Abstract
Differences in the direction and degree to which invasive alien and native plants are influenced by mycorrhizal associations could indicate a general mechanism of plant invasion, but whether or not such differences exist is unclear. Here, we tested whether mycorrhizal responsiveness varies by plant invasive status while controlling for phylogenetic relatedness among plants with two large grassland datasets. Mycorrhizal responsiveness was measured for 68 taxa from the Northern Plains, and data for 95 taxa from the Central Plains were included. Nineteen percent of taxa from the Northern Plains had greater total biomass with mycorrhizas while 61% of taxa from the Central Plains responded positively. For the Northern Plains taxa, measurable effects often depended on the response variable (i.e., total biomass, shoot biomass, and root mass ratio) suggesting varied resource allocation strategies when roots are colonized by arbuscular mycorrhizal fungi. In both datasets, invasive status was nonrandomly distributed on the phylogeny. Invasive taxa were mainly from two clades, that is, Poaceae and Asteraceae families. In contrast, mycorrhizal responsiveness was randomly distributed over the phylogeny for taxa from the Northern Plains, but nonrandomly distributed for taxa from the Central Plains. After controlling for phylogenetic similarity, we found no evidence that invasive taxa responded differently to mycorrhizas than other taxa. Although it is possible that mycorrhizal responsiveness contributes to invasiveness in particular species, we find no evidence that invasiveness in general is associated with the degree of mycorrhizal responsiveness. However, mycorrhizal responsiveness among species grown under common conditions was highly variable, and more work is needed to determine the causes of this variation.
Keywords: arbuscular mycorrhizal fungi, grassland, invasion ecology, mixed‐grass prairie, mycorrhizal responsiveness, phylogenetic signal, phylogeny, tallgrass prairie
1. INTRODUCTION
Many natural plant communities are either invaded or at risk of invasion by non‐native plant species (Pimentel, Lach, Zuniga, & Morrison, 2000). The negative consequences of such invasions have stimulated extensive research aimed at identifying the responsible mechanisms, so as to identify efficacious control/management strategies (Mack et al., 2000). Although several mechanisms are likely in play (Gurevitch, Fox, Wardle, Inderjit, & Taub, 2011), exotic species are generally thought to be less reliant on specialist mutualists than native taxa (Richardson, Allsopp, D'Antonio, Milton, & Rejmanek, 2000; Vogelsang & Bever, 2009) but whether this also extends to generalist mutualists is less known. Arbuscular mycorrhizal fungi (AMF) are obligate biotrophs that colonize about 75% of all plant species (Brundrett, 2009) with apparent low specificity (Lekberg & Waller, 2016). These ubiquitous fungi can provide increased nutrient uptake, drought tolerance, and pathogen protection to plants in return for carbon (Smith & Read, 2008). AMF could contribute to invasiveness if exotic plants benefitted more from these services than native plants, or if exotic plants were less dependent on AMF and/or could disrupt this mutualism in native plants. To determine whether exotic plants are less reliant on the mutualism with AMF, an important first step is to compare the mycorrhizal status and responsiveness (measured as the biomass difference between inoculated plants and non‐inoculated controls; Janos, 2007) of native and exotic plants.
Some have suggested that a weak mycorrhizal responsiveness may be a general mechanism of plant invasion (van der Putten, Klironomos, & Wardle, 2007; Vogelsang & Bever, 2009) because invasions often occur in disturbed habitats (Mooney & Hobbs, 2000) that tend to harbor lower AMF abundance (Abbott & Robson, 1991). Consistent with this hypothesis is the fact that some well‐studied noxious invaders in North America are either nonmycorrhizal or have a low mycorrhizal responsiveness (e.g., Busby, Gebhart, Stromberger, Meiman, & Paschke, 2011; Stinson et al., 2006) and that exotic plants in California are disproportionally from nonmycorrhizal families (Pringle et al., 2009). Also, careful comparisons of native and exotic populations of both St. John's Wort (Hypericum perforatum; Seifert, Bever, & Maron, 2009) and yellow star thistle (Centaurea solstilialis; Waller, Callaway, Klironomos, Ortega, & Maron, 2016) have documented more ruderal traits and a reduced AMF responsiveness in the exotic populations.
On the other hand, AMF can increase growth and competitiveness of spotted knapweed (Centaurea stoebe; Marler, Zabinski, & Callaway, 1999), which is one of the most invasive weeds in the intermountain west of the USA. This and another invasive forb (Euphorbia esula) can also increase AMF abundance and diversity compared to remnant native communities (Lekberg, Gibbons, Rosendahl, & Ramsey, 2013). And, contrary to California grasslands, exotic plants in Great Britain and Germany were more likely to be from families that were more dependent on AMF (i.e., greater proportion that were obligatory mycorrhizal and lower proportion that were facultatively mycorrhizal) than native plants (Hempel et al., 2013; Pringle et al., 2009). Thus, it is clear that interactions with AMF differ drastically among invasive plants and possibly across regions being invaded.
