ABSTRACT
Genes encoding proteins that carry out essential informational tasks in the cell, in particular where multiple interaction partners are involved, are less likely to be transferable to a foreign organism. Here, we investigated the constraints on transfer of a gene encoding a highly conserved informational protein, translation elongation factor Tu (EF-Tu), by systematically replacing the endogenous tufA gene in the Escherichia coli genome with its extant and ancestral homologs. The extant homologs represented tuf variants from both near and distant homologous organisms. The ancestral homologs represented phylogenetically resurrected tuf sequences dating from 0.7 to 3.6 billion years ago (bya). Our results demonstrate that all of the foreign tuf genes are transferable to the E. coli genome, provided that an additional copy of the EF-Tu gene, tufB, remains present in the E. coli genome. However, when the tufB gene was removed, only the variants obtained from the gammaproteobacterial family (extant and ancestral) supported growth which demonstrates the limited functional interchangeability of E. coli tuf with its homologs. Relative bacterial fitness correlated with the evolutionary distance of the extant tuf homologs inserted into the E. coli genome. This reduced fitness was associated with reduced levels of EF-Tu and reduced rates of protein synthesis. Increasing the expression of tuf partially ameliorated these fitness costs. In summary, our analysis suggests that the functional conservation of protein activity, the amount of protein expressed, and its network connectivity act to constrain the successful transfer of this essential gene into foreign bacteria.
KEYWORDS: EF-Tu, horizontal gene transfer, ancient genes, proteobacteria, tuf
IMPORTANCE
Horizontal gene transfer (HGT) is a fundamental driving force in bacterial evolution. However, whether essential genes can be acquired by HGT and whether they can be acquired from distant organisms are very poorly understood. By systematically replacing tuf with ancestral homologs and homologs from distantly related organisms, we investigated the constraints on HGT of a highly conserved gene with multiple interaction partners. The ancestral homologs represented phylogenetically resurrected tuf sequences dating from 0.7 to 3.6 bya. Only variants obtained from the gammaproteobacterial family (extant and ancestral) supported growth, demonstrating the limited functional interchangeability of E. coli tuf with its homologs. Our analysis suggests that the functional conservation of protein activity, the amount of protein expressed, and its network connectivity act to constrain the successful transfer of this essential gene into foreign bacteria.
INTRODUCTION
The complexity hypothesis assigns the function and the network interactivity of a gene product as two primary factors determining a gene’s capacity to successfully transfer and adapt in another genome (1–5). Consequently, genes involved in central informational tasks (i.e., replication, transcription, and translation) are expected to be less transferable to a foreign genome than genes involved in other activities (1, 6, 7).
Genomic replacement of an essential gene with another homolog may potentially disturb the gene product’s function depending on the degree of functional equivalence or compatibility between the two genes (8–11). Alterations in gene dosage resulting from the acquisition of a foreign gene may also perturb cellular homeostasis, resulting in a lower transcription rate or an altered global pattern of gene expression, network function, and organismal survivability (11–18).
Genes involved in the translation of mRNA into protein perform one of the most crucial informational tasks in the cell, and based on phylogenomic analysis, they are expected to be highly resistant to gene transfer (1, 19). There are examples demonstrating that some ribosomal protein genes can be integrated into foreign genomes under certain conditions (20–23). However, no study has systematically tested whether there is a direct correlation between organism fitness and the evolutionary distance between an essential endogenous gene and its substituted (ancestral or extant) homolog.
Here, we focus on the bacterial elongation factor Tu (EF-Tu) protein, encoded by tuf, one of the most ancient and highly conserved proteins. EF-Tu has an essential function in the translation machinery by delivering aminoacylated tRNA (aa-tRNA) molecules into the A site of the ribosome (24). EF-Tu is encoded by two genes in Escherichia coli, tufA and tufB, generated by an ancient duplication event thought to be specific to the proteobacterial lineage preceding the Cambrian period (25, 26). Expression of EF-Tu is primarily driven by tufA, with 66% of cellular EF-Tu expressed from the tufA gene (27). EF-Tu protein levels in the cell are correlated with cellular fitness and intrinsically regulated in order to maintain growth rate (28–30). EF-Tu belongs to the ancient protein repertoire of the cell, evolves slowly, and serves as a functional fossil by participating in ancient and conserved functions (31). It remains unclear whether tuf genes are replaceable by their ancient counterparts or homologs obtained from an extant organism. Answering this question would allow us to explore the limits of interchangeability for the E. coli tuf gene and to ascertain a pattern within and between bacterial lineages across time and divergence. We sought guidance from a methodology referred to as ancestral sequence reconstruction (32–37) and accessed reconstructed ancestral tuf variants (35) as well as modern tuf gene sequences in order to observe the patterns of interchangeability among multiple nodes along the EF-Tu phylogenetic tree. We utilized a set of foreign genes representing E. coli EF-Tu homologs from closely and distantly related bacteria, as well as phylogenetically inferred ancestral EF-Tu proteins dating from 0.7 to 3.6 billion years ago (bya), thus accessing interspecies (modern) and ancestral (paleogenetic) axes. We determined the fitness effects of the introduction of a foreign tuf gene into each strain by replacing the native E. coli tufA gene with foreign variants and asked whether these foreign genes could support cell viability when the tufB gene was removed from the chromosome. We examined the impact of the tuf gene replacements on growth rate and protein levels, as well as on protein function-structure, and assessed the extent of lateral and ancestral phylogenetic distances between the alien gene and host genome that yielded viable organisms.
RESULTS
Replacement of the tufA gene in E. coli reduces relative fitness.
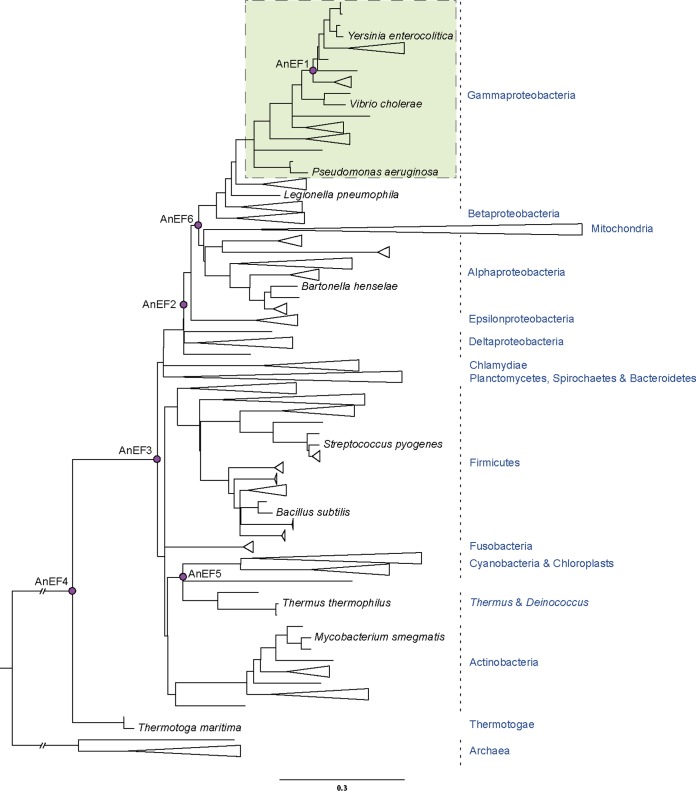
Using genetic recombineering, we generated a set of E. coli strains in which the tufA gene coding sequence was precisely replaced by the coding sequence of its ancestral and modern homologs (Fig. 1; see also Fig. S1 in the supplemental material). The 16 tuf homologs cover bacterial species from a wide span of taxa (Yersinia enterocolitica, Vibrio cholerae, Pseudomonas aeruginosa, Legionella pneumophila, Bartonella henselae, Streptococcus pyogenes, Bacillus subtilis, Thermus thermophilus, Mycobacterium smegmatis, and Thermotoga maritima) as well as six ancestral sequences that extend deep into the bacterial phylogenetic tree (Fig. 1). These six sequences represent ancestral nodes dating from approximately 0.7 bya back to the last common ancestor of bacterial tuf, dated to approximately 3.6 bya (35, 38). These homologs of tuf encode EF-Tu variants that range in amino acid identity from 93.9% (Y. enterocolitica) to 69.4% (T. maritima) relative to E. coli EF-Tu (Fig. S2). Nucleotide sequences of the ancestral tuf genes are shown in Table S1.
FIG 1 .
Phylogenetic tree indicating the node and taxa of the ancestral and modern tuf (EF-Tu) homologs. Pink circles represent the ancestral EF-Tu nodes. E. coli was genetically engineered to carry ancestral or modern homologs of tuf, encoding translation elongation factor EF-Tu, replacing the native E. coli tufA gene. A shaded box indicates the area of viability of EF-Tu gene exchange. The scale bar expresses units of amino acid substitutions per site. The tree was created with data from references 35 and 38.
