 autosomal genes in the male cohort that were up-regulated at least 2.5 fold in younger compared to older men (S3 Table). Based on an analysis of gene ontology pathways, one of these two genes was involved in cytokine activity while the other one encodes for an olfactory receptor that interacts with odorant molecules in the nose.
Enrichment analysis
To better understand the biological function of genes that were differentially expressed between men and women, sex-biased genes with a q-value < 0.05 and a fold change higher than 1.5 were submitted to IPA for functional annotation and identification of up-stream regulators and canonical pathways. The significant gene ontology (GO) terms associated with each group (male- or female-biased genes) are listed in Tables 3 and 4.
Table 3. Biological functions associated with female biased genes.
| Network | Top Functions | p Value | Genes |
|---|---|---|---|
| Diseases and disorders | |||
| 1 | Inflammatory response | 4.70E-03–1.08E-08 | 50 |
| 2 | Inflammatory disease | 4.67E-03–1.39E-07 | 31 |
| 3 | Cancer | 4.66E-03–2.35E-07 | 110 |
| 4 | Gastrointestinal disease | 4.67E-03–2.35E-07 | 101 |
| 5 | Organismal injury and abnormalities | 4.67E-03–2.35E-07 | 111 |
| Molecular and cellular functions | |||
| 1 | Cellular movement | 4.70E-03–3.85E-11 | 44 |
| 2 | Cell-to-cell signaling and interaction | 4.40E-03–1.93E-08 | 35 |
| 3 | Cell death and survival | 4.70E-03–4.11E-07 | 50 |
| 4 | Cell morphology | 3.34E-03–2.03E-06 | 28 |
| 5 | Cellular growth and proliferation | 4.69E-03–1.48E-05 | 49 |
| Physiological system development and function | |||
| 1 | Hematological system development and function | 4.70E-03–3.85E-11 | 40 |
| 2 | Immune cell trafficking | 4.70E-03–3.85E-11 | 33 |
| 3 | Cell-mediated immune response | 4.70E-03–2.85E-08 | 19 |
| 4 | Tissue morphology | 4.70E-03–1.63E-07 | 43 |
| 5 | Cardiovascular system development and function | 3.34E-03–2.10E-07 | 29 |
Table 4. Biological functions associated with male biased genes.
| Network | Top Functions | p Value | Genes |
|---|---|---|---|
| Disease and disorders | |||
| 1 | Respiratory disease | 1.33E-02–1.47E-04 | 6 |
| 2 | Connective tissue disorders | 4.58E-02–2.03E-03 | 5 |
| 3 | Skeletal and muscular disorders | 4.10E-02–2.03E-03 | 6 |
| 4 | Cancer | 4.58E-02–2.03E-03 | 5 |
| 5 | Muscular disorders | 4.10E-02–2.03E-03 | 6 |
| Molecular and cellular functions | |||
| 1 | Cell morphology | 4.80E-02–2.16E-04 | 13 |
| 2 | Cell cycle | 4.58E-02–3.74E-04 | 3 |
| 3 | Cell death and survival | 4.58E-02–7.28E-04 | 5 |
| 4 | Cell-to-cell signaling and interaction | 4.58E-02–2.23E-03 | 8 |
| 5 | Cellular assembly and organization | 4.58E-02–2.23E-03 | 7 |
| Physiological system development and function | |||
| 1 | Connective tissue development and function | 4.58E-02–2.16E-04 | 5 |
| 2 | Organismal development | 4.58E-02–2.16E-04 | 10 |
| 3 | Tissue morphology | 4.80E-02–2.16E-04 | 14 |
| 4 | Auditory and vestibular system development and function | 4.37E-02–2.23E-03 | 2 |
| 5 | Embryonic development | 4.37E-02–2.23E-03 | 9 |
This analysis revealed a female bias in the expression of 30 genes related to the immune system (Table 5). In contrast, male biased genes were related to respiratory disease and connective tissue disorders, but did not reach statistical significance (data not shown).