A recent systematic literature search and summary (Bunn, Ramsey, & Lekberg, 2015) reported no appreciable difference between invasive and native plants in their growth responses to AMF, suggesting that invasions do not select for directional shifts in AMF associations. While this meta‐analysis represents the most comprehensive summary of published studies to date, there are some important limitations to the inferences drawn from such analyses. Specifically, meta‐analyses cannot always account for a high level of experimental heterogeneity among the included studies. The Bunn et al. (2015) analysis, for example, included data for 55 invasive and 70 native species from 67 studies that varied in experimental conditions (e.g., soil chemistry, identity of AMF taxa, growth conditions), which are known to affect plant–AMF interactions (Johnson, Wilson, Bowker, Wilson, & Miller, 2010; Klironomos, 2003; Stahl & Smith, 1984). These experimental nuances are not easily incorporated into meta‐analysis as factors or covariates (Koricheva, Gurevitch, & Mengersen, 2013) and may have prevented detection of actual differences between invasive non‐native vs. native taxa that are apparent only with a dataset obtained from a common set of experimental conditions. Also, plant responses to AMF are often phylogenetically conserved (Anacker, Klironomos, Maherali, Reinhart, & Strauss, 2014; Reinhart, Wilson, & Rinella, 2012), and phylogenetic nonindependence can bias results if the pool of species in comparison groups is disproportionately from clades that are highly responsive (or unresponsive) to AMF.
To circumvent these limitations, we analyzed two relatively large datasets that approximated the total sample size of the Bunn et al. (2015) meta‐analysis where plants were grown under the same environmental conditions and where the effects of shared ancestry were controlled. We generated new mycorrhizal responsiveness data for 68 plant species from the Northern Plains (14 invasive: 54 noninvasive) and performed separate tests for a published dataset with 95 taxa (11 invasive: 84 noninvasive) from the Central Plains (Wilson & Hartnett, 1998). A previous analysis of the Central Plains dataset which did not control for phylogeny found that native grassland plants were more responsive to AMF than taxa from outside the region (fig. 3 in Pringle et al., 2009), and we tested whether the same result would be found if the effects of shared ancestry were included.
2. METHODS
2.1. Northern Plains mycorrhizal growth response experiment
AMF spores were originally derived by either extraction from field soil or trap cultures of plant root (or soil) samples from four sites in Custer County, Montana, USA. The spores were extracted using wet sieving and sucrose density gradient centrifugation (Brundrett, Melville, & Peterson, 1994). Spores were extracted into a water solution. To propagate the fungal inoculum, we used Sorghum sudanese as the plant host. Seeds of S. sudanese were surface sterilized (placed in 10% bleach solution for 10 min then thoroughly rinsed) and then sowed into aluminum trays with once‐autoclaved vermiculite. Eighteen pots (3.21 L) were planted with three 1‐week‐old S. sudanese seedlings and inoculated with water containing AMF spores. There were nine AMF morphotypes identified, and 8–50 AMF spores were used to inoculate two replicate pots per morphotype. Eighteen additional control pots with the same number of S. sudanese seedlings were sham inoculated (Tintjer & Rudgers, 2006) with aliquots of the microbial solution from which AMF spores had been aspirated. The sham inoculant was necessary to control for additional forms of microbial life (e.g., bacteria, saprobic fungi) in the water solution containing AMF spores. Pots were filled with once autoclaved field soil (Eapa fine loam, frigid Aridic Argiustolls) from a calcareous grassland site. Pots were watered three times a week for 6 months. At harvest, we discarded shoots and the top 1 cm of soil. The remaining soil, which was to be used as inoculant for the mycorrhizal growth response experiment, was homogenized (i.e., one AMF mixture and one control mixture) and roots were diced into 1 cm fragments on 6 February 2014. Experimental units (pots) were prepared on 7 February 2014 (details below). We note that the AMF inoculum, although representative of a mycorrhizal community for the Northern Plains, still represents a simplification of the many AMF communities in the region (Reinhart & Anacker, 2014).
To determine the invasive and noninvasive species to be used in the experiment, we consulted regional experts. Invasive status was based on classifications by the US Forest Service (http://www.fs.fed.us/database/feis/plants/weed/index.html). Although the species sampled included more noninvasive than invasive species, this ratio reflects the scarcity of invasive relative to noninvasive taxa in the flora. This ratio also permits an unbiased estimate of phylogenetic effects on variation in plant growth response to mycorrhizal inoculation. Seed for the 68 selected species was collected by the authors from field sites or donated by other researchers. However, cases of insufficient seed, low germination rates, or both, necessitated purchasing seed of some species from specialized native seed providers (Western Native Seed Company [Coaldale, Colorado, USA], WindRiver Seed [Manderson, Wyoming, USA], and Prairie Moon Nursery [Winona, Minnesota, USA]). Two varieties of invasive Agropyron cristatum were also included. Seed was surface sterilized, and seedlings were established in growth chambers with varying conditions determined to best facilitate their individual germination. Seeds were sown into once autoclaved vermiculite in aluminum trays. Since seedlings established at varying times, seedlings were transplanted from February 8 through March 3, 2014. [Note: AMF viability remained high as indicated by Allium cemuum (Fig. S1) which was one of the last species transplanted].