Foreign tuf genes were integrated into the bacterial genome in a stepwise fashion as shown. (A) Each of the foreign tuf genes was integrated into the E. coli chromosome (replacing native tufA from start to stop codons). (B) In each strain carrying a foreign tuf gene at the tufA locus, an attempt was made to remove the native E. coli tufB gene (replacing the coding sequence from start to stop codons). Download FIG S1, PDF file, 0.2 MB (250.8KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative fitness of strains carrying two tuf genes (tufA-foreign and tufB E. coli) as a function of percent identity to E. coli EF-Tu (indicated as 100%). Strains shown in green carry a foreign tuf gene that can support viability when the E. coli tufB gene has been deleted. Download FIG S2, PDF file, 0.2 MB (167.8KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nucleotide sequences of ancestral tuf genes. Download TABLE S1, PDF file, 0.1 MB (64.7KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
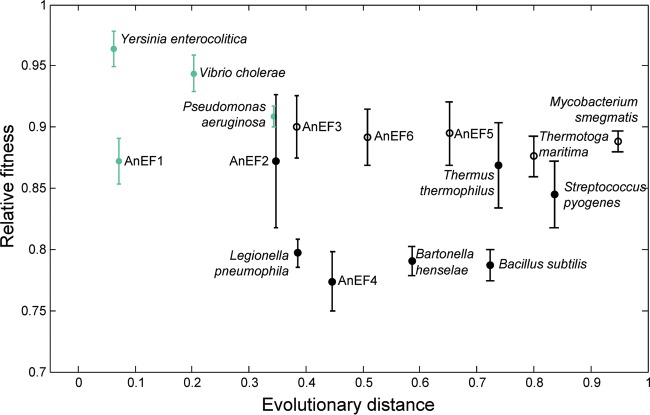
To construct a set of isogenic strains, the endogenous E. coli tufA gene was replaced with each of the tuf variants while the second endogenous tuf gene, tufB, remained intact in the genome. All engineered strains in which tufA was replaced with a foreign tuf gene retained viability (Fig. 2). The effects on relative fitness of the foreign tuf genes were determined by measuring the exponential growth rates in LB and relating them to that of the isogenic wild type (carrying native tufA and tufB), where relative fitness was set to 1.0. The relative fitness of each of the engineered constructs varied from 0.96 down to 0.77 (Fig. 2). The relative fitness of E. coli in which tufA was deleted from the chromosome was 0.7. The similarity in relative fitness between E. coli lacking tufA and some of the strains carrying foreign tuf genes raised the question of whether all of the foreign genes would be capable of supporting viability in the absence of a functioning tufB gene.
FIG 2 .
Correlation between relative fitness and evolutionary distance for bacterial strains carrying a foreign tuf gene. Relative fitness is shown as a function of evolutionary divergence (see Materials and Methods) of EF-Tu homologs from E. coli (1 indicates greatest difference from E. coli). Each E. coli strain carried a foreign tuf gene at the tufA location and an intact native tufB gene. Fitness was measured as exponential growth rate, relative to the E. coli wild type carrying tufA and tufB. Species names and ancestor notations (AnEF, ancestral EF-Tu) refer to the source of the foreign tuf gene sequence in each strain. Strains shown in green carry a foreign tuf gene that can support viability even when the E. coli tufB gene has been deleted. Strains shown in black carry foreign tuf genes that do not support viability in the absence of the E. coli tufB gene. Empty symbols represent strains where the tufB region was amplified.
Because the tufB gene is located between the rRNA operons rrnB and rrnE, in a region that is subject to frequent amplification (39), we asked whether tufB was amplified in any of the strains carrying a foreign tuf allele. Our expectation was that a less effective, or completely inactive, foreign tuf gene might select for genomes in which tufB was amplified as a fitness-compensatory mechanism. Using real-time quantitative PCR (RT-qPCR), we found that the tufB region was duplicated or triplicated in 5 of the 16 engineered strains (Fig. 2). The 5 strains in which the tufB region was amplified are those that carry the most distant tuf homologs, compatible with selection for improved fitness.
A subset of foreign tuf genes supports viability.
We next asked whether any of the foreign tuf genes could support viability in the absence of E. coli tufB, by attempting to remove the endogenous tufB gene from the chromosome of each of the 16 strains carrying a foreign tuf gene, as outlined in Fig. S1. In the absence of tufB, the only foreign tuf sequences that supported viability were those from Y. enterocolitica, V. cholerae, and P. aeruginosa and AnEF1, the youngest of the ancestral tuf genes at ca. 0.7 bya (Fig. 2), whereas the phylogenetically more distant tuf genes did not. We were interested to know how strains carrying the different viable tuf homologs were affected in each phase of the growth cycle. To do this, we measured length of lag phase, doubling time (DT) in exponential growth phase, and final optical density (OD) achieved in stationary phase. The parameters differed significantly between LB and DM25 (Davis minimal medium with 25 mg/liter glucose), with shorter lag times, shorter doubling times, and higher final ODs associated with growth in LB for each of the homologs (Table 1). The ranking of the parameters differed somewhat between LB and DM25. In rich medium, where growth rates are highest, the translation system, including EF-Tu, represents a major fraction of the bacterial mass, arguing that under these conditions any maladaptations will be more likely to reduce physiological fitness. In agreement, the strain carrying the native E. coli tuf gene had the shortest doubling time, shortest lag time, and maximum final OD in LB relative to strains carrying any of the other tuf homologs (Table 1). In the remainder of the text, we use doubling time in LB as a proxy for relative fitness. One striking indication that foreign tuf homologs can be maladapted is seen for tuf from V. cholerae, where the lag time in LB is 30- to 40-fold longer than that associated with any of the other tuf genes.
TABLE 1 .
Growth characteristics of strains harboring only one tuf gene
| tuf gene | Growth in medium, mean ± SDa |
|||||
|---|---|---|---|---|---|---|
| LB |
DM25 |
|||||
| Lag time (min) | Doubling time (min) | Max OD600 | Lag time (min) | Doubling time (min) | Max OD600 | |
| Escherichia coli | 6.8 ± 0.8 | 25.0 ± 0.3 | 1.39 ± 0.02 | 124.0 ± 3.0 | 81.7 ± 3.3 | 0.60 ± 0.01 |
| Yersinia enterocolitica | 8.2 ± 1.2 | 26.3 ± 0.4 | 1.39 ± 0.02 | 133.0 ± 8.6 | 80.0 ± 1.4 | 0.61 ± 0.01 |
| Vibrio cholerae | 300.2 ± 11.5 | 41.5 ± 2.2 | 1.38 ± 0.02 | 745.4 ± 36.1 | 134.5 ± 15.3 | 0.58 ± 0.05 |
| AnEF1 | 10.8 ± 1.5 | 48.4 ± 0.9 | 1.23 ± 0.02 | 73.1 ± 4.1 | 88.9 ± 3.2 | 0.53 ± 0.01 |
| Pseudomonas aeruginosa | 7.3 ± 5.5 | 66.9 ± 2.6 | 1.24 ± 0.02 | 87.7 ± 6.7 | 134.1 ± 3.0 | 0.63 ± 0.02 |
LB, Luria-Bertani broth; DM25, Davis minimal medium (25 mg/liter glucose). Each value represents the mean from 3 biological replicates.
Fitness correlates with EF-Tu protein levels and rate of protein synthesis.
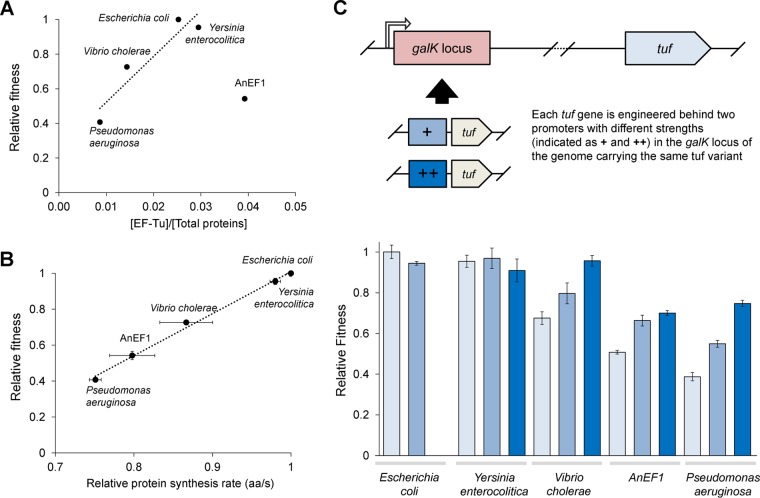
A key question is why the viable EF-Tu gene replacements reduce fitness. Two possibilities (which are not mutually exclusive) are that the foreign genes are suboptimally expressed (i.e., a concentration problem) and that they are suboptimal in their function and interaction with the protein synthesis machinery and cellular network (i.e., an activity or toxicity problem). To evaluate the possibility that the foreign genes were suboptimally expressed, we measured EF-Tu abundance by liquid chromatography-tandem mass spectrometry (LC-MS/MS) in each of the viable strains (Fig. 3A). This analysis demonstrated that the level of EF-Tu relative to total protein varied significantly between engineered strains carrying different foreign tuf genes (Fig. 3A). With the exception of the ancestral gene, AnEF1, there is a good correlation between relative fitness of the extant species’ genes and the concentration of EF-Tu (R2 value, 0.938). The mechanism resulting in these concentration differences is not known, but neither fitness nor concentration correlates with differences in codon usage between the tuf genes (Fig. S3). The proteomics data suggest that in most cases at least some of the reduction in relative fitness associated with foreign tuf genes is because bacteria do not produce an adequate level of EF-Tu to support fast growth. In the case of AnEF1, the reduced fitness may be more closely associated with reduced specific activity and/or toxicity.