Table 5. Female up-regulated genes in inflammatory and heart development pathways.
| Term | P-value | Adjusted P-value | Z-score | Combined Score | Genes |
|---|---|---|---|---|---|
| Regulation of leukocyte activation (GO:0002694) | 5.27207E-06 | 0.004336 | -2.5289 | 13.75908 | SPN; LAG3; CD83; VCAM1; CCL21; FOXF1; FES; CCL5; ZC3H12A; TNFAIP3; THY1; TNFRSF4 |
| Regulation of cytokine production (GO:0001817) | 4.10711E-05 | 0.011260 | -2.4979 | 11.20706 | SPN; NFKBIA; AFAP1L2; LAG3; CD83; ZC3H12A; CASP1; TNFAIP3; TNFRSF4; CX3CL1; MB21D1; MAPK13 |
| Taxis (GO:0042330) | 3.90027E-05 | 0.011260 | -2.3959 | 10.74952 | SPN; VCAM1; CCL21; CCL5; CCL4; CXCL1; SLIT3; TREM1; CX3CL1 |
| Chemotaxis (GO:0006935) | 3.90027E-05 | 0.011260 | -2.3933 | 10.73764 | SPN; VCAM1; CCL21; CCL5; CCL4; CXCL1; SLIT3; TREM1; CX3CL1 |
| Cell chemotaxis (GO:0060326) | 5.62173E-05 | 0.013211 | -2.2419 | 9.699928 | VCAM1; CCL21; CCL5; CCL4; CXCL1; TREM1; CX3CL1 |
| Negative regulation of immune system process (GO:0002683) | 0.00013572 | 0.020580 | -2.4380 | 9.467821 | SPN; NFKBIA; LAG3; CCL21; FOXF1; ZC3H12A; TNFAIP3; THY1; NBL1 |
| Regulation of inflammatory response (GO:0050727) | 0.000155026 | 0.021251 | -2.4038 | 9.257948 | SPN; FOXF1; CCL5; CASP1; TNFAIP3; CX3CL1; CFB; MAPK13 |
| Inflammatory response (GO:0006954) | 0.000111624 | 0.020402 | -2.3622 | 9.193863 | AOC3; AFAP1L2; VCAM1; CCL21; CCL5; CCL4; TNFAIP3; CXCL1; TNFRSF4; PTGES |
| Regulation of immune effector process (GO:0002697) | 0.000240717 | 0.030460 | -2.4394 | 8.51707 | SPN; LAG3; FOXF1; FES; TNFAIP3; TNFRSF4; CFB; MB21D1 |
| Regulation of lymphocyte activation (GO:0051249) | 0.000282146 | 0.032930 | -2.4716 | 8.436548 | SPN; LAG3; CD83; VCAM1; CCL21; CCL5; TNFAIP3; THY1; TNFRSF4 |
Similarly, analysis of canonical pathways showed significant female bias in the up-regulation of immune-related processes (Fig 3) including the role of IL-17A in arthritis, granulocyte adhesion and diapedesis, and cytokine production; whereas, the top canonical pathways regulated by male-biased genes were hepatic fibrosis and hepatic stellate cell activation (Table 6).
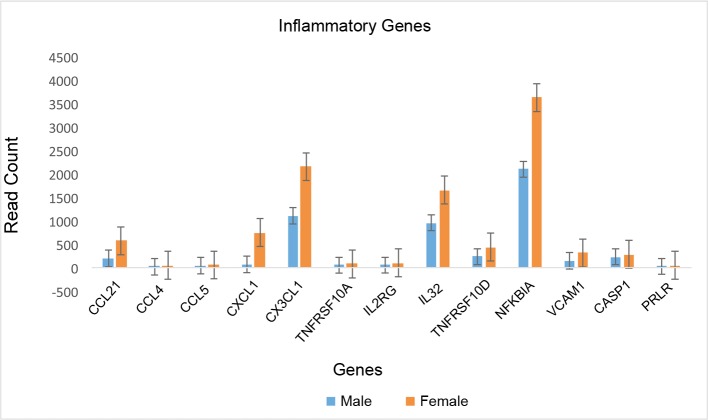
Fig 3. Female heart over expressed genes involve in inflammatory pathways.
The bar graphs show the read count and error bars represent standard deviation.