To establish the mycorrhizal growth response experiment, each species was grown in ten pots inoculated with AMF and ten pots inoculated with the (sham) control (e.g., Koide & Li, 1989). Seedlings of each plant species were planted into Deepots™ (500 ml, Steuwe and Sons, Tangent, Oregon, USA) mostly filled (~90% of pot volume, ca. 500 ml) with once autoclaved (2 hr) field soil (Eapa loam). The field soil was collected in 2013 from an upland plain in northern mixed‐grass prairie, mechanically sieved through a 6.5‐mm screen. The grassland soil was a well‐drained loam with pH of 7.3, 1.3% organic matter, 7.3 ppm NO3‐N, 16 ppm P [Bray test 1], and 306 ppm potassium. The soil series has a B horizon containing carbonates which may immobilize P (e.g., Lajtha & Schlesinger, 1988).
Deepots™ were filled with 59 ml of once autoclaved gravel (to obstruct drain holes), 340 ml of once autoclaved field soil, 50 ml of inoculum (AMF or control), and topped with once autoclaved field soil. All stages of the experiments were conducted in a growth room facility at Fort Keogh Livestock and Range Research Laboratory, which was modeled after the facility at the International Culture Collection of (Vesicular) Arbuscular Mycorrhizal Fungi at West Virginia University. The space housed three identically designed grow benches, each with two 400 W high‐output sodium vapor lights (photosynthetically active radiation average = 492 ± 57 [95% CI] μmol × m−2 × s−1). The growth room was maintained on a 12‐hr day and night temperature cycle with day temperature maintained between 20.0 and 22.2°C and night temperatures maintained between 16.7 and 18.9°C. These temperature ranges were similar to those used by Wilson and Hartnett (1998) while conducting their greenhouse experiment. Pots were carefully watered by hand to avoid splash transfer among pots. Pots were provided with a small quantity of reverse osmosis water every other day to ensure a relatively stable soil moisture environment throughout the week and one main watering event per week to ensure pots returned to field capacity at least once per week. Deepot™ racks were randomized weekly.
After 100 days since the time of planting, the plants were harvested. As we were also interested in determining the palatability of the shoots for another study, shoot material was freeze‐dried (Freezone 6, Labconco, Kansas City, MO, USA) to constant weight and weighed. We did not collect data for pots with dead plants. We retained two species in the study with moderate levels of mortality Andropogon gerardii (45%) and Panicum virgatum (30%), and mortality was evenly distributed across the two inoculant treatments (data not shown). Previous trials indicated estimates of root biomass could be inflated by residual clay soil that remained attached to washed root samples. To overcome this, roots were separated from the soil by hand, dried to constant mass, weighed, combusted at 450°C for 4 hr, cooled to 60°C, and reweighed to determine the proportion of ash‐free dry mass (AFDM) after combusting the samples. Total plant biomass was then determined by summing the shoot dry mass with root weight (root dry mass × proportion AFDM). Plant biomass data for analyses included total biomass, shoot mass, and root mass ratio (root mass × total biomass−1).
To assess the infectivity of AMF inoculum, we quantified root infection by AMF (%) on a subset of species (McGonigle, Miller, Evans, Fairchild, & Swan, 1990) at the conclusion of the mycorrhizal growth response experiment. We also sequenced the DNA of pooled root samples and identified 11 AMF virtual taxa (VT) based on MaarjAM (Öpik et al., 2010). The two most abundant taxa matched with Funneliformis mosseae (VT67) and Claroideoglomus (VT402), whereas the rest matched with Glomus (VTs 130, 135, 140, 165, 172, 332, 416), Claroideoglomus (VT193) and Acaulospora (VT21). For a brief explanation of methods used for DNA extraction, amplification, sequencing, and bioinformatics, please see Appendix S1 in Supporting Information.
Methods information for the Central Plains mycorrhizal responsiveness experiment can be found in Wilson and Hartnett (1998).
2.2. Phylogenetic reconstruction
To analyze data while simultaneously controlling for the effects of shared ancestry among plant species, we used Reinhart et al.'s (2012) phylogeny (represented as a phylogram) for the Central Plains dataset, and also constructed a new phylogeny for the Northern Plains' taxa using gene sequence data. To construct the latter phylogeny, we searched the GenBank database for four gene sequences often used to construct angiosperm phylogenies: matK, rbcL, ITS1, and trnL. Multiple gene sequences are necessary to differentiate plant species because some sequences represent conservative coding regions (e.g., rbcL) and others represent more rapidly evolving portions (e.g., matK) (Reinhart et al., 2012). When a sequence was not available for the exact same species for a marker, we used sequences available for species of the same genus, and when possible of species also native to North America. This was performed for eight of 68 species. We included one representative from an early diverging angiosperm lineage as an outgroup species, Magnolia grandiflora (L.). Sequences of these 69 species were aligned using MUSCLE version 3.7 (Edgar, 2004). We identified the best‐fit maximum‐likelihood models of nucleotide substitution for each gene using Akaike Information Criterion using MrModeltest version 2.3 (Nylander, 2004).
