Abstract
Human C-reactive protein (CRP), a classical human acute-phase plasma protein, is not only a sensitive systemic inflammatory marker but also an independent risk predictor of cardiovascular diseases. However, existing heterologous expression systems for expressing CRP is not efficient and cost-effective for large-scale industrial production of CRP to meet the growing market demand for CRP. This study aims to improve the secretion of recombinant CRP by Pichia pastoris via optimizing signal peptides, promoters and carbon sources. The CRP genes with encoding four different signal peptides were designed and synthesized. The genes were cloned into pPICZαA or pPICZ B, respectively via splicing by overlap extension polymerase chain reaction (SOE-PCR) technology and expressed in P. pastoris X-33, regulated by the alcohol oxidase I promoter (pAOX1). The CRP led by the α-mating factor secretion signal peptide (α-MF) was secreted at the highest level in these signal peptides. Then, a constitutive construct and expression of the CRP genes were achieved by switching to the glyceraldehyde-3-phosphate dehydrogenase promoter (pGAP). Subsequently, different carbon sources and at different concentrations were used to further improve the secretion of CRP. The expression of CRP with the α-MF driven by the pGAP gave the highest yield of secreted CRP, about 3 mg/l of culture on the optimized culture conditions. The purified recombinant CRP exhibited good immunoreactivity determined by ELISA with anti-human CRP monoclonal antibody. The efficient engineering strategy established in this work might provide potential uses in large-scale industrial production of human CRP in the future.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0917-0) contains supplementary material, which is available to authorized users.
Keywords: C-reactive protein, Pichia pastoris, Signal peptide, Promoter, Carbon source, Secretory expression
Introduction
C-reactive protein (CRP) is a highly conserved plasma protein discovered by Tillett and Francis (1930). It was named CRP because of its ability to precipitate Streptococcus pneumonia C-polysaccharide (Oliveira et al. 1977). CRP is a pentameric protein composed of five identical 23-kDa subunits (Thompson et al. 1999). A large number of epidemiological studies have demonstrated that CRP is not only a sensitive systemic inflammatory marker but also an independent risk predictor of cardiovascular diseases (Munk and Larsen 2009; Pepys and Hirschfield 2003; Volanakis 2001). Furthermore, the new development of high-sensitivity assays for CRP has enabled CRP to be more widely used in diagnosis and differential diagnosis of various diseases (e.g., inflammation, autoimmune disorders, metabolic syndrome, malignancies) and the predictive stratification of cardiovascular events (Adukauskiene et al. 2016; Avabratha et al. 2009; McWhorter et al. 2004; Mirhafez et al. 2016), thus resulting in an ever-growing demand for CRP in clinical applications or researches. However, the current methods used to prepare CRP are less than ideal.
Currently, there are two main approaches to obtain CRP: one is the direct purification of human CRP from malignant ascite or serum of the patient (Kindmark and Williams 1989), and the other is via genetic expression of the protein in a heterologous expression system (Agrawal et al. 2002; Dortay et al. 2011; Kilpatrick et al. 2012; Marnell et al. 1995; Tanaka et al. 2002). Although direct purification of CRP from the serum has met with success, the limited sources, complex operation procedures and low yield have limited the application of direct purification of CRP from the serum or malignant ascites. In recent years, a growing number of heterologous proteins have been produced by genetic engineering methods, and the recombinant form of CRP has been obtained from expression performed in Escherichia coli (Tanaka et al. 2002), animal cells (Agrawal et al. 2002), insect cells (Marnell et al. 1995) and yeast (Dortay et al. 2011). However, the E. coli expression system, a prokaryotic expression system, lacks the ability to correctly fold eukaryotic proteins because of the lack of post-translational modifications, such as disulfide bond formation, phosphorylation, glycosylation, thus affecting the solubility, stability, immunoreactivity, the biological activities and other physical and chemical properties of the expressed proteins (Daly and Hearn 2005). In addition, endotoxin generated by E. coli may be carried to the final product, which practically limits the medical application of the purified recombinant proteins in the future. The insect cell and mammalian cell, eukaryotic expression systems, can be used to overcome these problems, but they have other problems, such as complex genetic manipulations, complicated cell culture procedures, costly production, difficult purification procedures and other issues (Agrawal et al. 2002; Marnell et al. 1995).
In contrast, yeast, a unicellular and lower eukaryote, possesses both prokaryotic and eukaryotic expression advantages and is more ideal for the expression of exogenous proteins (Cereghino and Cregg 2000). The recombinant CRP has been generated in yeast cells, including Kluyveromyces lactis (Dortay et al. 2011) and P. pastoris GS115 (Kilpatrick et al. 2012), but productions were too low to meet the needs of industrial production. Now it remains a huge challenge to acquire the high-yield CRP with good immune activity at sufficiently low cost.
In this work, a series of engineering strategies, including the use of different secretory signal peptides, promoters and carbon sources, were taken to enhance the expression of human CRP in a secreted form in P. pastoris. The high-level secretory expression of CRP in P. pastoris X-33 was achieved through the use of appropriate signaling sequence (α-MF), promoter (GAP) and carbon source (1% glucose (w/v)). The attachment of a 6× His tag to the C-terminus of CRP greatly simplified the purification of the protein, allowing 97% purity to be achieved in a single purification step, and the purified CRP exhibited good immunoreactivity determined by ELISA.