FIG 3 .
Fitness characteristics of strains carrying single tuf genes. (A) Relative fitness of strains carrying a single tuf gene, at the tufA locus, as a function of EF-Tu protein produced. EF-Tu concentration was normalized to the total protein concentration. Species names indicate the species origin of the only tuf gene present (linear regression involves only extant sequences; R2 = 0.938). (B) Relative protein synthesis rate (amino acids/second) of the ribosome in each constructed bacterial strain (R2 = 0.994), determined by the time that it takes to produce β-galactosidase activity. Relative fitness of strains carrying a single tuf gene at the tufA locus correlates with the rate of protein synthesis. Results are means for three biological replicates with error bars representing standard deviation values. (C) Relative fitness of strains expressing each of the five tuf genes at three different levels: no additional tuf (light blue), additional tuf expression under promoter J23105 (medium blue), and additional tuf expression under promoter J23100 (dark blue). Promoter J23105 expresses the gene at a medium level (represented by +), whereas promoter J23100 expresses the gene at a higher level (represented by ++). Results are means for three biological replicates with error bars representing standard deviation values.
Codon adaptation index analysis. Species names indicate the species origin of the only tuf gene present. (A) Codon adaptation index as a function of the relative fitness of the strains. (B) Codon adaptation index as a function of the EF-Tu concentration normalized to the total protein content. Codon adaptation index was calculated with the CAI Calculator 2 using the Sharp and Li equation (P. M. Sharp and W.-H. Li, Nucleic Acids Res 15:1281–1295, 1987). Download FIG S3, PDF file, 0.3 MB (359.2KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
As an additional assay of the suboptimal functionality of foreign tuf genes as a possible reason for reduced fitness, we asked whether they supported a similar rate of protein synthesis as E. coli tufA. We constructed a set of isogenic strains suitable for measuring protein synthesis step time, using β-galactosidase as a model protein. Step time varied between the 5 different tuf genes, with the highest rate associated with tuf from E. coli and the lowest rate associated with tuf from P. aeruginosa (Fig. 3B). The data showed a very good correlation between protein synthesis step time and relative fitness (R2 value, 0.994).
Given the observed correlation between EF-Tu amount and relative fitness, we asked whether increasing the amount of EF-Tu would ameliorate the fitness costs. To test this, we engineered strains carrying these five tuf genes, placing an additional copy of each gene into the chromosome under the control of either of two different constitutive promoters, J23105 and J23100 (where J23100 is the stronger promoter [see Materials and Methods]). This created a set of isogenic strains putatively expressing each of the five tuf genes at three different levels. The exponential growth rate in LB was measured for each strain (Fig. 3C). For the three tuf genes associated with significantly reduced fitness relative to E. coli tufA (V. cholerae, AnEF1, and P. aeruginosa), overexpression was associated with a significant reduction in doubling time (higher growth rate) and in the case of V. cholerae reached a growth rate almost as high as that supported by the single copy of tufA in E. coli. In contrast, overexpression of Y. enterocolitica tuf, where a single copy already supported a growth rate close to that of E. coli tufA, did not further increase the growth rate (Fig. 3C). These data support the conclusion that an effectively low level of EF-Tu is one cause of the reduced fitness associated with foreign tuf genes.
Overall, these experiments show that the differences in relative fitness cost associated with different foreign tuf genes in E. coli correlate both with the amount of EF-Tu protein (measured by MS and overproduced from additional genes) and with the relative rate of protein synthesis supported by each tuf gene.
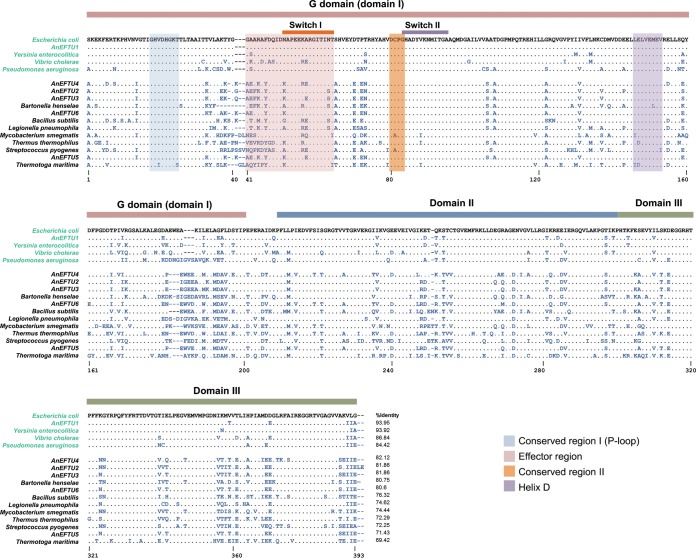
Conservation of EF-Tu residues correlates with viability in E. coli.
The established function of EF-Tu is to deliver aminoacylated tRNAs into the A site on mRNA-programmed ribosomes in order to drive rapid and accurate protein synthesis. This function requires that EF-Tu must have a sequence and structure that can efficiently interact with GTP, EF-Ts, each of the elongator tRNAs, and the ribosome. EF-Tu has a highly conserved structure which must be capable of undergoing major structural rearrangements during its functional cycle (40, 41). All 17 EF-Tu sequences were aligned with the EF-Tu sequence from E. coli (Fig. 4) to aid visualization of amino acid differences between EF-Tus associated with viability and those associated with nonviability. The four viable foreign EF-Tus (green in Fig. 4) show a bias toward a higher conservation of amino acid identity (84% to 94%), relative to E. coli EF-Tu, than the 12 nonviable EF-Tus (82% to 69%). It is interesting that there is only a small difference in total percent identity between several of the nonviable EF-Tus (AnEF2, AnEF3, AnEF4, AnEF6, and B. henselae, 80 to 82% similarity) and the most distantly related viable EF-Tu (P. aeruginosa, 84% similarity), suggesting that the loss of viability in these cases might be associated with changes in a few important residues of EF-Tu.
FIG 4 .
Alignment of EF-Tu protein sequences encoded by foreign tuf variants. EF-Tu sequences were aligned using Clustal Omega. Sequences labeled in green indicate that the foreign tuf gene supports viability as the only tuf gene in the genome. Sequences labeled in black indicate that the strain requires E. coli tufB for viability.
Connectivity analysis of EF-Tu reveals an extensive interaction network.
Several studies suggest that protein connectivity in a given network might modulate its activity in the cell (42–44). To examine this, we retrieved an E. coli interactome from the HitPredict database (45) and measured the connectivity of EF-Tu in this network. The connectivity of EF-Tu was quantified by measuring its so-called “degree centrality.” Degree centrality is defined as the number of interactions that a given node has in its network. The average degree centrality of all proteins in the E. coli interactome is 12, whereas the degree centrality of EF-Tu is much higher at 172. Relative to all proteins in the interactome, EF-Tu ranks among the top 10 most connected proteins in E. coli (Table S2). Interestingly, the set of its first interaction partners is enriched in essential proteins (P = 3.66 × 10−21) (Fig. S4). Accordingly, this analysis suggests the possibility that, in addition to affecting the specific activity of EF-Tu in protein synthesis, additional deleterious effects on fitness might be due to the foreign tuf homologs disturbing the extensive interaction network of EF-Tu.
The interaction partners of EF-Tu and all interactions among these first neighbors of EF-Tu. Nodes represent proteins (indicated by the relevant gene names), and each purple line represents an interaction. To easily distinguish tufA in the network, it has been colored cyan. Essential proteins are rectangular and colored blue in this subnetwork. Protein-protein interactions of E. coli were retrieved from the HitPredict database (A. Patil, K. Nakai, and H. Nakamura, Nucleic Acids Res 39:D744–D749, 2011, https://doi.org/10.1093/nar/gkq897), and the Cytoscape visualization tool was used to draw the network (B. Demchak et al., F1000Res 3:151, 2014, https://doi.org/10.12688/f1000research.4492.2). Download FIG S4, PDF file, 1 MB (1MB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The elongation factor EF-Tu is among the top 10 most connected proteins in the overall E. coli interactome. The proteins listed below are ranked according to their degree of connectivity using the UniProt database. The mean average degree of interaction in the whole interactome is 12. Download TABLE S2, PDF file, 0.1 MB (75.3KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
We generated E. coli strains in which the E. coli tufA gene was replaced by ancestral and modern homologs of tuf, from a broad spectrum of species and ancestral nodes. The origins of the ancestral EF-Tu sequences ranged in age from the Precambrian era, approximately 0.7 bya, back to the last universal common bacterial EF-Tu ancestor, approximately 3.6 bya (Fig. 1). We showed that homologs of EF-Tu encoded by tuf genes from within the gammaproteobacteria, including one of the reconstructed ancestral node sequences, AnEF1, are functionally active in E. coli and support viability when present as the only tuf gene in the chromosome (shown in green in Fig. 2). In contrast, more distantly related homologs and ancestral sequences were unable to support viability as the sole tuf gene. Among the four viable homologs, there was a good correlation (R2 = 0.9371, P = 0.032) between phylogenetic distance from E. coli EF-Tu and the magnitude of reduced growth fitness for the three homologs from extant species (Fig. 2). The exception was the reconstructed ancestral node sequence, AnEF1, where the decrease in relative fitness was much greater than that predicted by phylogenetic distance (Fig. 2). It could be argued that our definition of viability (ability to construct strains carrying the foreign tuf gene and in which the native tufB gene was removed) risks excluding some foreign genes because of technical difficulties in constructing the strains. However, the correlation that we observed between reduced relative fitness and reduced amounts of EF-Tu for the viable foreign genes is consistent with the hypothesis that even more distantly related foreign genes might be inviable because they produce an even smaller amount of EF-Tu to support growth. In addition, the observation that many of the “inviable” tuf genes were associated with duplication of the E. coli tufB region suggests that such strains were under significant selective pressure to compensate for low levels of EF-Tu, consistent with an inadequate level of active EF-Tu associated with the foreign tuf gene.