Using the concatenated sequence alignments, we performed a partitioned Bayesian Inference, estimating the posterior probability distribution of all possible phylogenies using a Markov Chain Monte Carlo algorithm (i.e., Metropolis algorithm) implemented in MrBayes version 3.1.2 (Huelsenbeck & Ronquist, 2001). To guide the search, we created a topological constraint tree from a published super tree (R20100428; Angiosperm Phylogeny Group, 2009), pruned to the taxa used in our experiment using Phylomatic (Webb & Donoghue, 2004). Two independent Markov Chains were run, each with four heated chains for 5 million generations. The final average standard deviation of split frequencies was 0.002, indicating good convergence of the two independent runs. We sampled runs every 500 generations and used a burnin of 5,000 trees to generate a majority rule consensus phylogeny. We transformed this, and the prior Central Plains, phylograms using penalized likelihood and maximum likelihood with “chronos” in APE version 3.2 in R. In order to create a time calibrated tree with branch lengths in millions of years (MYA), calibration points for seven nodes based on Bell, Soltis, and Soltis (2010) were used: Asteraceae (31–48 MYA), Plantaginaceae (34–51), Caryophyllales (79–96), Fabaceae (47–75), Malpighiales (88–91), Brassicaceae (21–42), and Poaceae (17–38). The newick‐formatted phylograms are provided in Appendix S2.
2.3. Statistical analysis
We tested the effects of treatment (AMF vs. control) on response variables for each plant species with two‐sample t tests that assumed either equal or unequal variances using Proc ttest in SAS version 9.4 (SAS Institute, Inc., Cary, NC, USA) as originally done by Wilson and Hartnett (1998).
We expressed plant growth response to AMF inoculation using two common effect size metrics: log response ratio (LRR; ln[response variable with AMF/response variable without AMF] and standardized effect size (SES; standardized mean difference between the AMF inoculated and control, relative to the pooled population variances). In these formulations, positive values indicate that plant growth was stimulated by AMF, whereas negative values indicate that plant growth was suppressed by AMF. Mycorrhizal responsiveness was calculated for plant total biomass and shoot biomass for the Northern Plains dataset and total biomass for the Central Plains dataset using metafor R‐library with measure = “ROM” or “SMDH.”
While Bunn et al. (2015) reported no appreciable difference between invasive and native taxa, they interpreted the magnitude of responses for native taxa tended to be greater than those for invasive taxa. To help address this, we tested whether the variances of SES (Northern Plains) and LRR (Central Plains) varied by invasive status with F tests for equality of variances using the “var.test” function in R.
To test whether mycorrhizal responsiveness (LRR or SES) varied by grassland plant invasive status, we conducted a phylogenetic ANOVA (phylo‐ANOVA hereafter) (Garland, Dickerman, Janis, & Jones, 1993) with the phylANOVA function implemented in the R‐package phytools (Revell, 2012). We also tested whether mycorrhizal responsiveness and invasive status were phylogenetically conserved, by calculating a phylogenetic signal. Phylogenetic signal of continuous data (mycorrhizal responsiveness) was assessed with Abouheif's Cmean (function adouheif.moran with method = “oriAbouheif” in adephylo R‐library) and Blomberg's K (function phylosig with method = “K” in phytools R‐library) and categorical data (invasive status) with the D statistic (function phylo.d in caper R‐library). For the K statistic, values close to 1 indicate that traits follow a Brownian motion (BM) model of evolution, K > 1 indicate that taxa are more similar than expected (trait conservatism), and when K < 1, closely related taxa are more dissimilar than expected (trait divergence) under BM evolution (Blomberg, Garland, & Ives, 2003). Because Blomberg's K relies on branch length information, we also used Abouheif's Cmean test, which provides an estimate of phylogenetic signal in a quantitative trait solely based on tree topology (Abouheif, 1999). For the D statistic, values close to 0 mean that the trait follows a Brownian motion model of evolution, whereas when D = ~1, the trait evolves randomly with respect to the phylogeny. When D < 0, then the trait is conserved relative to a BM model of evolution, whereas when D > 1, the trait is more divergent than expected.