Table 6. Five top canonical pathways for female and male biased genes.
| Female-biased top canonical pathways | p-value | Overlap |
| Granulocyte adhesion and diapedesis | 5.17E-05 | 4.0% (7/177) |
| Agranulocyte adhesion and diapedesis | 7.81E-05 | 3.7% (7/189) |
| Differential regulation of cytokine production in macrophages and T helper cells by IL-17A and IL-17F | 1.20E-04 | 16.7% (3/18) |
| Role of IL-17A in arthritis | 2.11E-04 | 7.4% (4/54) |
| Differential regulation of cytokine production in intestinal epithelial cells by IL-17A and IL- 17F | 2.55E-04 | 13.0% (3/23) |
| Male-biased top canonical pathways | p-value | Overlap |
| Hepatic fibrosis / hepatic stellate cell activation | 7.31E-04 | 2.2% (4/183) |
| Pancreatic adenocarcinoma signaling | 1.71E-03 | 2.8% (3/106) |
| Glutathione biosynthesis | 6.68E-03 | 33.3% (1/3) |
| ILK Signaling | 8.12E-03 | 1.6% (3/185) |
| LPS/IL-1 mediated inhibition of RXR function | 1.30E-02 | 1.4% (3/220) |
To verify these findings, we used another public domain enrichment analysis tool, Enrichr (http://amp.pharm.mssm.edu/Enrichr/) [17], to analyze the function of sex-biased genes. GO analysis using Enrichr revealed that genes up-regulated in females heart were involved mainly in biological processes related to immune function including regulation of leukocyte activation, cytokine production and cell adhesion; whereas, no genes were significantly up-regulated in males heart (Table 7).
Table 7. Top biological functions, molecular functions and cellular components of female biased genes.
| Female-Biased Genes | P-value | Adjusted p-value | Z-score |
|---|---|---|---|
| Biological function | |||
| Regulation of cell adhesion (GO:0030155) | 1.185E-06 | 0.0019494 | -2.4626837 |
| Regulation of leukocyte activation (GO:0002694) | 5.2721E-06 | 0.00433628 | -2.528899 |
| Regulation of cell activation (GO:0050865) | 1.0921E-05 | 0.00598837 | -2.5255131 |
| Regulation of cytokine production (GO:0001817) | 4.1071E-05 | 0.01126033 | -2.4979698 |
| Mesodermal cell differentiation (GO:0048333) | 0.00013762 | 0.02058018 | -2.7762975 |
| Molecular function | |||
| Chemokine activity (GO:0008009) | 2.2994E-05 | 0.00738119 | -2.4379607 |
| CCR chemokine receptor binding (GO:0048020) | 0.00029438 | 0.0314983 | -3.1141924 |
| Chemokine receptor binding (GO:0042379) | 5.4359E-05 | 0.0087247 | -2.2605308 |
| Cellular component | |||
| Side of membrane (GO:0098552) | 1.8363E-05 | 0.00131292 | -2.2865759 |
| External side of plasma membrane (GO:0009897) | 2.9874E-05 | 0.00142397 | -2.2381773 |
| Extracellular space (GO:0005615) | 1.3675E-05 | 0.00131292 | -2.2091901 |
Similarly, KEGG metabolic pathway analysis indicated that genes were only up-regulated in female hearts. These genes were located specifically in the pathways of cytokine cytokine receptor interaction (p value = 0.00005, adjusted p value = 0.0023, Z score = -2.08). Similarly, MGI Mammalian Phenotype indicated that mutations of genes in female over-expressed genes were associated with disease in immune system; whereas, there were no significant male over-expressed genes (Table 8). Taken together, genes that were overexpressed in female heart tissue were highly enriched in genes involved in inflammation.