The correlation between phylogenetic distance from E. coli and relative growth fitness for the viable homologs raises the question of whether the underlying cause of the reduced fitness is a reduction in the specific activity of the foreign EF-Tus and/or a reduction in the amount of EF-Tu produced. By measuring EF-Tu protein concentration as a function of total protein concentration for each viable strain, including E. coli carrying only tufA, we observed a strong correlation between relative growth fitness and EF-Tu concentration for each of the four EF-Tus from extant species (Fig. 3). The slope of the correlation suggests that most of the reduced fitness associated with the EF-Tu homologs from the extant species could be attributed to a reduced level of EF-Tu rather than a reduced activity. Consistent with this, previous experiments have shown that reductions in growth rate are associated with reductions in cellular EF-Tu concentrations (46). Once again, the exception to this correlation is found for the reconstructed ancestral AnEF1, where a very high concentration of EF-Tu was associated with a low relative fitness (Fig. 3A), consistent with AnEF1 having a low specific activity. This conclusion is supported by the results of in vitro translation, where in a system containing only T. thermophilus components (tRNAs, EF-Ts, and ribosomes) AnEF1 supported protein synthesis but with only 30% of the activity of native E. coli EF-Tu, demonstrating that AnEF1 can participate in peptide synthesis, albeit in a diminished fashion relative to its modern counterpart EF-Tu (47). To address the significance of the correlation between protein amount and relative fitness, we also overexpressed each of the five viable tuf genes (Fig. 3C). We found, in each case where a foreign tuf gene was associated with reduced fitness, that the growth rate could be significantly increased by overexpressing the gene. We also found that the step time of protein synthesis varied between the different tuf genes and correlated with the relative fitness of each strain. Taken together, these data support the hypothesis that suboptimal expression is a significant cause of the reduced growth fitness associated with the foreign tuf genes. However, the data do not rule out the possibility that at least some of the effects are associated with a poor integration of the foreign EF-Tu into the network of interactions that support optimal growth. In principle, reduced fitness associated with a change in specific activity (for example, a weakened interaction) could also be reversed by an increase in the intracellular concentration of EF-Tu. To tease apart the relative contributions to fitness of intracellular EF-Tu concentration from the specific activity of each foreign EF-Tu will require extensive biochemical analysis and is beyond the scope of this paper.
The overexpression of EF-Tu, by recombineering an additional copy at a second chromosomal location, helped answer a question concerning the significance of tufB region amplification in many of the strains carrying “inviable” foreign tuf genes (Fig. 2). The strains with tufB amplified have relatively high fitness values, but their foreign tuf genes were subsequently found to be inviable. The question is whether the amplification (which probably includes all genes in a 40-kb region between rrnB and rrnE) is selected because it increases the copy number of tuf genes in the cell or because it increases the copy number of some other gene in the amplified region that increases relative fitness. By constructing strains in which the expression of low-fitness foreign tuf genes was individually increased, without increasing the copy number of any other genes, and finding that relative fitness increased as a result, we could conclude that at least in these cases an increase in tuf expression was sufficient to increase fitness. It seems reasonable to conclude that the amplification of the region containing tufB was most probably also selected because it increased the intracellular concentration of EF-Tu, thus improving fitness.
Why, given that the tuf gene regulatory regions are identical in all strains, do the foreign tuf coding sequences cause a reduction in the level of EF-Tu produced? Possible explanations include that the nucleotide sequence introduced with the foreign genes affects tuf mRNA half-life as previously shown for a mutant of tuf (48) or that the altered nucleotide sequence reduces translation efficiency, possibly by effects mediated through altered codon usage or by affecting transcription-translation coupling, as recently shown for tufB (30, 49). No correlation was found between differences in the codon usage of the foreign tuf genes and the amounts of EF-Tu produced (see Fig. S3 in the supplemental material).
An important question is why some of the not-too-distant foreign homologs are unable to support viability. At the level of total amino acid similarity, there is very little separating viable from nonviable EF-Tu sequences, with the boundary falling at approximately 84% similarity to E. coli EF-Tu (Fig. S2). Given the very high level of conservation of EF-Tu, it is possible that the nonviability of some foreign EF-Tus might be related to the alteration of just one or a few critically important residues. Indeed, many single-amino-acid substitutions in EF-Tu have been shown to generate protein variants that do not support viability (50), including one single-amino-acid substitution in EF-Tu that permits ternary complex formation but abolishes translation activity by preventing ternary complex interaction with the ribosome (46). To facilitate an assessment of the amino acid differences between viable and nonviable EF-Tus, their amino acid sequences were aligned (Fig. 4). Of the 393 residues in E. coli EF-Tu, there were 116 residues that were identical in all of the viable EF-Tu homologs but differed in at least one of the 12 nonviable homologs. Variation at one or more of these residues might explain the difference between EF-Tu viability and nonviability in E. coli. The 116 residues were distributed between each of the three structural domains of EF-Tu, with 48 in the G domain, 38 in domain 2, and 30 in the C-terminal domain 3 (Fig. 4 and S5). Several of the 116 variant residues lie within functionally important regions of EF-Tu, including those involved in coordinating GTP hydrolysis, interaction with EF-Ts, and interaction with the ribosome (51–53). For example, in each of the nonviable EF-Tus there were from 3 to 8 variant residues in regions of the G domain that are known to be functionally important for coordinating the hydrolysis of GTP on EF-Tu during protein synthesis (Fig. S5). These variant residues are located in the conserved regions I and II, the effector region, and the switch I and switch II loops of the G domain of EF-Tu (Fig. S5). The altered residues potentially affecting GTP hydrolysis are at V20, G41, A43, R44, F46, N51, N63, T64, C81, and V88 (Fig. 4). In addition, there are alterations in the P loop, the switch II region, and parts of domain 3 that are involved in interactions with EF-Ts and in helix D of EF-Tu (residues 144 to 156), which is involved in interactions with EF-Ts and protein L7/L12 on the ribosome (52, 53). We can only speculate on the exact reason for the nonviability of these EF-Tu homologs. It seems unlikely that it is directly related to defects in binding or hydrolyzing GTP, given that this process involves highly conserved residues and structures and that the EF-Tus from extant organisms must be capable of supporting viability, including GTP binding and hydrolysis, in their natural system. Similarly, each of the ancestral homologs can support in vitro translation, albeit at a low efficiency, arguing that they also can bind and hydrolyze GTP. A similar line of reasoning could also rule out interactions with aa-tRNAs as the cause of nonviability. Perhaps the most plausible reason for nonviability is defective interactions with EF-Ts and/or the ribosome. It seems reasonable to suggest that EF-Tu has coevolved with EF-Ts and the ribosome to modulate the efficiency of these interactions in each species. We suggest accordingly that the cause of nonviability for distantly related EF-Tus is not that they cannot function as enzymes capable of forming a ternary complex and hydrolyzing GTP but rather that they are defective in one or more of the other important interactions made by EF-Tu, namely, with EF-Ts, with the mRNA-programmed ribosome, and possibly even interactions outside protein synthesis involving one or more members of EF-Tu’s extensive protein interaction network (Fig. S4). Coevolution of EF-Tu with its interaction partners would create a barrier to transfer for EF-Tus beyond a certain threshold.
Structural representation of EF-Tu-GDP. EF-Tu is shaded gray, and residues that are conserved in all viable homologs but changed in at least one of the nonviable homologs are colored. (A) Changes within the G domain are colored red, those in domain II are colored yellow, and those in domain III are colored blue. (B) A close-up view of the G domain where the change in the P loop is shown in green, changes in the effector/switch I region are shown in orange, the change in the switch II region is shown in purple, and changes in the D-helix are shown in blue. GDP is shown in red, and the Mg ion is shown as a cyan sphere. Graphic analyses were performed with the UCSF Chimera package (E. F. Pettersen et al., J Comput Chem 25:1605–1612, 2004, https://doi.org/10.1002/jcc.20084) using the Protein Data Bank identifier 1EFC (H. Song, M. R. Parsons, S. Rowsell, G. Leonard, and S. E. Phillips. J Mol Biol 285:1245–1256, 1999). Download FIG S5, PDF file, 0.8 MB (852.7KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We previously hypothesized that evolutionary novelties are more likely to be shared between a descendant and its ancient homolog than between two currently existing protein homologs (54). Accordingly, replacing an existing gene with its ancient homolog may have a smaller negative fitness impact on the organism relative to exchanging the native gene with a currently existing homolog. However, functional divergence occurring through time could result in ancestral sequences being so maladapted to the new host cell that a functional organism is all but precluded (55). This limitation does not apply only to ancestral genes, and it has been suggested that as the number of edges connecting a protein within its protein-protein interaction network increases, the probability that a protein could be successfully replaced with a homolog will decrease even if there is a functional equivalence between the endogenous gene and the homolog (2). While a careful assessment of candidate ancestral protein properties prior to integration is helpful, in most cases, studying gene-triggered genomic perturbations experimentally through the integration of ancestral genes offers a valuable and complementary alternative to existing methodologies that use extant homologous proteins (56–59).