Materials and methods
Materials
The yeast expression vectors pPICZαA, pPICZ B, pGAPZαA and P. pastoris X-33 yeast strain (wild-type) were purchased from Invitrogen (China). Escherichia coli JM109 competent cell, dNTPs, high-fidelity pyrobest DNA polymerase, T4 DNA ligase and restriction endonucleases were purchased from Takara (China). Tryptone, yeast extract, glucose, methanol and glycerol were purchased from Sangon Biotech (China). Zeocin was obtained from Invitrogen. Mouse anti-human CRP monoclonal antibody and goat anti-mouse IgG antibody conjugated with HRP were purchased from Sigma-Aldrich (China). Other reagents were all of analytical grade and were obtained from Sigma-Aldrich.
Cell cultures media
Pichia pastoris was cultured in different media depending on the purpose of the experiment. The media used were YPD (1% yeast extract, 2% peptone, 2% glucose), YPDS (1% yeast extract, 2% peptone, 2% glucose, 1 M sorbitol), BMMY (1% yeast extract, 2% peptone, 100 mM potassium phosphate (pH 6.0), 1.34% YNB, 10−5% biotin, and 0.5% methanol), BMGY (1% yeast extract, 2% peptone, 100 mM potassium phosphate (pH 6.0), 1.34% YNB, 10−5% biotin, and 1% glycerol), BMDY (1% yeast extract, 2% peptone, 100 mM potassium phosphate (pH 6.0), 1.34% YNB, 10−5% biotin, and 2% glucose), and BMY (1% yeast extract, 2% peptone, 100 mM potassium phosphate (pH 6.0), 1.34% YNB, 10−5% biotin).
Construction of plasmids for CRP expression
The DNA sequence of the human CRP gene was obtained from GenBank (Accession No: X56214.1). Four different constructs carrying the human CRP gene were previously prepared in Professor Xuejun Hu’s laboratory via SOE-PCR technology (Nelson and Fitch 2011). Each construct contained a different signal sequence at the 5′ end of the CRP gene and a stretch of nucleotides coding for 6× His at the 3′ end of the CRP gene. The four CRP-expressing constructs (Supplementary Fig. 1(a–d), which were designated as pPICZαA/CRP, pPICZ B/CRN + CRP, pPICZ B/SUC2 + CRP and pPICZαA-op/CRP contained the α-mating factor secretion signal sequence (α-MF) (Murasugi and Tohma-Aiba 2001), native secretory signal sequence of CRP (CRN), invertase signal sequence (SUC2) (Li et al. 2002) and α-mating factor signal sequence optimized by mutant (α-MF-op) (Rakestraw et al. 2009), respectively. The expression of CRP from these constructs required induction by methanol, and thus, they were referred to the inducible constructs or system.
A constitutive expression system was also prepared by excising the CRP DNA fragment from pPICZαA/CRP via restriction digestion with XhoI and XbaI, followed by subcloning into the constitutive expression vector pGAPZαA to yield pGAPZαA/CRP (Supplementary Fig. 1e). In this case, the expression of CRP was controlled by the pGAP, and was, therefore, designated as the constitutive construct or system.
All constructs were transformed into E. coli JM109, and the plasmids were extracted and further verified by enzymatic cleavage. The DNA sequence and the orientation of the CRP insert in these plasmids were then confirmed by DNA sequencing using GenScript Biotechnology (China).
Transformation of P. pastoris
Each of the CRP construct was linearized separately with SacI (in the case of inducible construct) or with BspHI (for the constitutive construct) and then transformed into P. pastoris via electroporation. Once inside the yeast cells, the linearized DNA was integrated into the yeast chromosomal DNA via homologous recombination. For control purpose, P. pastoris cells were transformed with the corresponding linearized empty plasmid. Transformation was carried out using the EasySelect Pichia Expression Kit (Invitrogen) according to the manufacturer’s instruction. The yeast transformants were spread onto YPDS selective medium containing 200 μg of Zeocin/ml and incubated at 28 °C for about three days. To obtain pure clones, ten colonies were randomly selected from every transformation and streaked onto YPDS medium containing 500 μg of Zeocin/ml. These positive clones were marked for further studies.
Small-scale expression of CRP
Small-scale inducible expression of CRP was performed as follows. The selected positive clones were inoculated into separate 50-ml conical centrifuge tubes containing 10 ml of BMGY medium, and then incubated overnight at 28 °C with shaking (220 rpm) until the cultures reached an OD600 of 2–6 (approximately 16–18 h). The cultures were then centrifuged at 1500×g for 5 min at room temperature and each cell pellet was resuspended in 10 ml of BMMY induction medium to induce the expression of CRP at 28 °C in a shaking incubator (220 rpm). As part of the induction mechanism, methanol was added to the culture every 24 h to a final concentration of 0.5% (v/v). The cells were allowed to grow for 96 h and 120-μl of samples were withdrawn every 24 h for analysis of protein expression. The samples were centrifuged at 1500×g for 5 min and the supernatants were analyzed by SDS-PAGE and western blot. Protein levels were compared using the Quantity-one 1-D analysis software.
For small-scale constitutive expression, 10 positive colonies were selected and each inoculated into 10 ml of YPD and cultured overnight at 28 °C in a shaking incubator (220 rpm) until the OD600 reached 6. Then, 0.5 ml of the overnight culture was inoculated into 10 ml of fresh YPD in 50-ml conical tube and the expression of CRP was carried out at 28 °C with shaking (220 rpm). Glucose was added to the culture to a final concentration of 2% (w/v) at every 24 h. At the desired time point, 0.15 ml of the culture was withdrawn and the culture was replaced with an equal amount of fresh medium. Sampling was performed at every 3 h for 48 h and then at every 24 h until 168 h. The samples were subjected to western blot analysis to determine the level of CRP expression.