3. RESULTS
For the Northern Plains dataset, we confirmed that the roots of plants grown in pots with AMF inoculant were colonized by AMF (% root length colonized; range 4%–87%) while roots in controls (sham‐inoculant) were not (Fig. S1). Most Northern Plains species (71%–80%; three response variables) did not respond to inoculation with AMF and had similar total and shoot biomass and root mass ratio when grown with AMF versus sham‐inoculant controls. Nineteen percent of plants (13 of 69 comparisons) had greater total biomass when grown with AMF than controls (Figure 1a), whereas seven taxa had lower total biomass when grown with AMF relative to the sham‐inoculant control (Figure 1a). Eleven taxa allocated less biomass to roots than shoots when grown with AMF compared to controls (Figure 2). However, three taxa (Bromus tectorum, Buchloe dactyloides, and Festuca idahoensis) allocated appreciably more to roots than shoots when grown with AMF (Figure 2). The SES effect sizes for invasive and noninvasive taxa (Figure 1) also had equal variances (total biomass, F 14,53 = 0.93, p = .93; shoot biomass, F 14,53 = 1.40, p = 0.37).
Figure 1.
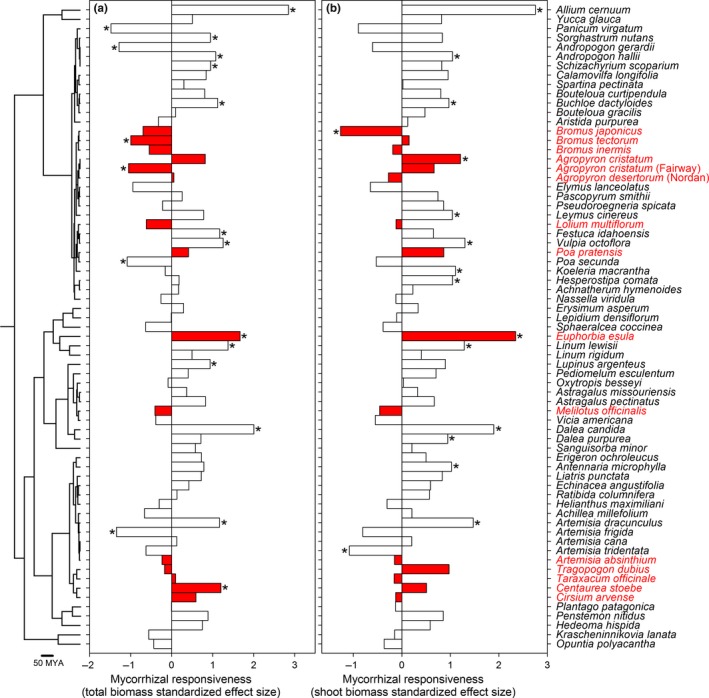
Molecular phylogeny and mycorrhizal responsiveness measures for 68 grassland plant species from the Northern Plains. Taxa labels and bars shaded red indicate taxa classified as invasive. Effect sizes were based on total plant biomass (a) and shoot biomass (b). Positive standardized effect size measurements indicate plants were larger with arbuscular mycorrhizal fungi than sham‐inoculant controls. We tested whether plants inoculated with AMF had significantly different biomass from controls by two‐sample t‐test (*p ≤ .05)
Figure 2.
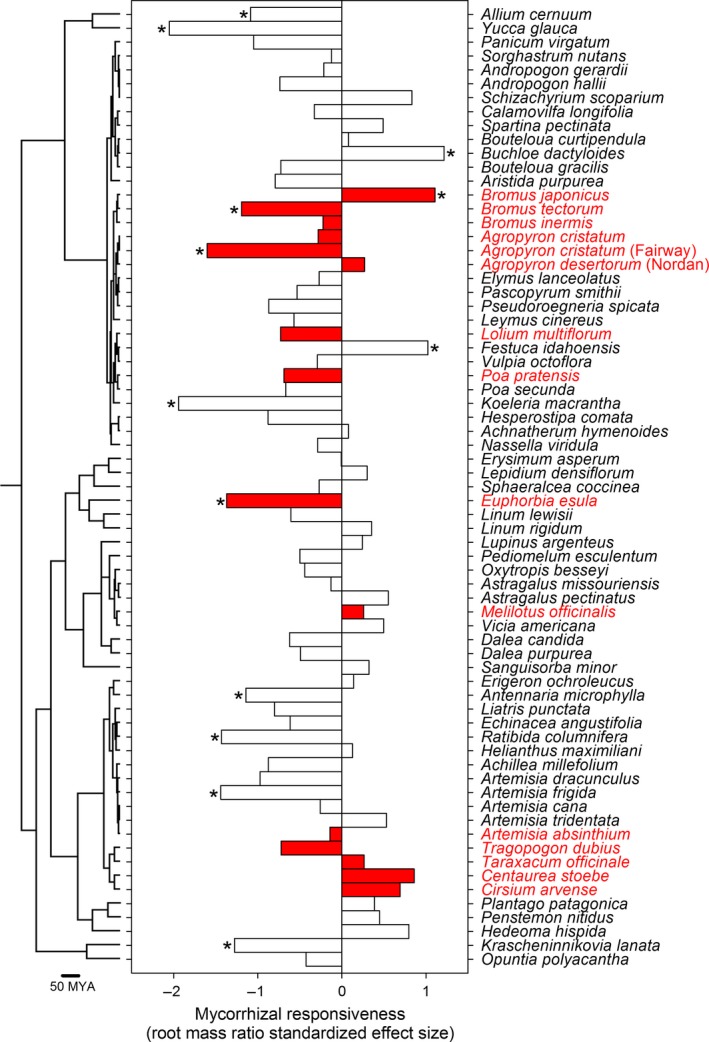
Molecular phylogeny and plant allometry (i.e., root mass ratio) for 68 plant species when grown with arbuscular mycorrhizal fungi (AMF) versus sham‐inoculant controls. Taxa labels and bars shaded red were classified as invasive. Negative standardized effect size measurements indicate that plants grown in soil with AMF allocated more resources to shoot than root biomass. We tested whether plants inoculated with AMF had significantly different root mass ratios from controls by two‐sample t‐test (*p ≤ .05)