Table 8. 5 top MGI mammalian phenotype for female biased genes.
| Female-Biased Genes | P-value | Adjusted P-value | Z-score |
|---|---|---|---|
| Abnormal innate immunity | 0.0002497 | 0.016636772 | -1.6699686 |
| Abnormal adaptive immunity | 8.7305E-05 | 0.009906402 | -1.3827691 |
| Abnormal immune cell | 9.2153E-05 | 0.009906402 | -1.3722959 |
Transcription factor binding site analysis
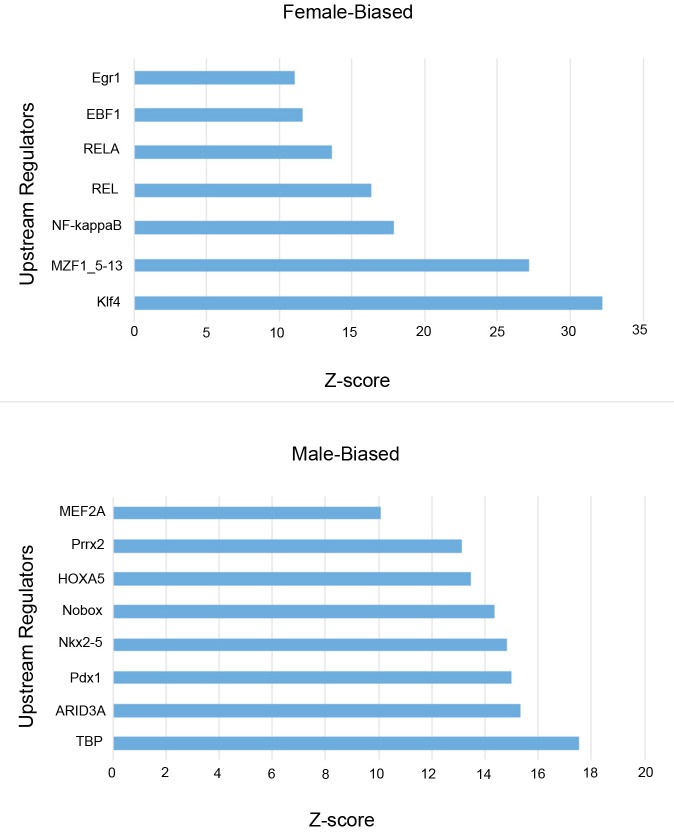
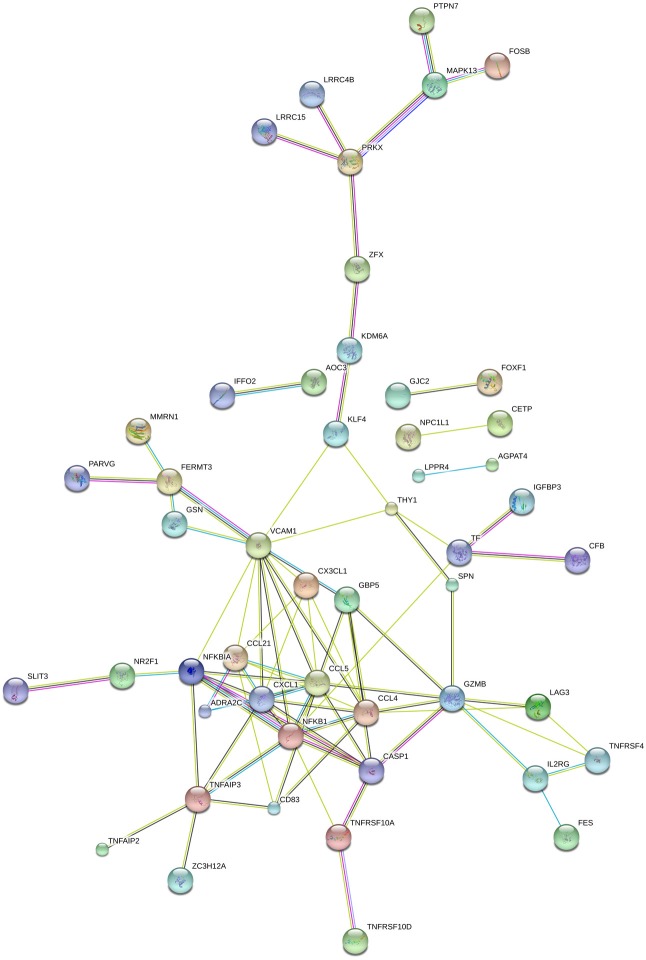
To predict transcription factors that may be involved in controlling sex-biased gene expression, we searched for conserved transcription factor binding sites (TFBS) in the 5 kb of DNA sequence up and downstream of the transcription start sites of sex-biased genes defined previously. Using the web-based promoter analysis program, oPOSSUM-3 and the JASPAR core motifs [18, 19], we identified potential binding sites for 7 transcription factors for female over expressed genes and 8 transcription factor for male over expressed genes (S4 Table, Fig 4) (Z-score > 10; Fisher score> 7). We tested if the genes encoding these TFs were expressed at detectable levels in the human heart tissue using publicly available RNA-Seq data (http://medicalgenomics.org/). Two transcription factors, KLF4 and NF-kappa B, were expressed at detectable levels in human heart tissues. These transcription factors regulate the expression of several genes involved in the regulation of the immune system that were differentially expressed between male and female donor hearts (S4 Table, Fig 5).