Optimization of carbon sources for CRP expression
P. pastoris harboring pPICZαA/CRP (inducible) or pGAPZαA/CRP (constitutive) that showed the best expression was streaked onto a new fresh YPDS plate containing 100 μg of Zeocin/ml and incubated at 30 °C for approximately 3 days until the appearance of single colonies. In the case of inducible expression, single colonies were picked and cultured in 2-l side baffled flasks, containing 80 ml of BMGY medium. For constitutive expression, single colonies were picked and cultured in 2-l side baffled flasks, containing 240 ml of BMGY medium. The flasks were incubated at 28 °C until the culture reached an OD600 of 4.05. Aliquots (10 ml) of the culture were then dispensed into 50-ml conical centrifuge tubes. The tubes were centrifuged at 1500×g for 5 min and the cell pellets were each resuspended in 10 ml BMY medium and the expression of CRP was initiated as follows.
In the case of the inducible clone, different concentrations (0.5, 1, 1.5, 2, 2.5, 3 and 3.5% (v/v)) of methanol were used to induce the expression of CRP. As for the constitutive clone, different carbon sources (glucose, glycerol and methanol) and at various concentrations (0.5, 1, 1.5, 2, 2.5, 3 and 3.5% (w/v or v/v)) were used to initiate the expression of CRP. In both cases, the expression of CRP was carried out at 28 °C with shaking (220 rpm). Samples were taken from individual tubes of the cultures at the desired time points and the expression of CRP in the culture supernatant was analyzed by western blot and the target band was quantified by densitometry using Quantity-one 1-D analysis software.
Large-scale expression of CRP
Large-scale expression of CRP was carried out using only the constitutive system under optimal conditions. The expression was performed in 2-l side baffled flask using 200-ml culture. After 48 h of incubation under the optimum medium, the CRP was purified from the culture supernatant as described below.
Purification of CRP
The secreted CRP was purified from 200 ml of culture supernatant by affinity chromatography. First, the culture supernatant was dialyzed for overnight against 20 mM sodium phosphate buffer (pH 7.4) containing 0.5 M NaCl. After dialysis, the purification was performed using a 1-ml HisTrap HP-affinity column (GE Healthcare Bio Sciences AB, Sweden) according to the manufacturer’s recommendation. The column was eluted with a linear gradient of imidazole from 100 to 500 mM in the same phosphate buffer at a flow rate of 1 ml/min, and 1-ml fractions were collected. Peak fractions were subjected to SDS-PAGE using 12% gel. Protein bands in the gel were visualized by Coomassie staining and the identity of CRP was also confirmed by western blot. The purified protein was quantified using Bradford protein assay method with BSA as standard.
Detection of the immunoreactivity of CRP
The immunoreactivity of the purified CRP was assayed by ELISA. The purified CRP was diluted with 0.01 M PBS (pH 7.4) and used to coat a 96-well plate (Polysorp Nunc, USA) using 100 μl/well. The plate was washed six times with 0.01 M PBS containing 0.05% Tween20 (PBST), and then blocked with the same buffer containing 5% nonfat milk, first at 37 °C for 1 h and then at 4 °C for overnight. After that, it was again washed six times with PBST. Next, 100 µl of a 1:6000 dilution of mouse anti-human CRP monoclonal antibody (Sigma-Aldrich, China) was added to each of the CRP-coated well and the plate was incubated at 37 °C for 2 h. After washing the plate with PBST, 100 µl of goat anti-mouse IgG antibody conjugated to HRP (1:5000 in 0.01 M PBS) was added to each of the CRP-coated well. The plate was incubated at 37 °C for 1 h. After another eight washes, 100 µl of fresh TMB (3,3′,5,5′-Tetramethylbenzidine, Sigma Chemicals, USA) was added to the well and the color reaction was allowed to occur in the dark at room temperature for 20 min, and then terminated by addition of 50 µl stop buffer (2 M H2SO4). The optical density (OD) at 450 nm was determined for all test wells using a microplate reader (Multiskan GO, Thermo, USA). The above experiment was repeated three times to ensure consistency of the results. For control, the plates were coated with just PBS instead of CRP and subjected to the same procedure.
Results
Construction of plasmids for the expression of human CRP
Five CRP expression constructs were made using commercially available yeast expression vectors and PCR-based technology. Four of the constructs allowed the expression of CRP to be induced by methanol while the remaining one allowed CRP to be expressed in a constitutive manner. Furthermore, in both inducible and constitutive constructs, CRP was fused to a signal sequence to allow the protein to be expressed as a secretory form (Supplementary Fig. 1(a–e)). Restriction analysis and DNA sequencing confirmed that all constructs contained the correct sequence of CRP plus signal sequence (data not shown.)