Most species (71%–79%) classified as invasive for the Northern Plains dataset did not grow appreciably larger or smaller with AMF than controls (Figure 1). Two prominent invasive forbs (Centaurea stoebe and Euphorbia esula) produced greater total biomass when grown in soil with AMF than control soils (Figure 1a). One invasive grass (Agropyron cristatum) and forb (E. esula) produced greater shoot biomass in pots with AMF than controls (Figure 1b). Two invasive grass species (A. cristatum [Fairway] and B. tectorum), however, had lower total biomass in AMF inoculated vs. control pots (Figure 1a), and one invasive grass (B. japonicus) had lower shoot biomass in AMF inoculated vs. control pots (Figure 1b).
In the Central Plains dataset, by contrast, 61% (58 of 95) of grassland plant species had greater total biomass with AMF than controls (Figure 3) (Wilson & Hartnett, 1998). Only one taxon (B. inermis) had reduced growth in AMF inoculated vs. control soils (Figure 3). For the Central Plains portion, most species (64%) classified as invasive did not grow appreciably larger or smaller with AMF than controls (Figure 3). Three (Cirsium vulgare, Eragrostis spectabilis, and Lespedeza cuneata) of eleven invasive species had greater total biomass with AMF than without, and one invasive grass (B. inermis) had less total biomass with AMF than without (Figure 3). The LRR effect sizes for invasive and noninvasive taxa (Figure 3) had equal variances (total biomass, F 10,83 = 0.71, p = 0.58).
Figure 3.
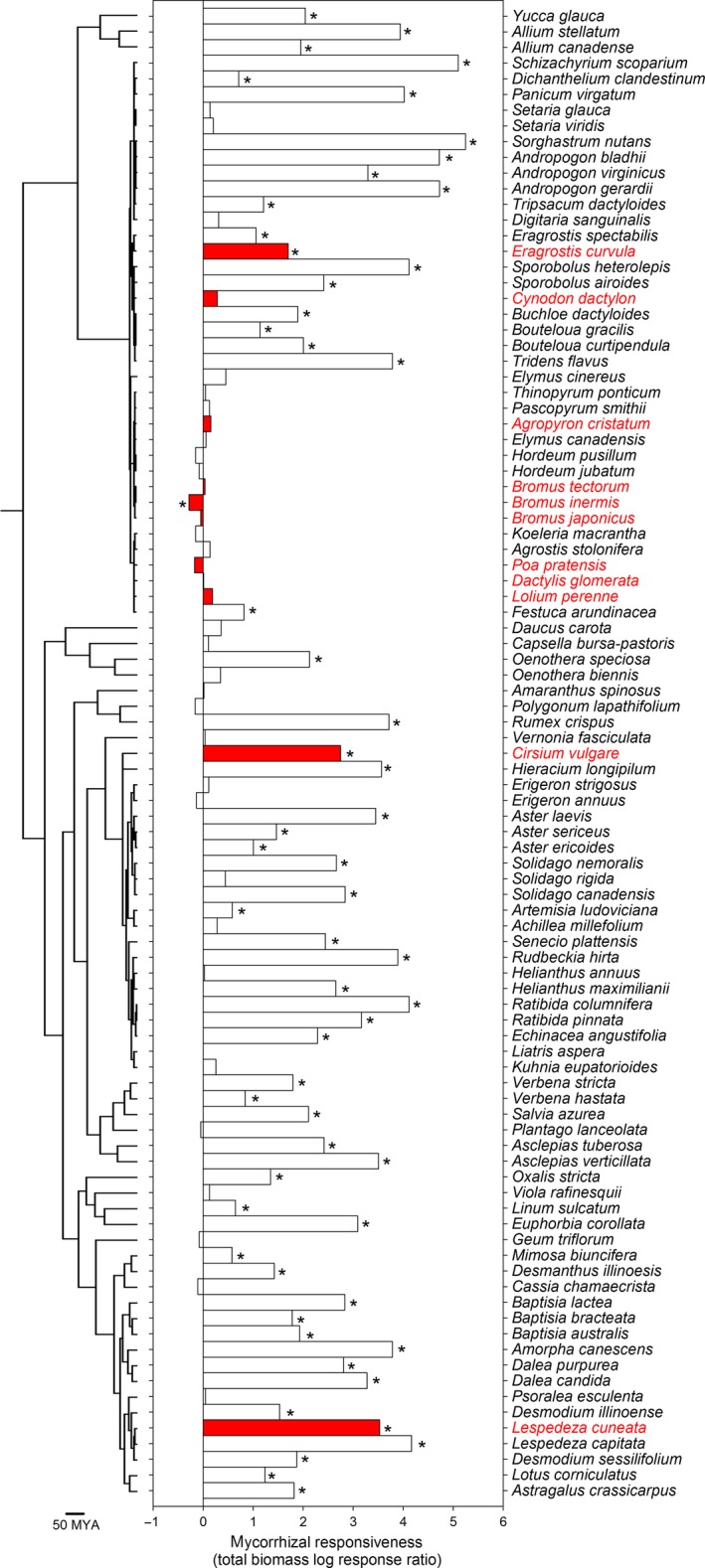
Molecular phylogeny and mycorrhizal responsiveness for 95 grassland plant species from the Central Plains (Wilson & Hartnett, 1998). Taxa labels and bars shaded red indicate taxa classified as invasive. Effect sizes were based on log response ratios (ln[mean biomass of plants with AMF/mean biomass of plants without AMF]) of total plant biomass data. Positive log response ratios indicate plants were larger with arbuscular mycorrhizal fungi than controls. Wilson and Hartnett (1998) tested whether plants inoculated with AMF had significantly different biomass from controls by two‐sample t‐test (*p ≤ .05)
We found no evidence (p ≥ .16) that plants classified as invasive interacted differently with AMF than noninvasive taxa for the Northern and Central Plains datasets while controlling for phylogenetic similarity (Table 1). Analyses for the Northern Plains portion included two effect size measurements (i.e., log response ratio and standardized effect size) for each of three response variables (i.e., total biomass, shoot biomass, and root mass ratio) (Table 1). The Central Plains dataset was more limited in terms of effect size measurements but there were still no appreciable differences among invasive and noninvasive taxa (Table 1).