Fig 4. Transcription factor binding sites.
Transcription factor binding site analysis comparing male-biased and female-biased genes in human LV tissue.
Fig 5. Associations between DEGs and predicted TFs.
Diagram of the network of predicted associations between DEGs and predicted TFs (KLF4 and NF-kappa B). The network nodes are proteins. The edges represent the predicted functional associations.
Discussion
There is very limited information about the sex and age related differences in expression profile of human heart tissue in healthy people. We analyzed the gene expression profiles of 29 men and 17 women without a history of heart disease and identified 178 differentially expressed genes between the sexes. The majority of these genes demonstrated modest absolute fold-changes of 1.5–2.0 (77.5%) with greatest enrichment on autosomes 8 and 11 as well as the Y chromosome. A breakdown of these genes into biological processes revealed an over-representation of genes involved in inflammatory response in female heart tissue.
Our results are consistent with findings from a recent study on the expression of sexually dimorphic genes expressed in different human tissues, such as muscle, blood and brain, where more than 70% of sexually dimorphic genes displayed less than 1.2-fold change in expression. Greater fold changes (>2.0 fold) were seen in sex chromosome-linked genes. Our findings are also similar to those reported in a small microarray study (n = 6) of human explanted donor hearts (left ventricle) that identified 16 sexually dimorphic genes with more than 2-fold change with most differences linked to sex chromosomes [20].
Consistent with other reports in non-cardiac tissue [21], one of the important pathways differentially regulated in male and female heart in our cohort is the adaptive and innate immune system, which has been found to contribute to the development and progression of cardiovascular disease [22, 23] In our analysis, we found that male and female hearts differed significantly in the transcription of sex-linked and autosomal genes involved in the regulation of the immune system. Similar to findings from a previous small microarray study in healthy donor hearts (21), our cohort showed differential expression of two sex-linked genes DDX3Y and EIF1AY that encode for human male-specific minor histocompatibility antigens that contribute to inflammatory disease [16]. Of the autosomal genes related to inflammation, the greatest bias was found in females in the expression of TNFAIP3, a gene involved in immune and inflammatory responses signaled by such cytokines as TNF-alpha and IL-1 beta, or directly by pathogens via Toll-like receptors (TLRs) through terminating NF-kappaB activity [24]. The other important inflammatory genes with greater than 2-fold change in expression levels were CCL4, CX3CL1 and VCAM1[25–28]. Both CCL4 and CX3CL1 are chemokines that regulate the migration and adhesion of monocytes and lymphocyte and, thus, play an important role in left ventricular remodeling in response to injury [29]. VCAM-1, on the other hand, is expressed on the cytokine-activated endothelium, attracting neutrophils to damaged myocardium and mediating ischemia reperfusion injury [30]. Finally, we found that the most important transcription factors enriched in female heart were KLF4 (Kruppel-like factor 4) and NF-kappaB. While the role of KLF4, a regulator of pro-inflammatory signaling in the heart, has been well established in the development of vascular disease including atherosclerosis [31–33], a recent study has also shown that KLF4 is important in regulating cardiac hypertrophy and cardiac mitochondrial homeostasis [34, 35]. In addition to KLF4, we found that NF-kappaB was differentially expressed. NF-kappaB controls multiple processes, including immunity, inflammation, cell survival, differentiation and proliferation, and regulates cellular responses to stress, hypoxia, stretch and ischemia. NF-kappaB has been shown to influence numerous cardiovascular diseases including atherosclerosis, cardiac hypertrophy, and heart failure [36]. Although several studies have shown that estrogen and androgen receptors control different cardiovascular related pathways in a sex-specific manner, such as the synthesis of collagen and matrix-metalloproteinase [37–40], we did not find any differences in hormone receptor gene expression in heart tissue between male and females in our cohort. It appears that like other tissues, sex specific differences mediated by sex hormones is not related to differential gene expression of hormone receptors[41].