Effects of different signal peptides on the expression of CRP
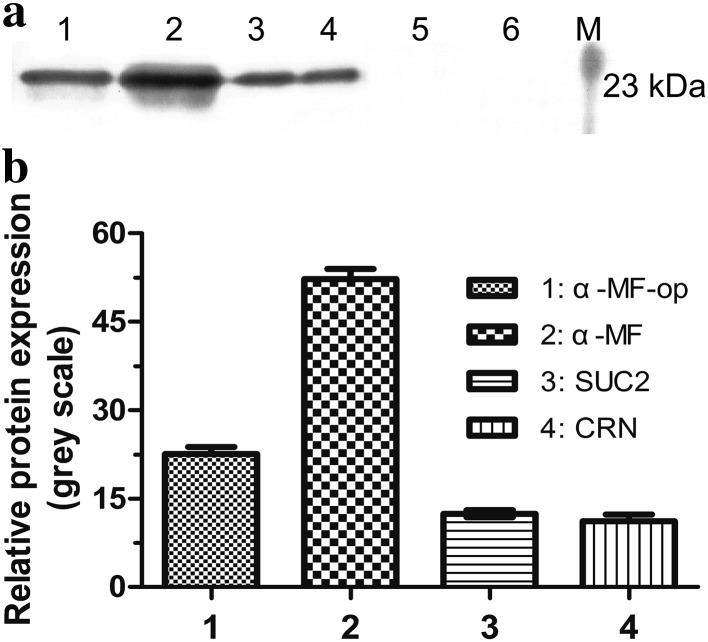
Expressions of CRP from the different constructs in P. pastoris X-33 under methanol induction were examined by western blot. The results indicated that among the four different signal sequences, the CRP with α-MF yielded the best expression level (Fig. 1). No band was observed for the culture supernatant of P. pastoris carrying empty plasmid pPICZαA or pPICZ B.
Fig. 1.
Expression of CRP fused to different signal peptides. Culture of P. pastoris transformed with the empty plasmid pPICZαA or pPICZ B or one of these two plasmids with CRP insert fused to a different signal sequence (α-MF-op, α-MF, SUC2 or CRN) was induced with methanol for 120 h and the samples of the culture supernatant were taken and analyzed for CRP expression by western blot. Lane M protein marker, lane 1 pPICZαA-op/CRP, lane 2 pPICZαA/CRP, lane 3 pPICZ B/SUC2 + CRP, lane 4 pPICZ B/CRN + CRP, lane 5 pPICZαA, lane 6 pPICZ B. a Representative blot is shown. b The plot shows the intensity of each band of the blot in gray scale. Data are the means ± SDs from three experiments
Small-scale constitutive expression of CRP in YPD medium
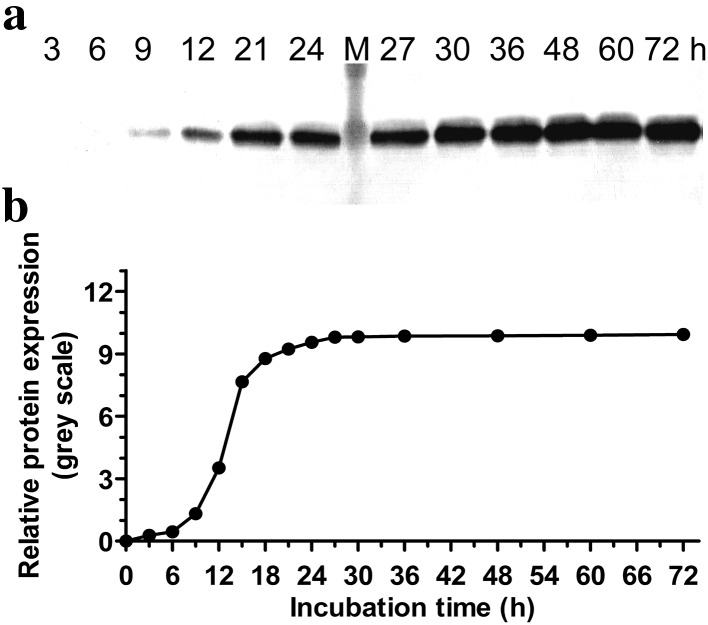
Constitutive expressions of CRP in YPD medium were examined for a number of different clones. The clone that gave the most intense CRP band, as examined by western blot, was selected and the expression of CRP over time was determined (Fig. 2). As expected, the target band was absent in the case of P. pastoris carrying the empty plasmid.
Fig. 2.
Time course analysis of constitutive CRP expression. P. pastoris transformed with pGAPZαA/CRP was cultured in YPD medium and the expression of CRP was monitored at the indicated time points. Lane M protein marker. a Representative blot is shown. b The plot below the blot shows the intensities of the bands in the blot. Data are the averages of three experiments
Effects of carbon sources on the expression of CRP
The effect of different methanol concentrations on the expression of CRP driven by the pAOX1 was investigated. Maximum expression of CRP was obtained in the presence of 0.5% (v//v) methanol (Supplementary Fig. 2).