How can we identify the specific historical constraints on replacement? We observe that only the ancient EF-Tu representing an ancestor within the gammaproteobacteria, AnEF1 (0.7 bya), and the modern EF-Tu homologs from extant gammaproteobacteria are viable. In contrast, the last common ancestor of the alpha-, beta-, and gammaproteobacteria, AnEF6 (1.3 bya), is nonviable. Accordingly, we speculate that mutational substitutions in EF-Tu occurring between 1.6 bya and 0.7 bya influenced the replaceability of tuf genes. These mutations may constrain tuf replaceability by disturbing EF-Tu’s functional interaction with other cellular components, ultimately impacting its participation in protein synthesis. Thus, extensive mutational remodeling of interaction partners may be necessary in order to engineer even older ancient tuf genes inside the bacteria.
Conclusions.
We show that foreign tuf genes encoding EF-Tu proteins exhibit suboptimal functionality and reduced fitness when introduced into another host. The suboptimality of the foreign tuf genes most likely results from disturbances in interactions directly important for protein synthesis, but suboptimal EF-Tu protein levels and disturbance of other potentially important interactions in the network of EF-Tu might also play a role. The observation that the only tuf homologs that supported viability belong to the gammaproteobacterial taxon, or an associated ancestral node within the gammaproteobacteria, suggests that there is a relatively stringent “transferability cutoff,” i.e., a point in the phylogeny beyond which functional divergence is too great for replacement. For EF-Tu protein, this transferability zone is within the ancestral and modern gammaproteobacterial taxon, unlike some ribosomal proteins where constraints on replaceability are less stringent (22, 60).
Future efforts may involve identifying protein sites that interfere with organismal-level function and epistatically inhibit an ancient protein’s function in a descendant organism. Our experiments suggest that a protein like EF-Tu, which is highly conserved and involved in multiple highly conserved interactions, is so highly optimized and finely tuned in the host organism that it is essentially irreplaceable by distantly related foreign genes. The degree to which epistatic interactions constrain EF-Tu replaceability and functionality in the cell needs to be studied more to deepen our understanding of the design principles of complex biological systems and to allow us to introduce alterations in modern organisms by genetic engineering and gene replacements.
MATERIALS AND METHODS
Media and growth conditions.
In general, liquid and solid media used were Luria-Bertani (LB) medium (per liter, 10 g NaCl [5 g in the case of low-salt LB], 5 g yeast extract, and 10 g tryptone) and LA (LB with 1.5% agar) plates. Where indicated, growth in minimal medium was made in Davis minimal (DM) medium [per liter, 7 g K2HPO4, 2 g KH2PO4, 1 g (NH4)2SO4, 0.5 g sodium citrate]. For sacB counterselections, the LB and LA media used had no NaCl, and LA medium was also supplemented with 5% sucrose. All incubations were made at 37°C (unless stated otherwise), and liquid cultures were shaken at 200 rpm for aeration. The antibiotics used (Sigma-Aldrich, Sweden) had the following final concentrations: 50 mg/liter kanamycin and 15 mg/liter tetracycline.
Growth parameter measurements.
To measure doubling time, lag time, and maximum OD at 600 nm (OD600) of the strains in LB and DM25 (DM with 25 g/liter glucose), 3 independent cultures of each strain were grown overnight in LB. For each culture, 2 aliquots of 400 µl were washed with NaCl2 to remove any traces of the medium and then resuspended in 400 µl of LB or DM25, respectively. Subsequently, 280 µl of a 100-fold dilution of every aliquot was added to 3 wells of the microtiter honeycomb plate, as technical replicates. Control wells were filled with 280 µl of only LB or DM25. Bacterial growth was monitored by measuring the rate of increase in optical density at OD600 using a Bioscreen C machine (Oy Growth Curves Ab Ltd., Finland) and growing the cultures in the honeycomb microtiter plate. Plates were incubated for 24 h at 37°C with continuous shaking, and readings of OD600 were taken at 5-min intervals. All data points were corrected by subtracting the OD600 of the corresponding control wells (medium only) at every time point and then converted to log values. The doubling time (DT) of each strain during exponential growth was calculated over an interval of 50 min (10 time points) in the linear region of the curve by calculating the slope of the interval using the following equation:
| (1) |
Lag times were estimated over the same time interval using the following equation:
| (2) |
Maximum OD600 was defined as the maximum optical density over 24 h of monitored growth. Relative fitness, defined in terms of doubling time in LB, was calculated by comparing each independent doubling time measurement to the average of doubling time measurements of the strain carrying E. coli tufA.
Bacterial strains.
All strains used in this study are derived from sequenced wild-type E. coli K-12 strain MG1655 (61), unless stated otherwise. A list of the strains used in these experiments is shown in Table S3 in the supplemental material.
Strains and genetic markers used in this work and their origin. Download TABLE S3, PDF file, 0.1 MB (92.6KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strain construction.
The genetic marker (TP22-amilCP_opt-kan-sacB-T0) was inserted into the chromosome to replace either tufA or tufB by double-stranded DNA lambda-red recombineering (62, 63). The lambda-red genes were induced in each strain from the temperature-sensitive pSIM5-tet plasmid by incubation of an overday culture (OD600 of 0.3) at 43°C for 15 min. After cooling for 10 min in ice, the cells were made electrocompetent by washing in ice-cold water three times. Electroporation of TP22-amilCP_opt-kan-sacB-T0 PCR product was done with a Gene Pulser (Bio-Rad, USA) by mixing 50 µl of electrocompetent cells and 100 ng of the cassette, with settings of 1.8 kV, 25 µF, and 200 Ω. Cells were recovered in 1 ml of low-salt LB at 30°C overnight with agitation, and after recovery, 100 µl of the culture was spread in LA plates containing kanamycin for selection of recombinants.
Genetic markers were moved between strains by phage-mediated (P1 virA) transduction. Lysates of the strains carrying the marker were made by mixing 1 ml of overnight culture containing 5 mM CaCl2 with 100 µl of the P1 virA lysate previously made on E. coli MG1655. The bacterium-phage mix was incubated for 10 min, and then 4 ml of soft agar (LB medium plus 0.8% agar plus 5 mM CaCl2) was added; this mixture was spread over an LA plate and incubated overnight. To release the bacteriophages, the soft agar was mixed with 4 ml and vortexed. The resultant slurry was centrifuged for 15 min at 5,000 rpm, and the supernatant was filtered through an 0.2-µm filter. Markers were transduced into the desired recipient strain by mixing 100 µl of the lysate with 500 µl of an overnight culture containing 5 mM CaCl2. After 10 min of incubation, 100 µl of the mixture was spread onto a selective plate.
Insertions of alien tuf genes were made by double-stranded DNA lambda-red recombineering to replace the previously inserted counterselectable TP22-amilCP_opt-kan-sacB-T0 marker. The accession numbers and/or references for the foreign tuf genes are given in Table S4 in the supplemental material. To make clean deletions, markers were deleted by single-stranded lambda-red recombineering with counterselection for sucrose resistance (64), following the same steps as described above for the double-stranded DNA lambda-red recombineering.
List of organisms and plasmids serving as EF-Tu sources. Download TABLE S4, PDF file, 0.1 MB (72.6KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To construct the strains expressing an extra copy of the tuf, gene, two promoters of different strengths (J23105 and J23100) (iGEM Registry of Standard Biological Parts, Cambridge, MA; http://partsregistry.org) transcriptionally fused to a cat-sacB cassette at the galK locus were transduced into strains carrying the relevant single tuf gene. Coding sequences for the tuf genes were amplified from each of the strains carrying a single tuf gene and engineered by recombineering behind each of the two promoters replacing the cat-sacB cassette. Constructs were confirmed by PCR and DNA sequencing (Macrogen Europe Laboratory, Amsterdam, The Netherlands).
Ancestral gene reconstruction.
Ancestral sequences used in this study originated from the study performed by Gaucher et al. (35). Briefly, the EF-Tu sequences were retrieved from GenBank database, and the phylogenetic tree was constructed with MrBayes (65). Ancestral sequences were calculated with PAML (66).
PCR and oligonucleotides.