There are several limitations to this study. Because of the size of our cohort, we could not perform eQTL analysis. As additional samples become available, these analyses could be performed. We also did not include diseased hearts in our cohort. This is beyond the scope of this study and has been performed previously [42]. It would also be important to further validate our findings using quantitative real-time PCR as well as evaluating the sex differences on the proteomic level. Nevertheless, this study is the first to evaluate RNAseq data from normal male and female hearts and contributes to a better understanding of how differences in gene expression may affect cardiovascular health in men and women.
Conclusion
In conclusion, gene expression in the human heart displayed evidence of sexual dimorphism similar to other somatic tissues. Notably, the female biased genes are highly enriched for genes involved in inflammatory response while no such pattern was seen for the male-biased genes, suggesting that the differences in cardiovascular disorder susceptibility between males and females are likely rooted from the sex-biased gene expression of the immune system.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We would like to thank the GTEX Consortium for access to the data.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the American Heart Association (10SDG4280129) to Dr. Patricia Nguyen; Stanford University School of Medicine to Ivy Nguyen; and U.S. Department of Veterans Affairs (HSRD 10-032) to Dr. Patricia Nguyen. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Maas A, Appelman YEA. Gender differences in coronary heart disease. Netherlands Heart Journal. 2010;18(12):598–602. PubMed PMID: PMC3018605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bairey Merz CN, Moriel M, Rozanski A, Klein J, Berman DS. Gender-related differences in exercise ventricular function among healthy subjects and patients. American Heart Journal. 1996;131(4):704–9. doi: http://dx.doi.org/10.1016/S0002-8703(96)90274-4 [DOI] [PubMed] [Google Scholar]
- 3.Ostadal B, Kolar F. Cardiac adaptation to chronic high-altitude hypoxia: Beneficial and adverse effects. Respiratory Physiology & Neurobiology. 2007;158(2–3):224–36. doi: http://dx.doi.org/10.1016/j.resp.2007.03.005 [DOI] [PubMed] [Google Scholar]
- 4.Regitz-Zagrosek V, Oertelt-Prigione S, Seeland U, Hetzer R. Sex and Gender Differences in Myocardial Hypertrophy and Heart Failure. Circulation Journal. 2010;74(7):1265–73. doi: 10.1253/circj.CJ-10-0196 [DOI] [PubMed] [Google Scholar]
- 5.Miller VM, Best PJ. Implications for reproductive medicine: sex differences in cardiovascular disease. Sexuality, reproduction & menopause. 2011;9(3):21. [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer S, van der Meer P, van Tintelen JP, van den Berg MP. Sex differences in cardiomyopathies. European Journal of Heart Failure. 2014;16(3):238–47. doi: 10.1002/ejhf.15 [DOI] [PubMed] [Google Scholar]
- 7.Czubryt MP, Espira L, Lamoureux L, Abrenica B. The role of sex in cardiac function and diseaseThis paper is one of a selection of papers published in this Special Issue, entitled Young Investigator's Forum. Canadian Journal of Physiology and Pharmacology. 2006;84(1):93–109. doi: 10.1139/y05-151 [DOI] [PubMed] [Google Scholar]
- 8.Vassalle C, Simoncini T, Chedraui P, Pérez-López FR. Why sex matters: the biological mechanisms of cardiovascular disease. Gynecological Endocrinology. 2012;28(9):746–51. doi: 10.3109/09513590.2011.652720 [DOI] [PubMed] [Google Scholar]
- 9.Mathur P, Ostadal B, Romeo F, Mehta JL. Gender-Related Differences in Atherosclerosis. Cardiovascular Drugs and Therapy. 2015;29(4):319–27. doi: 10.1007/s10557-015-6596-3 [DOI] [PubMed] [Google Scholar]