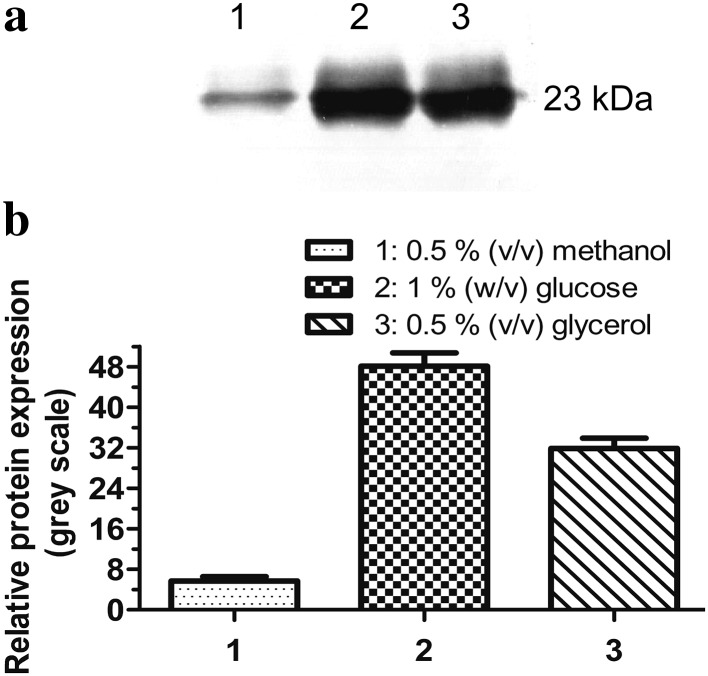
Different carbon sources were used to examine their effects on the constitutive expression of CRP driven by the pGAP. Optimum expression was obtained at a carbon concentration of 0.5% (v/v) in the cases of methanol and glycerol, and 1% (w/v) in the case of glucose. However, glucose (1% (w/v)) appeared to yield the best expression among the three different carbon sources (Fig. 3).
Fig. 3.
Effects of different carbon sources on constitutive CRP expression. P. pastoris transformed with pGAPZαA/CRP was cultured in BMY medium containing 0.5% (v/v) methanol (lane 1), 1% (w/v) glucose (lane 2) or 0.5% (v/v) glycerol (lane 3) for 18 h and the expression of CRP was analyzed by western blot. a Representative blot is shown. b The plot shows the intensity of each band of the blot in gray scale. Data are the means ± SDs from three experiments
In addition, differences in CRP expression were also noted when the expression of CRP from the constitutive construct was carried out in different media containing the same carbon source. In this case, notable increase in CRP expression was detected in BMY medium containing 1% (w/v) glucose compared to YPD medium (data not shown). Thus, BMY medium containing 1% (w/v) glucose was the optimum medium for the constitutive expression of CRP.
Comparison of the effect of two expression systems on the target protein secretion
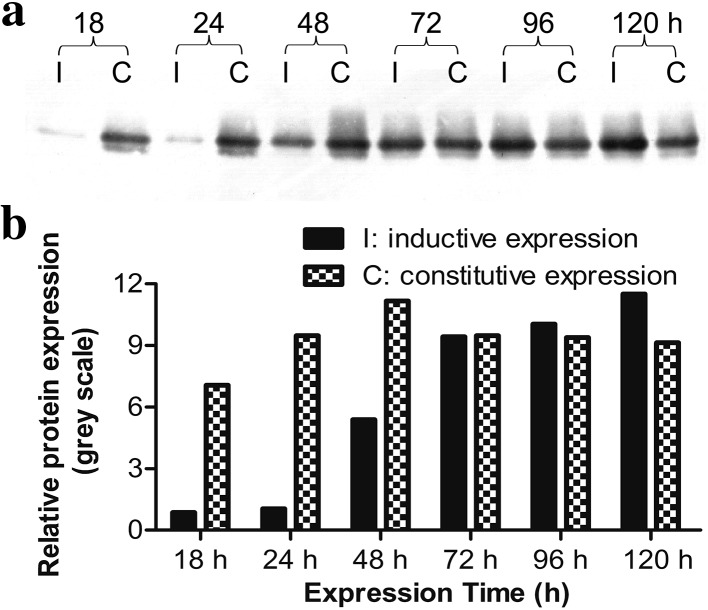
Under the same culture condition but with different carbon sources, analysis of time-dependent expression of CRP revealed notable difference between the two expression systems (Fig. 4). The constitutive construct appeared to yield better expression of CRP, at least in the earlier stage of the expression (up to 48 h) when 1% glucose was used as a carbon source (Fig. 4). On the other hand, the expression of CRP from the inducible construct was very low during the first 48 h after induction with 0.5% methanol, but continued to increase. Further details are shown in Fig. 4.
Fig. 4.
Changes in CRP expression between inducible and constitutive expression systems. P. pastoris carrying pPICZαA/CRP (inducible) was cultured in BMY medium containing 0.5% (v/v) methanol, whereas P. pastoris carrying pGAPZαA/CRP (constitutive) was cultured in BMY medium containing 1% (w/v) glucose. Samples of the cultures were collected at the indicated time points and the expression of CRP was analyzed by western blot. I inductive expression; C constitutive expression. a Representative blot is shown. b The plot below the blot shows the intensities of the bands in the blot. Data are the averages of three experiments
Analysis of the purified CRP via SDS-PAGE and western blot
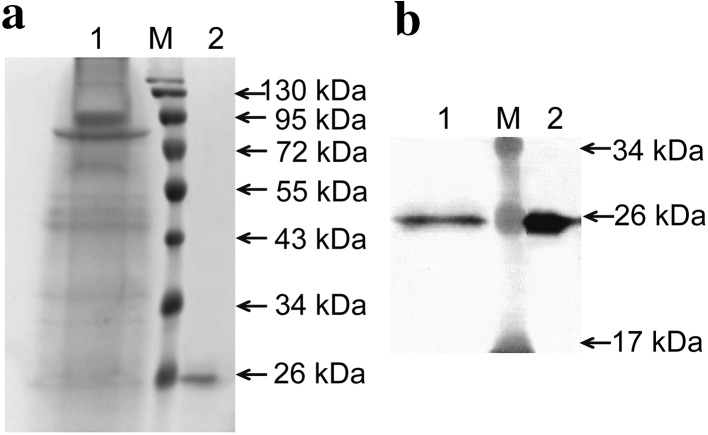
Large-scale expression of CRP was performed using P. pastoris transformed with pGAPZαA/CRP. After 48 h of incubation in optimum medium, the purification of recombinant CRP was achieved from the culture supernatant by HisTrap HP-affinity column chromatography, which yielded a single species according to SDS-PAGE analysis (Fig. 5a). The purified CRP was about 97% purity. Due to short incubation time and secretory expression, the proteins in the culture medium including target protein and non-target proteins were less. The target band was almost undetectable in the supernatant of the culture medium prior to purification detected by SDS-PAGE. Subsequently, the identity of CRP was performed by western blot. The results were shown in Fig. 5b and the target band was confirmed by western blot at the expected location. The protein concentration in the final preparation was about 3 mg/l estimated by Bradford protein assay.