PCR was performed on an S1000 Thermal Cycler (Bio-Rad, USA). Oligonucleotides were designed with the software CLC Main workbench 7 (CLC bio, Denmark) using the genome of E. coli MG1655 as reference. For generation of the TP22-amilCP_opt-kan-sacB-T0 cassette, PCR was performed using Phusion High-Fidelity PCR master mix with HF buffer (New England Biolabs, USA) and with the following cycling conditions: 98°C for 30 s and 30 cycles of 98°C for 10 s, 55°C for 30 s, 72°C for 4 min, and 72°C for 7 min. For routine diagnostic PCR, Fermentas PCR master mix (Thermo Scientific, USA) was used with the following cycling conditions: 95°C for 5 min and 30 cycles of 95°C for 30 s, annealing temperature (TA) for 30 s, 72°C for elongation time (ET), and 72°C for 5 min. TA varied depending on the pair of primers used, and ET was based on the length of the expected product (30 s per kilobase). Oligonucleotides for construction of strains, PCR, and sequencing are shown in Table S5.
Oligonucleotide primers used in this study. Download TABLE S5, PDF file, 0.1 MB (64KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Preparation of genomic DNA and real-time quantitative PCR.
Genomic DNA prepared using the MasterPure DNA purification kit (Epicentre Biotechnologies, USA) was used to run real-time quantitative PCR. One microliter genomic DNA (gDNA) (diluted 1:10, 1:100, 1:1,000, and 1:10,000), 10 μl PerfeCTa SYBR green FastMix (Quanta Biosciences), 0.6 μl of 10 μM forward and reverse primers, and double-distilled water (ddH2O) were added to a final reaction volume of 20 μl. The Eco real-time PCR system (Illumina) was used for running the PCR. Oligonucleotides amplifying the rpoB and purD genes were used to quantify the amplification status of the rrnB-rrnE region. The cysG and indT genes were used as controls. The oligonucleotide sequences used as RT-qPCR primers are listed in Table S5.
Whole-genome sequencing and analysis.
Genomic DNA was prepared using the MasterPure DNA purification kit (Epicentre Biotechnologies, USA). To create libraries of paired-end fragments, the Nextera XT sample preparation kit (Illumina, USA) was used according to the instructions from the manufacturer. Sequencing was performed on the Illumina MiSeq instrument, generating 250-bp paired-end reads. Whole-genome sequencing data were analyzed using the CLC Genomic Workbench software (CLC bio, Denmark).
Local DNA sequencing.
Local sequencing of PCR-amplified products was performed at the Macrogen Europe sequencing facilities (Amsterdam, The Netherlands), and data were analyzed using the CLC Main Workbench 7 software (CLC bio, Denmark).
Protein synthesis rate measurements (step time).
Liquid cultures were initiated from overnight cultures grown in LB to mid-log phase at 37°C with shaking by diluting 1:100 in 20 ml DM, with 0.2% glycerol. Before induction, a time zero sample (200 μl) was taken and added to 300 μl ice-cold chloramphenicol (0.5 mg/ml in 1:1 H2O-ethanol). Expression of lacZ from the F′23 plasmid was induced by the addition of 200 ml IPTG (isopropyl-β-d-thiogalactopyranoside; 0.1 M; final concentration, 1 mM). Samples (200 μl) were taken every 20 s after induction for 300 s and added to 300 μl chloramphenicol solution. Cells were pelleted by centrifugation (3 min, 12,000 × g) and resuspended in 300 μl Z buffer (0.06 M Na2HPO4⋅2H2O, 0.04 M NaH2PO4⋅H2O, 0.1 M KCl, 0.001 M MgSO4⋅7H2O, 0.05 M β-mercaptoethanol). To each sample, 100 μl chloroform and 50 μl 0.1% SDS were added. The tubes were vortexed and left on ice for 20 min to allow the chloroform to sink before 200 μl of each sample was added to a honeycomb plate with 40 μl o-nitrophenyl-β-d-thiogalactopyranoside (ONPG; 4 mg/ml) added per well. The plate was incubated with shaking in a Bioscreen C machine (Oy Growth Curves AB Ltd.), and absorbance at 420 nm and 540 nm was measured. Background absorbance (ONPG in Z buffer without cells) and absorbance at time zero were subtracted, and the data were plotted with √[(OD420) − (1.75 × OD540)] as a function of time. The intercept with the x axis of the induced curve is the step time, the time that it takes to produce the first β-galactosidase activity. To calculate the protein synthesis rate in amino acids/second, the length of the β-galactosidase (1,024 amino acids) is divided by the step time of each strain. The relative protein synthesis rate was determined in each case by comparing the protein synthesis rate to the average of all measurements of the strain carrying E. coli tuf.
Statistical analysis.
All statistical analyses were performed using GraphPad Prism v6.0c (GraphPad Software, Inc., USA). The significance of differences between fitness costs was calculated using an unpaired two-tailed t test.
Proteomics: cell lysis, digestion, and labeling procedures.
All cells were placed into Covaris microTUBE-15 (Woburn, MA) microtubes with Covaris TPP buffer. Samples were lysed in a Covaris S220 focused ultrasonicator instrument with 125-W power over 180 s with 10% maximum peak power. Lysed cells were digested via filter-aided sample preparation (FASP) digest according to the FASP protocol for trypsin digestion, followed by high-pressure liquid chromatography (HPLC) purification. We used Promega sequencing-grade trypsin-LysC (V5073; Madison, WI) overnight at 38°C. Each sample was submitted for a single LC-MS/MS experiment that was performed on an LTQ Orbitrap Elite (Thermo Fisher) equipped with a Waters (Milford, MA) NanoAcquity HPLC pump or Orbitrap Lumos (Thermo Fisher, San Jose, CA) equipped with EasyLC1000 (Thermo Fisher, San Jose, CA). Peptides were separated onto a 100-µm-inner-diameter microcapillary trapping column packed first with approximately 5 cm of C18 Reprosil resin (5 µm, 100 Å; Dr. Maisch GmbH, Germany), followed by an analytical column with ~20 cm of Reprosil resin (1.8 µm, 200 Å; Dr. Maisch GmbH, Germany). Separation was achieved by applying a gradient from 5 to 27% acetonitrile (ACN) in 0.1% formic acid over 90 min at 200 nl min−1. Electrospray ionization was enabled by applying a voltage of 1.8 kV using a homemade electrode junction at the end of the microcapillary column and spraying from fused silica pico tips (New Objective, MA). The LTQ Orbitrap Elite/Lumos was operated in data-dependent mode for the mass spectrometry methods. The mass spectrometry survey scan was performed in the Orbitrap in the range of 395 to 1,800 m/z at a resolution of 6 × 104, followed by the selection of the 20 most intense ions (TOP20) for collision-induced dissociation (CID)–MS2 fragmentation in the ion trap using a precursor isolation width window of 2 m/z, an AGC (automatic gain control) setting of 10,000, and a maximum ion accumulation of 200 ms. Singly charged ion species were not subjected to CID fragmentation. Normalized collision energy was set to 35 V and an activation time of 10 ms. Ions in a 10-ppm m/z window around ions selected for MS2 were excluded from further selection for fragmentation for 60 s. The same TOP20 ions were subjected to an HCD (Higher-energy collisional dissociation) MS2 event in the Orbitrap part of the instrument. The fragment ion isolation width was set to 0.7 m/z, the AGC was set to 50,000, the maximum ion time was 200 ms, normalized collision energy was set to 27 V, and an activation time of 1 ms for each HCD MS2 scan was used.
Mass spectrometry analysis.
Raw data were submitted for analysis in Proteome Discoverer 2.1.0.81 (Thermo Scientific) software. Assignment of MS/MS spectra was performed using the Sequest HT algorithm by searching the data against a protein sequence database including all entries from the user database and our E. coli K-12 database as well as other known contaminants such as human keratins and common lab contaminants. Sequest HT searches were performed using a 20-ppm precursor ion tolerance and requiring each peptide’s N/C termini to adhere with trypsin protease specificity, while allowing up to two missed cleavages. Six-plex tandem mass tags (TMTs) on peptide N termini and lysine residues (+229.162932 Da) were set as static modifications, while methionine oxidation (+15.99492 Da) was set as a variable modification. An MS2 spectrum assignment false discovery rate (FDR) of 1% on the protein level was achieved by applying the target-decoy database search. Filtering was performed using a 64-bit Percolator. For quantification, an 0.02 m/z window was centered on the theoretical m/z value of each of the six reporter ions and the intensity of the signal closest to the theoretical m/z value was recorded. Reporter ion intensities were exported in a result file of the Proteome Discoverer 2.1 search engine as an Excel table.
Evolutionary divergence.
Two independent approaches were utilized in order to estimate the evolutionary distance between sequences, both leading to the same evolutionary distance estimate output. (i) For MEGA software, the analyses involved 18 sequences. Analyses were conducted using the Poisson correction model (67). All positions containing gaps and missing data were eliminated. There were a total of 385 positions in the final data set. Evolutionary analyses were conducted in MEGA7 (68). (ii) The branch length distances were also calculated via a custom script that used ETE software (69). The custom Python ETE v.3 Python library script is provided in the supplemental material.
Computational methods.
Computational analyses include (i) the pairwise and multiple sequence alignments, (ii) protein structure analysis, and (iii) protein interaction network analysis. Clustal Omega (70) was used to perform multiple sequence alignment with default parameters. Pairwise sequence alignments of the wild-type EF-Tu against each ancient and modern EF-Tu protein were performed by EMBOSS Needle of Clustal Omega with default parameters, which uses the Needleman-Wunsch alignment algorithm.