- 10.The GC. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45(6):580–5. doi: 10.1038/ng.2653 PubMed PMID: PMC4010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Consortium GT. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science (New York, NY). 2015;348(6235):648–60. doi: 10.1126/science.1262110 PubMed PMID: PMC4547484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635 PubMed PMID: PMC3530905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative Genomics Viewer. Nature biotechnology. 2011;29(1):24–6. doi: 10.1038/nbt.1754 PubMed PMID: PMC3346182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. doi: 10.1093/bioinformatics/btu638 PubMed PMID: PMC4287950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550 doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miklos DB, Kim HT, Miller KH, Guo L, Zorn E, Lee SJ, et al. Antibody responses to H-Y minor histocompatibility antigens correlate with chronic graft-versus-host disease and disease remission. Blood. 2005;105(7):2973–8. doi: 10.1182/blood-2004-09-3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14(1):128 doi: 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon AT, Arenillas DJ, Hunt RW, Wasserman WW. oPOSSUM-3: Advanced Analysis of Regulatory Motif Over-Representation Across Genes or ChIP-Seq Datasets. G3: Genes|Genomes|Genetics. 2012;2(9):987–1002. doi: 10.1534/g3.112.003202 PubMed PMID: PMC3429929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Research. 2014;42(Database issue):D142–D7. doi: 10.1093/nar/gkt997 PubMed PMID: PMC3965086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isensee J, Witt H, Pregla R, Hetzer R, Regitz-Zagrosek V, Ruiz Noppinger P. Sexually dimorphic gene expression in the heart of mice and men. Journal of Molecular Medicine (Berlin, Germany). 2008;86(1):61–74. doi: 10.1007/s00109-007-0240-z PubMed PMID: PMC2755745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hewagama A, Patel D, Yarlagadda S, Strickland FM, Richardson BC. Stronger inflammatory/cytotoxic T cell response in women identified by microarray analysis. Genes and immunity. 2009;10(5):509–16. doi: 10.1038/gene.2009.12 PubMed PMID: PMC2735332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansson GK. Inflammation, Atherosclerosis, and Coronary Artery Disease. New England Journal of Medicine. 2005;352(16):1685–95. doi: 10.1056/NEJMra043430 . [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Ruiz I. Immune system and cardiovascular disease. Nat Rev Cardiol. 2016;13(9):503–. doi: 10.1038/nrcardio.2016.127 [DOI] [PubMed] [Google Scholar]
- 24.Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-kappaB activation. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(13):6721–5. PubMed PMID: PMC39093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skelton RJP, Costa M, Anderson DJ, Bruveris F, Finnin BW, Koutsis K, et al. SIRPA, VCAM1 and CD34 identify discrete lineages during early human cardiovascular development. Stem Cell Research. 2014;13(1):172–9. https://doi.org/10.1016/j.scr.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 26.Chang T-T, Chen J-W. Emerging role of chemokine CC motif ligand 4 related mechanisms in diabetes mellitus and cardiovascular disease: friends or foes? Cardiovascular Diabetology. 2016;15(1):117 doi: 10.1186/s12933-016-0439-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apostolakis S, Spandidos D. Chemokines and atherosclerosis: focus on the CX3CL1/CX3CR1 pathway. Acta Pharmacol Sin. 2013;34(10):1251–6. doi: 10.1038/aps.2013.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ley K, Huo Y. VCAM-1 is critical in atherosclerosis. Journal of Clinical Investigation. 2001;107(10):1209–10. PubMed PMID: PMC209304. doi: 10.1172/JCI13005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nemska S, Monassier L, Gassmann M, Frossard N, Tavakoli R. Kinetic mRNA Profiling in a Rat Model of Left-Ventricular Hypertrophy Reveals Early Expression of Chemokines and Their Receptors. PloS one. 2016;11(8):e0161273 doi: 10.1371/journal.pone.0161273 ; PubMed Central PMCID: PMC4985150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowden RA, Ding ZM, Donnachie EM, Petersen TK, Michael LH, Ballantyne CM, et al. Role of alpha4 integrin and VCAM-1 in CD18-independent neutrophil migration across mouse cardiac endothelium. Circulation research. 2002;90(5):562–9. . [DOI] [PubMed] [Google Scholar]