Fig. 5.
Purification of recombinant CRP expressed in P. pastoris X-33. After 48 h of constitutive expression in optimum medium, the recombinant CRP was purified from the culture supernatant by HisTrap HP-affinity column chromatography. a The purified recombinant CRP was detected via SDS-PAGE analysis. b The purified recombinant CRP was confirmed via western blot analysis. Lane M protein marker. Lane 1 culture supernatant prior to purification. Lane 2 eluate fraction from affinity chromatography
Detection of the immunity reactivity of purified CRP
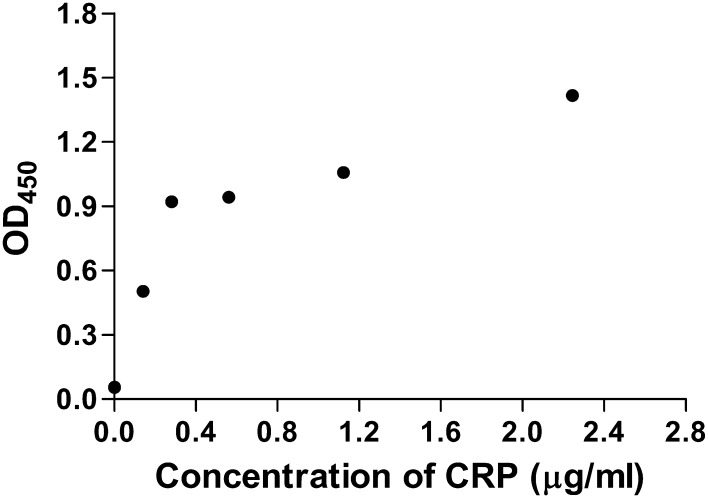
The purified CRP was able to bind with mouse anti-human CRP monoclonal antibody as revealed by ELISA test (Fig. 6), indicating that it had good immunoreactivity. In addition, the purified CRP showed no significant loss in its immunoreactivity when stored at 4 °C at pH 7.4 for more than 8 weeks.
Fig. 6.
Immunoreactivity of purified CRP as detected by ELISA. Purified CRP was diluted in 0.01 M PBS to different concentrations (2.245, 1.123, 0.561, 0.281, 0.141 µg/ml) and subjected to immunoreactivity assay as described in “Materials and methods”. Data are the means ± SDs from three experiments
Discussion
In recent years, the number of human CRP used for clinical diagnosis or research applications has increased dramatically. Production of human CRP has a remarkable demand in the market. Nonetheless, as mentioned previously, the current method used in the preparation of CRP is less than ideal.
The P. pastoris, an efficient expression system for the expression and secretion of heterologous proteins, can offer many advantages for the efficient and economical production of human CRP, including the capability of post-translational modifications, the simplicity of genetic manipulation and cell culture (Li et al. 2016; Wang et al. 2012). In particular, there is still a lot of room for improvement of the secretion of heterologous proteins in P. pastoris via a series of engineering strategies (Daly and Hearn 2005; Damasceno et al. 2012). In this study, we managed to achieve the high-level secretion of CRP from P. pastoris X-33 to meet the needs of industrial production via optimizing secretion signal sequences, promoters and culture conditions.
Efficient expression of heterologous proteins in secretory forms is preferred for industrial production of recombinant protein. The high-level secretion of the target protein in P. pastoris can be achieved through the use of appropriate signaling sequence, thus simplifying their purification procedure (Li et al. 2002; Murasugi and Tohma-Aiba 2001). The α-factor secretion signal sequence (α-MF) is the most widely used efficient signal sequence for expressing proteins in a secretory form in P. pastoris (Murasugi and Tohma-Aiba 2001), but the signal peptide can also be a native signal peptide (Guo and Ma 2008), a heterologous signal peptide (Hitzeman et al. 1990) or an optimized signal peptide (Rakestraw et al. 2009). There is no one signal peptide that can work equally well for the production of all proteins, and therefore, the most suitable signal peptide for human CRP needs to be specifically identified by experiments. In this work, four secretory signal peptides (α-MF, CRN, SUC2 or α-MF-op) were employed to improve secretion efficiency of human CRP expressed in P. pastoris X-33. And the α-MF signal peptide was the best one for secreting CRP driven by the AOX1 promoter. It facilitated a fivefold higher secretory expression of CRP than did the CRN signal peptide (Fig. 1).