The protein interactome of E. coli was retrieved from the HitPredict (45) database, and the network analysis was performed with the network package of Python (71). The subnetwork of EF-Tu interaction partners was visualized with Cytoscape (72).
ACKNOWLEDGMENTS
We thank S. Andersson, B. K. Hammer, E. A. Gaucher, and L. Guy for providing the DNA sequences for some of the EF-Tu homologs used in this study.
This work was supported by a NASA Astrobiology Postdoctoral Fellowship (B.K.); John Templeton Foundation grant number 58562 (B.K.); grants from the Swedish Research Council (D.H. and D.I.A.), the Knut and Alice Wallenberg Foundation, and the RiboCORE project (D.I.A. and D.H.); and Carl Trygger Foundation grant number CTS16:194 (D.H.).
N.T. thanks the TUBITAK-Marie Curie Co-funded Brain Circulation Scheme (114C026) and the Young Scientist Award Programme (BAGEP) of the Turkish Science Academy for support.
The opinions expressed in this publication are those of the authors and do not necessarily reflect the views of any particular organization. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Kacar B, Garmendia E, Tuncbag N, Andersson DI, Hughes D. 2017. Functional constraints on replacing an essential gene with its ancient and modern homologs. mBio 8:e01276-17. https://doi.org/10.1128/mBio.01276-17.
REFERENCES
- 1.Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. 2007. Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318:1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- 2.Jain R, Rivera MC, Lake JA. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci U S A 96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitahara K, Miyazaki K. 2013. Revisiting bacterial phylogeny: natural and experimental evidence for horizontal gene transfer of 16S rRNA. Mob Genet Elements 3:e24210. doi: 10.4161/mge.24210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wellner A, Lurie MN, Gophna U. 2007. Complexity, connectivity, and duplicability as barriers to lateral gene transfer. Genome Biol 8:R156. doi: 10.1186/gb-2007-8-8-r156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen O, Gophna U, Pupko T. 2011. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol Biol Evol 28:1481–1489. doi: 10.1093/molbev/msq333. [DOI] [PubMed] [Google Scholar]
- 6.Jain R, Rivera MC, Moore JE, Lake JA. 2003. Horizontal gene transfer accelerates genome innovation and evolution. Mol Biol Evol 20:1598–1602. doi: 10.1093/molbev/msg154. [DOI] [PubMed] [Google Scholar]
- 7.Smith MW, Feng DF, Doolittle RF. 1992. Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem Sci 17:489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- 8.Kaçar B, Gaucher EA. 2013. Experimental evolution of protein-protein interaction networks. Biochem J 453:311–319. doi: 10.1042/BJ20130205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen P, Huang HV. 1986. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112:441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blank D, Wolf L, Ackermann M, Silander OK. 2014. The predictability of molecular evolution during functional innovation. Proc Natl Acad Sci U S A 111:3044–3049. doi: 10.1073/pnas.1318797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bershtein S, Serohijos AW, Bhattacharyya S, Manhart M, Choi JM, Mu W, Zhou J, Shakhnovich EI. 2015. Protein homeostasis imposes a barrier on functional integration of horizontally transferred genes in bacteria. PLoS Genet 11:e1005612. doi: 10.1371/journal.pgen.1005612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. 2008. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol 190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A 102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zotenko E, Mestre J, O’Leary DP, Przytycka TM. 2008. Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput Biol 4:e1000140. doi: 10.1371/journal.pcbi.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coulomb S, Bauer M, Bernard D, Marsolier-Kergoat MC. 2005. Gene essentiality and the topology of protein interaction networks. Proc Biol Sci 272:1721–1725. doi: 10.1098/rspb.2005.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito K, Uno M, Nakamura Y. 1998. Single amino acid substitution in prokaryote polypeptide release factor 2 permits it to terminate translation at all three stop codons. Proc Natl Acad Sci U S A 95:8165–8169. doi: 10.1073/pnas.95.14.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lind PA, Andersson DI. 2013. Fitness costs of synonymous mutations in the rpsT gene can be compensated by restoring mRNA base pairing. PLoS One 8:e63373. doi: 10.1371/journal.pone.0063373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karcagi I, Draskovits G, Umenhoffer K, Fekete G, Kovács K, Méhi O, Balikó G, Szappanos B, Györfy Z, Fehér T, Bogos B, Blattner FR, Pál C, Pósfai G, Papp B. 2016. Indispensability of horizontally transferred genes and its impact on bacterial genome streamlining. Mol Biol Evol 33:1257–1269. doi: 10.1093/molbev/msw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 20.Brochier C, Philippe H, Moreira D. 2000. The evolutionary history of ribosomal protein RpS14: horizontal gene transfer at the heart of the ribosome. Trends Genet 16:529–533. doi: 10.1016/S0168-9525(00)02142-9. [DOI] [PubMed] [Google Scholar]
- 21.Lind PA, Berg OG, Andersson DI. 2010. Mutational robustness of ribosomal protein genes. Science 330:825–827. doi: 10.1126/science.1194617. [DOI] [PubMed] [Google Scholar]
- 22.Lind PA, Tobin C, Berg OG, Kurland CG, Andersson DI. 2010. Compensatory gene amplification restores fitness after inter-species gene replacements. Mol Microbiol 75:1078–1089. doi: 10.1111/j.1365-2958.2009.07030.x. [DOI] [PubMed] [Google Scholar]
- 23.Knöppel A, Näsvall J, Andersson DI. 2016. Compensating the fitness costs of synonymous mutations. Mol Biol Evol 33:1461–1477. doi: 10.1093/molbev/msw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller DL, Weissbach H. 1977. Factors involved in the transfer of aminoacyl-tRNA to the ribosome, p 323–373. In Weissbach H, Petska S (ed), Molecular mechanisms of protein biosynthesis. Academic Press, New York, NY. [Google Scholar]
- 25.Lathe WC III, Bork P. 2001. Evolution of tuf genes: ancient duplication, differential loss and gene conversion. FEBS Lett 502:113–116. doi: 10.1016/S0014-5793(01)02639-4. [DOI] [PubMed] [Google Scholar]
- 26.Baldauf SL, Palmer JD, Doolittle WF. 1996. The root of the universal tree and the origin of eukaryotes based on elongation factor phylogeny. Proc Natl Acad Sci U S A 93:7749–7754. doi: 10.1073/pnas.93.15.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdulkarim F, Hughes D. 1996. Homologous recombination between the tuf genes of Salmonella typhimurium. J Mol Biol 260:506–522. doi: 10.1006/jmbi.1996.0418. [DOI] [PubMed] [Google Scholar]
- 28.Tubulekas I, Hughes D. 1993. Growth and translation elongation rate are sensitive to the concentration of EF-Tu. Mol Microbiol 8:761–770. doi: 10.1111/j.1365-2958.1993.tb01619.x. [DOI] [PubMed] [Google Scholar]
- 29.Kacar B, Xueliang G, Sanyal S, Gaucher EA. 2017. Experimental evolution of modern Escherichia coli harboring an ancient elongation factor gene. J Mol Evol 84:69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandis G, Bergman JM, Hughes D. 2016. Autoregulation of the tufB operon in Salmonella. Mol Microbiol 100:1004–1016. doi: 10.1111/mmi.13364. [DOI] [PubMed] [Google Scholar]
- 31.Hsiao C, Mohan S, Kalahar BK, Williams LD. 2009. Peeling the onion: ribosomes are ancient molecular fossils. Mol Biol Evol 26:2415–2425. doi: 10.1093/molbev/msp163. [DOI] [PubMed] [Google Scholar]
- 32.Pauling L, Zuckerkandl E, Henriksen T, Lövstad R. 1963. Chemical paleogenetics. Molecular “restoration studies” of extinct forms of life. Acta Chem Scand 17(Suppl):S9–S16. doi: 10.3891/acta.chem.scand.17s-0009. [DOI] [Google Scholar]
- 33.Thornton JW. 2004. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat Rev Genet 5:366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 34.Benner SA, Sassi SO, Gaucher EA. 2007. Molecular paleoscience: systems biology from the past. Adv Enzymol Relat Areas Mol Biol 75:1–132. doi: 10.1002/9780471224464.ch1. [DOI] [PubMed] [Google Scholar]
- 35.Gaucher EA, Govindarajan S, Ganesh OK. 2008. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature 451:704–707. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- 36.Kacar B. 2016. Rolling the dice twice: evolving reconstructed ancient proteins in extant organisms, p 264–276. In Ramsey G, Pence CH (ed), Chance in evolution from Darwin to contemporary biology. University of Chicago Press, Chicago, IL. [Google Scholar]
- 37.Tortolero-Luna G, Mitchell MF. 1995. The epidemiology of ovarian cancer. J Cell Biochem 59:200–207. doi: 10.1002/jcb.240590927. [DOI] [PubMed] [Google Scholar]
- 38.Battistuzzi FU, Feijao A, Hedges SB. 2004. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol 4:44. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson P, Roth J. 1981. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc Natl Acad Sci U S A 78:3113–3117. doi: 10.1073/pnas.78.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H, Chan YL, Wool IG. 2009. The identification of the determinants of the cyclic, sequential binding of elongation factors tu and g to the ribosome. J Mol Biol 386:802–813. doi: 10.1016/j.jmb.2008.12.071. [DOI] [PubMed] [Google Scholar]