Table 1.
Phylogenetic ANOVA results for differences in effect size (log response ratio [LRR] and standardized effect size [SES]) by invasive status (invasive vs. noninvasive) of grassland plants from the Northern and Central Plains. Effect sizes differentiate plant responses when grown with arbuscular mycorrhizal fungi versus a sham‐inoculant control
| Biomass data | Effect size | Northern Plains | Central Plainsa |
|---|---|---|---|
| Total biomass | LRR | F = 1.33, p = .23 | F = 3.75, p = .43 |
| SES | F = 2.02, p = .16 | No data | |
| Shoot biomass | LRR | F = 1.31, p = .24 | No data |
| SES | F = 1.28, p = .23 | No data | |
| Root mass ratio | LRR | F = 0.14, p = .72 | No data |
| SES | F = 0.17, p = .66 | No data |
Data from Wilson and Hartnett (1998).
Invasive status was nonrandomly distributed over the phylogeny of taxa from the Northern Plains (D‐statistic = 0.31, p < .001; Figure 1) and Central Plains (D‐statistic = 0.32, p < .001; Figure 3) and followed a Brownian model of evolution in both the Northern (p = .161) and Central (p = .176) Plains. Invasive taxa were primarily from two plant clades (i.e., Asteraceae, Poaceae), which contained a mix of invasive and noninvasive taxa. Overall, we observed little evidence that effects of AMF on plants from the Northern Plains were strongly dependent on phylogeny. For the Northern Plains dataset, we did not detect evidence that AMF responsiveness was phylogenetically conserved using log response ratios for total biomass (Abouheif's Cmean, p = .24, K statistic, K = 0.02, p = .90) and shoot biomass (Abouheif's Cmean, p = .20, K statistic, K = 0.02, p = .82) or standardized effect sizes for total biomass (Abouheif's Cmean, p = .06, K statistic, K = 0.03, p = .64; Figure 1), shoot biomass (Abouheif's Cmean, p = .24, K statistic, K = 0.03, p = .75), and root mass ratios (Abouheif's Cmean, p = .13, K statistic, K = 0.04, p = .38; Figure 2). There was, however, a phylogenetical signal for log response ratios of root mass ratio data (Abouheif's Cmean, p = .009, K statistic, K = 0.07, p = .03). In contrast to the Northern Plains dataset, we observed a phylogenetic signal for log response ratios of total biomass data (Abouheif's Cmean, p = .001, K statistic, K = 0.16, p = .004; Figure 3) for the Central Plains. In both datasets K < 1, indicating that related taxa tended to have more divergent responses to AMF than expected.