Promoter is one of the most important factors for protein expression in any expression system. The pAOX1 and pGAP are both strong promoters which have been used to drive the expression of numerous heterologous genes in P. pastoris (Daly and Hearn 2005). The pAOX1 is a tightly regulated promoter induced by methanol (Kim et al. 2009). The pGAP is a constitutive promoter, but not suitable for the production of proteins that may be toxic to the P. pastoris cells (Mao et al. 2015; Varnai et al. 2014). In this work, the two promoters were both employed to optimize the expression of CRP in P. pastoris X-33: the highest expression level of CRP in P. pastoris driven by the pGAP was comparable to that obtained with the pAOX1, and the expression of CRP under the control of the pGAP reached a peak much quicker than the expression driven by the pAOX1 (Fig. 4). The results indicated that the pGAP was more efficient than the pAOX1 for the expression of CRP in P. pastoris.
In addition, compared to inducible promoter pAOX1, more carbon sources can be utilized for heterologous gene expression driven by the pGAP in P. pastoris and the strength of constitutive expression level is dependent on the carbon source used for cells culture (Shahidan et al. 2011). In this study, various carbon sources have been tested for the expression of CRP, such as glucose, glycerol and methanol containing different concentrations (0.5, 1, 1.5, 2, 2.5, 3 and 3.5% (v/v)), and BMY medium containing 1% (w/v) glucose was found to result in the highest expression level of CRP under the control of the pGAP (Fig. 3), thus eliminating the hazards caused by methanol. Equal importantly, the pGAP-based constitutive expression system for CRP production required fewer incubation steps, and the product could be easily purified from the medium, allowing about 97% purity to be achieved by one step purification procedure. Therefore, the pGAP-based constitutive expression system would be more suitable for the large-scale industrial production of CRP.
In fact, there are many other factors, such as the host strains, gene dosages and the fermentation parameters, which can influence the expression of CRP in P. pastoris (Daly and Hearn 2005; Liu et al. 2012). Thus, it is possible to further increase the secretory expression of CRP via further strategies mentioned above in the future.
Conclusions
This work established an efficiently secreted expression strategy in P. pastoris for the production of human immunoreactive CRP, and this is the first report that the high-level secretory expression of the human CRP in P. pastoris was successfully achieved using the pGAP-based constitutive expression system, thus making the large-scale production of CRP safer and more efficient. The engineering strategy established in this work might provide potential uses in industrial production of human CRP in the future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by Grants from Shandong Provincial Natural Science Foundation, China (No. ZR2015PH034) and National Natural Science Foundation of China (No. 31370937).
Compliance with ethical standards
Conflict of interest
Authors declare no financial or commercial conflict of interest to this work.
Footnotes
Junming Li and Chengming Sun contributed equally to this work.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0917-0) contains supplementary material, which is available to authorized users.
References
- Adukauskiene D, Ciginskiene A, Adukauskaite A, Pentiokiniene D, Slapikas R, Ceponiene I. Clinical relevance of high sensitivity C-reactive protein in cardiology. Medicina (Kaunas) 2016;52:1–10. doi: 10.1016/j.medici.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Simpson MJ, Black S, Carey MP, Samols D. A C-reactive protein mutant that does not bind to phosphocholine and pneumococcal C-polysaccharide. J Immunol. 2002;169:3217–3222. doi: 10.4049/jimmunol.169.6.3217. [DOI] [PubMed] [Google Scholar]
- Avabratha KS, Rau AT, Venkataravanamma P, Rau A. Significance of C-reactive protein during febrile neutropenia in pediatric malignancies. Indian Pediatr. 2009;46:797–799. [PubMed] [Google Scholar]
- Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- Daly R, Hearn MT. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J Mol Recognit. 2005;18:119–138. doi: 10.1002/jmr.687. [DOI] [PubMed] [Google Scholar]
- Damasceno LM, Huang CJ, Batt CA. Protein secretion in Pichia pastoris and advances in protein production. Appl Microbiol Biotechnol. 2012;93:31–39. doi: 10.1007/s00253-011-3654-z. [DOI] [PubMed] [Google Scholar]
- Dortay H, Schmockel SM, Fettke J, Mueller-Roeber B. Expression of human c-reactive protein in different systems and its purification from Leishmania tarentolae. Protein Expr Purif. 2011;78:55–60. doi: 10.1016/j.pep.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Guo J-P, Ma Y. High-level expression, purification and characterization of recombinant Aspergillus oryzae alkaline protease in Pichia pastoris. Protein Expr Purif. 2008;58:301–308. doi: 10.1016/j.pep.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Hitzeman RA, et al. Use of heterologous and homologous signal sequences for secretion of heterologous proteins from yeast. Methods Enzymol. 1990;185:421–440. doi: 10.1016/0076-6879(90)85037-O. [DOI] [PubMed] [Google Scholar]