- 41.Nilsson J, Nissen P. 2005. Elongation factors on the ribosome. Curr Opin Struct Biol 15:349–354. doi: 10.1016/j.sbi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 42.Hahn MW, Conant GC, Wagner A. 2004. Molecular evolution in large genetic networks: does connectivity equal constraint? J Mol Evol 58:203–211. doi: 10.1007/s00239-003-2544-0. [DOI] [PubMed] [Google Scholar]
- 43.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. 2002. Evolutionary rate in the protein interaction network. Science 296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 44.Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. 2005. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol Biol Evol 22:1345–1354. doi: 10.1093/molbev/msi122. [DOI] [PubMed] [Google Scholar]
- 45.Patil A, Nakai K, Nakamura H. 2011. HitPredict: a database of quality assessed protein-protein interactions in nine species. Nucleic Acids Res 39:D744–D749. doi: 10.1093/nar/gkq897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tubulekas I, Hughes D. 1993. A single amino acid substitution in elongation factor Tu disrupts interaction between the ternary complex and the ribosome. J Bacteriol 175:240–250. doi: 10.1128/jb.175.1.240-250.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y, Asahara H, Gaucher EA, Chong S. 2012. Reconstitution of translation from Thermus thermophilus reveals a minimal set of components sufficient for protein synthesis at high temperatures and functional conservation of modern and ancient translation components. Nucleic Acids Res 40:7932–7945. doi: 10.1093/nar/gks568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammarlöf DL, Hughes D. 2008. Mutants of the RNA-processing enzyme RNase E reverse the extreme slow-growth phenotype caused by a mutant translation factor EF-Tu. Mol Microbiol 70:1194–1209. doi: 10.1111/j.1365-2958.2008.06472.x. [DOI] [PubMed] [Google Scholar]
- 49.Brandis G, Hughes D. 2016. The selective advantage of synonymous codon usage bias in salmonella. PLoS Genet 12:e1005926. doi: 10.1371/journal.pgen.1005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abdulkarim F, Tuohy TM, Buckingham RH, Hughes D. 1991. Missense substitutions lethal to essential functions of EF-Tu. Biochimie 73:1457–1464. doi: 10.1016/0300-9084(91)90178-4. [DOI] [PubMed] [Google Scholar]
- 51.Kavaliauskas D, Nissen P, Knudsen CR. 2012. The busiest of all ribosomal assistants: elongation factor Tu. Biochemistry 51:2642–2651. doi: 10.1021/bi300077s. [DOI] [PubMed] [Google Scholar]
- 52.Thirup SS, Van LB, Nielsen TK, Knudsen CR. 2015. Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu. J Struct Biol 191:10–21. doi: 10.1016/j.jsb.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 53.Kothe U, Wieden HJ, Mohr D, Rodnina MV. 2004. Interaction of helix D of elongation factor Tu with helices 4 and 5 of protein L7/12 on the ribosome. J Mol Biol 336:1011–1021. doi: 10.1016/j.jmb.2003.12.080. [DOI] [PubMed] [Google Scholar]
- 54.Kacar B, Gaucher EA. 2012. Towards the recapitulation of ancient history in the laboratory: combining synthetic biology with experimental evolution, p 11–18. In Artificial life XIII: proceedings of the Thirteenth International Conference on the Simulation and Synthesis of Living Systems. MIT Press, Cambridge, MA. [Google Scholar]
- 55.Copley SD. 2003. Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Curr Opin Chem Biol 7:265–272. doi: 10.1016/S1367-5931(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 56.Pál C, Papp B, Pósfai G. 2014. The dawn of evolutionary genome engineering. Nat Rev Genet 15:504–512. doi: 10.1038/nrg3746. [DOI] [PubMed] [Google Scholar]
- 57.Acevedo-Rocha CG, Fang G, Schmidt M, Ussery DW, Danchin A. 2013. From essential to persistent genes: a functional approach to constructing synthetic life. Trends Genet 29:273–279. doi: 10.1016/j.tig.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hobbs JK, Prentice EJ, Groussin M, Arcus VL. 2015. Reconstructed ancestral enzymes impose a fitness cost upon modern bacteria despite exhibiting favourable biochemical properties. J Mol Evol 81:110–120. doi: 10.1007/s00239-015-9697-5. [DOI] [PubMed] [Google Scholar]
- 59.Johnsen PJ, Levin BR. 2010. Adjusting to alien genes. Mol Microbiol 75:1061–1063. doi: 10.1111/j.1365-2958.2010.07075.x. [DOI] [PubMed] [Google Scholar]
- 60.Condon C, Squires C, Squires CL. 1995. Control of rRNA transcription in Escherichia coli. Microbiol Rev 59:623–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2:20006.0007. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ellis HM, Yu D, DiTizio T, Court DL. 2001. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci U S A 98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. 2004. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- 66.Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 67.Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins, p 97–166. In Bryson V, Vogel HJ (ed), Evolving genes and proteins. Academic Press, New York, NY. [Google Scholar]
- 68.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huerta-Cepas J, Dopazo J, Gabaldón T. 2010. ETE: a python Environment for Tree Exploration. BMC Bioinformatics 11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hagberg A, Schult D, Swart P. 2008. Exploring network structure, dynamics, and function using NetworkX, p 11–15. In Varoquaux G, Vaught T, Millman J (ed), Proceedings of the 7th Python in Science Conference. [Google Scholar]
- 72.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Foreign tuf genes were integrated into the bacterial genome in a stepwise fashion as shown. (A) Each of the foreign tuf genes was integrated into the E. coli chromosome (replacing native tufA from start to stop codons). (B) In each strain carrying a foreign tuf gene at the tufA locus, an attempt was made to remove the native E. coli tufB gene (replacing the coding sequence from start to stop codons). Download FIG S1, PDF file, 0.2 MB (250.8KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative fitness of strains carrying two tuf genes (tufA-foreign and tufB E. coli) as a function of percent identity to E. coli EF-Tu (indicated as 100%). Strains shown in green carry a foreign tuf gene that can support viability when the E. coli tufB gene has been deleted. Download FIG S2, PDF file, 0.2 MB (167.8KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nucleotide sequences of ancestral tuf genes. Download TABLE S1, PDF file, 0.1 MB (64.7KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Codon adaptation index analysis. Species names indicate the species origin of the only tuf gene present. (A) Codon adaptation index as a function of the relative fitness of the strains. (B) Codon adaptation index as a function of the EF-Tu concentration normalized to the total protein content. Codon adaptation index was calculated with the CAI Calculator 2 using the Sharp and Li equation (P. M. Sharp and W.-H. Li, Nucleic Acids Res 15:1281–1295, 1987). Download FIG S3, PDF file, 0.3 MB (359.2KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The interaction partners of EF-Tu and all interactions among these first neighbors of EF-Tu. Nodes represent proteins (indicated by the relevant gene names), and each purple line represents an interaction. To easily distinguish tufA in the network, it has been colored cyan. Essential proteins are rectangular and colored blue in this subnetwork. Protein-protein interactions of E. coli were retrieved from the HitPredict database (A. Patil, K. Nakai, and H. Nakamura, Nucleic Acids Res 39:D744–D749, 2011, https://doi.org/10.1093/nar/gkq897), and the Cytoscape visualization tool was used to draw the network (B. Demchak et al., F1000Res 3:151, 2014, https://doi.org/10.12688/f1000research.4492.2). Download FIG S4, PDF file, 1 MB (1MB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The elongation factor EF-Tu is among the top 10 most connected proteins in the overall E. coli interactome. The proteins listed below are ranked according to their degree of connectivity using the UniProt database. The mean average degree of interaction in the whole interactome is 12. Download TABLE S2, PDF file, 0.1 MB (75.3KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structural representation of EF-Tu-GDP. EF-Tu is shaded gray, and residues that are conserved in all viable homologs but changed in at least one of the nonviable homologs are colored. (A) Changes within the G domain are colored red, those in domain II are colored yellow, and those in domain III are colored blue. (B) A close-up view of the G domain where the change in the P loop is shown in green, changes in the effector/switch I region are shown in orange, the change in the switch II region is shown in purple, and changes in the D-helix are shown in blue. GDP is shown in red, and the Mg ion is shown as a cyan sphere. Graphic analyses were performed with the UCSF Chimera package (E. F. Pettersen et al., J Comput Chem 25:1605–1612, 2004, https://doi.org/10.1002/jcc.20084) using the Protein Data Bank identifier 1EFC (H. Song, M. R. Parsons, S. Rowsell, G. Leonard, and S. E. Phillips. J Mol Biol 285:1245–1256, 1999). Download FIG S5, PDF file, 0.8 MB (852.7KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and genetic markers used in this work and their origin. Download TABLE S3, PDF file, 0.1 MB (92.6KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of organisms and plasmids serving as EF-Tu sources. Download TABLE S4, PDF file, 0.1 MB (72.6KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotide primers used in this study. Download TABLE S5, PDF file, 0.1 MB (64KB, pdf) .
Copyright © 2017 Kacar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.