- 31.Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, et al. Krüppel-like factor 4 regulates macrophage polarization. The Journal of Clinical Investigation. 2011;121(7):2736–49. doi: 10.1172/JCI45444 PubMed PMID: PMC3223832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma N, Lu Y, Zhou G, Liao X, Kapil P, Anand P, et al. Myeloid KLF4 deficiency augments atherogenesis in ApoE-/- mice. Arteriosclerosis, thrombosis, and vascular biology. 2012;32(12):2836–8. doi: 10.1161/ATVBAHA.112.300471 PubMed PMID: PMC3574634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, et al. Kruppel-like Factor 4 Regulates Endothelial Inflammation. Journal of Biological Chemistry. 2007;282(18):13769–79. doi: 10.1074/jbc.M700078200 [DOI] [PubMed] [Google Scholar]
- 34.Yoshida T, Yamashita M, Horimai C, Hayashi M. Kruppel-like factor 4 protein regulates isoproterenol-induced cardiac hypertrophy by modulating myocardin expression and activity. The Journal of biological chemistry. 2014;289(38):26107–18. doi: 10.1074/jbc.M114.582809 ; PubMed Central PMCID: PMC4176239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao X, Zhang R, Lu Y, Prosdocimo DA, Sangwung P, Zhang L, et al. Kruppel-like factor 4 is critical for transcriptional control of cardiac mitochondrial homeostasis. J Clin Invest. 2015;125(9):3461–76. doi: 10.1172/JCI79964 ; PubMed Central PMCID: PMC4588311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Heiden K, Cuhlmann S, Luong Le A, Zakkar M, Evans Paul C. Role of nuclear factor κB in cardiovascular health and disease. Clinical Science. 2010;118(10):593–605. doi: 10.1042/CS20090557 [DOI] [PubMed] [Google Scholar]
- 37.Dubey RK, Gillespie DG, Jackson EK, Keller PJ. 17β-Estradiol, Its Metabolites, and Progesterone Inhibit Cardiac Fibroblast Growth. Hypertension. 1998;31(1):522–8. doi: 10.1161/01.hyp.31.1.522 [DOI] [PubMed] [Google Scholar]
- 38.Mahmoodzadeh S, Pham TH, Kuehne A, Fielitz B, Dworatzek E, Kararigas G, et al. 17β-Estradiol-induced interaction of ERα with NPPA regulates gene expression in cardiomyocytes. Cardiovascular Research. 2012;96(3):411–21. doi: 10.1093/cvr/cvs281 [DOI] [PubMed] [Google Scholar]
- 39.Petrov G, Regitz-Zagrosek V, Lehmkuhl E, Krabatsch T, Dunkel A, Dandel M, et al. Regression of Myocardial Hypertrophy After Aortic Valve Replacement. Faster in Women? 2010;122(11 suppl 1):S23–S8. doi: 10.1161/circulationaha.109.927764 [DOI] [PubMed] [Google Scholar]
- 40.Zhou L, Shao Y, Huang Y, Yao T, Lu L-M. 17β-Estradiol inhibits angiotensin II-induced collagen synthesis of cultured rat cardiac fibroblasts via modulating angiotensin II receptors. European Journal of Pharmacology. 2007;567(3):186–92. http://dx.doi.org/10.1016/j.ejphar.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 41.Chen C-Y, Lopes-Ramos CM, Kuijjer ML, Paulson JN, Sonawane AR, Fagny M, et al. Sexual dimorphism in gene expression and regulatory networks across human tissues. bioRxiv. 2016. 10.1101/082289.
- 42.Boheler KR, Volkova M, Morrell C, Garg R, Zhu Y, Margulies K, et al. Sex- and age-dependent human transcriptome variability: Implications for chronic heart failure. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(5):2754–9. doi: 10.1073/pnas.0436564100 PubMed PMID: PMC151413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.