- Kilpatrick EL, Liao WL, Camara JE, Turko IV, Bunk DM. Expression and characterization of 15 N-labeled human C-reactive protein in Escherichia coli and Pichia pastoris for use in isotope-dilution mass spectrometry. Protein Expr Purif. 2012;85:94–99. doi: 10.1016/j.pep.2012.06.019. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Lee JA, Kim YH, Song BK. Optimization of the functional expression of Coprinus cinereus peroxidase in Pichia pastoris by varying the host and promoter. J Microbiol Biotechnol. 2009;19:966–971. doi: 10.4014/jmb.0901.018. [DOI] [PubMed] [Google Scholar]
- Kindmark CO, Williams JC. Purification of human C-reactive protein by barium sulfate and preparative agarose electrophoresis. APMIS. 1989;97:891–896. doi: 10.1111/j.1699-0463.1989.tb00494.x. [DOI] [PubMed] [Google Scholar]
- Li J, Xu H, Bentley WE, Rao G. Impediments to secretion of green fluorescent protein and its fusion from Saccharomyces cerevisiae. Biotechnol Prog. 2002;18:831–838. doi: 10.1021/bp020066t. [DOI] [PubMed] [Google Scholar]
- Li P, et al. High-level secretory expression and purification of recombinant human interleukin 1 beta in Pichia pastoris. Protein Pept Lett. 2016;23:763–769. doi: 10.2174/0929866523666160530184936. [DOI] [PubMed] [Google Scholar]
- Liu Z, Tyo KE, Martinez JL, Petranovic D, Nielsen J. Different expression systems for production of recombinant proteins in Saccharomyces cerevisiae. Biotechnol Bioeng. 2012;109:1259–1268. doi: 10.1002/bit.24409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Teng D, Wang X, Zhang Y, Jiao J, Cao X, Wang J. Optimization of expression conditions for a novel NZ2114-derived antimicrobial peptide-MP1102 under the control of the GAP promoter in Pichia pastoris X-33. BMC Microbiol. 2015;15:57. doi: 10.1186/s12866-015-0389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnell L, Mold C, Volzer MA, Burlingame RW, Du Clos TW. Expression and radiolabeling of human C-reactive protein in baculovirus-infected cell lines and Trichoplusia ni larvae. Protein Expr Purif. 1995;6:439–446. doi: 10.1006/prep.1995.1059. [DOI] [PubMed] [Google Scholar]
- McWhorter VC, Ford LC, Butch AW. Analytical performance of the Synchron LX20 Pro, BN trade mark II and IMMAGE high sensitivity C-reactive protein assays and concordance in cardiovascular risk stratification. Clin Chim Acta. 2004;347:71–79. doi: 10.1016/j.cccn.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Mirhafez SR, et al. Serum high-sensitivity C-reactive protein as a biomarker in patients with metabolic syndrome: evidence-based study with 7284 subjects. Eur J Clin Nutr. 2016;70:1298–1304. doi: 10.1038/ejcn.2016.111. [DOI] [PubMed] [Google Scholar]
- Munk PS, Larsen AI. Inflammation and C-reactive protein in cardiovascular disease. Tidsskr Nor Laegeforen. 2009;129:1221–1224. doi: 10.4045/tidsskr.08.0011. [DOI] [PubMed] [Google Scholar]
- Murasugi A, Tohma-Aiba Y. Comparison of three signals for secretory expression of recombinant human midkine in Pichia pastoris. Biosci Biotechnol Biochem. 2001;65:2291–2293. doi: 10.1271/bbb.65.2291. [DOI] [PubMed] [Google Scholar]
- Nelson MD, Fitch DH. Overlap extension PCR: an efficient method for transgene construction. Methods Mol Biol. 2011;772:459–470. doi: 10.1007/978-1-61779-228-1_27. [DOI] [PubMed] [Google Scholar]
- Oliveira EB, Gotschlich EC, Liu TY. Primary structure of human C-reactive protein. Proc Natl Acad Sci USA. 1977;74:3148–3151. doi: 10.1073/pnas.74.8.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepys MB, Hirschfield GM. C-reactive protein: a critical update. J Clin Invest. 2003;111:1805–1812. doi: 10.1172/JCI200318921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakestraw JA, Sazinsky SL, Piatesi A, Antipov E, Wittrup KD. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae. Biotechnol Bioeng. 2009;103:1192–1201. doi: 10.1002/bit.22338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahidan NH, Rahman RNZA, Leow TC, Rosfarizan M, Basri M, Salleh AB. The effect of carbon sources on the expression level of thermostable L2 lipase in Pichia pastoris. Afr J Biotech. 2011;10:13528–13535. doi: 10.5897/AJB11.1550. [DOI] [Google Scholar]
- Tanaka T, Horio T, Matuo Y. Secretory production of recombinant human C-reactive protein in Escherichia coli, capable of binding with phosphorylcholine, and its characterization. Biochem Biophys Res Commun. 2002;295:163–166. doi: 10.1016/S0006-291X(02)00622-8. [DOI] [PubMed] [Google Scholar]
- Thompson D, Pepys MB, Wood SP. The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure. 1999;7:169–177. doi: 10.1016/S0969-2126(99)80023-9. [DOI] [PubMed] [Google Scholar]
- Tillett WS, Francis T. Serological reactions in pneumonia with a non-protein somatic fraction of pneumococcus. J Exp Med. 1930;52:561–571. doi: 10.1084/jem.52.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai A, Tang C, Bengtsson O, Atterton A, Mathiesen G, Eijsink VG. Expression of endoglucanases in Pichia pastoris under control of the GAP promoter. Microb Cell Fact. 2014;13:57. doi: 10.1186/1475-2859-13-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volanakis JE. Human C-reactive protein: expression, structure, and function. Mol Immunol. 2001;38:189–197. doi: 10.1016/S0161-5890(01)00042-6. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhu M, Zhang A, Yang F, Chen P. Synthesis and secretory expression of hybrid antimicrobial peptide CecA-mag and its mutants in Pichia pastoris. Exp Biol Med (Maywood) 2012;237:312–317. doi: 10.1258/ebm.2011.011153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.