4. DISCUSSION
Overall, using phylogenetically controlled statistical tests, we found no evidence that invasive grassland plant species responded differently to inoculation with AMF relative to other plant taxa in either the Northern or Central Plains dataset. Plant invasive status in both datasets was nonrandomly distributed across the phylogeny; invasive plants were disproportionally from two plant families (Asteraceae and Poaceae; Figures 1, 2, 3). Although there was a significant phylogenetic signal for mycorrhizal responsiveness with the Central Plains dataset (Figure 3), the overall pattern (K < 1) indicated that closely related taxa were more likely to differ than expected from a Brownian random walk model of evolution (Ackerly, 2009). Others have also reported that invasive plant taxa occur predominantly in a few plant clades (Lim, Crawley, de Vere, Rich, & Savolainen, 2014) and that mycorrhizal responsiveness exhibited greater divergence than convergence (K < 1) for grassland taxa (Anacker et al., 2014). The relatively weak effects of phylogeny, and the finding that mycorrhizal responsiveness is more likely to be divergent than conserved suggest that predicting mycorrhizal responsiveness in different ecosystems may be enhanced by considering key functional traits associated with root function or key genes related to mycorrhizal interactions, rather than phylogenies constructed with neutral molecular markers (e.g., matK, rbcL) used to identify plants.
Our findings are consistent with a recent meta‐analysis that compared invasive non‐natives to natives, albeit without the inclusion of phylogenetic information (Bunn et al., 2015), but contrary to a prior analysis of the Central Plains dataset (Fig. 3 in Pringle et al., 2009). In the latter case, inclusion of phylogenetic information helped to partition variance due to invasive status and phylogenetic relatedness which was not possible by Pringle et al. (2009). When phylogenetic relatedness was not controlled (i.e., standard ANOVA), results for the Central Plains dataset continued to provide marginally significant evidence that invasive taxa were less responsive to AMF (ANOVA, F 1,93 = 3.745, p = .056) similar to related comparisons by Pringle et al. (2009). Phylogenetic analyses, however, indicated a nonrandom phylogenetic signal for AMF responsiveness with the Central Plains dataset and enabled us to differentiate whether AMF responsiveness varied by invasive status and/or phylogeny. Nevertheless, phylogeny was a weak overall predictor of mycorrhizal responsiveness for the Central Plains dataset (Reinhart et al., 2012) partly because related taxa tended to have greater trait divergence (K < 1) than convergence (K > 1). Trait divergence may be a result of related (and neighboring) plants associating with divergent AMF communities (Reinhart & Anacker, 2014; Veresoglou & Rillig, 2014) and/or other factors that contribute to niche partitioning among related plant species.
The Northern Plains region is known to support some of North America's most invasive plant taxa, including Bromus tectorum, Centaurea stoebe, and Euphorbia esula. Among the 14 invasive species in the Northern Plains dataset, three (Agropyron cristatum, C. stoebe, and E. esula) responded positively to AMF and three (A. cristatum [Fairway], B. japonicus, and B. tectorum) responded negatively to AMF. These divergent responses likely contributed to the lack of differences between invasive and other taxa. These findings support the hypothesis that a multitude of factors (e.g., anthropogenic disturbance, plant–microbe interactions, traits) contribute to invasiveness (Hierro et al., 2016; e.g., Maron et al., 2013; Müller, Horstmeyer, Rönneburg, van Kleunen, & Dawson, 2016).
It is not too surprising that on average invasive taxa are not less responsive to AMF as invasiveness may be facilitated by either decreased or increased mycorrhizal responsiveness. For example, the invasive grass B. tectorum is known to reduce AMF abundance (Busby, Stromberger, Rodriguez, Gebhart, & Paschke, 2013; Lekberg et al., 2013) and responded either negatively or neutrally to AMF (Figures 1 and 3). Other invasive grasses (A. cristatum, B. inermis) also appear capable of reducing AMF diversity (Jordan, Aldrich‐Wolfe, Huerd, Larson, & Muehlbauer, 2012) and had varying responses to AMF (Figures 1 and 3). Some invasive plants are thought to degrade AMF communities (Hale & Kalisz, 2012; Pakpour & Klironomos, 2015; Stinson et al., 2006; Vogelsang & Bever, 2009; Zhang et al., 2010) which may contribute to soil legacy effects that negatively impact the resilience of resident plant communities (Jordan, Larson, & Huerd, 2008; Perkins & Nowak, 2012, 2013). In contrast, some invasive forbs (i.e., C. stoebe and E. esula) increased the abundance and diversity of AMF communities (Lekberg et al., 2013), responded positively to AMF (Figure 1), and AMF enhanced the invasiveness of C. stoebe (Callaway, Mahall, Wicks, Pankey, & Zabinski, 2003; Marler et al., 1999). Therefore, mycorrhizal responsiveness facilitates the invasion of particular plant species, but weak mycorrhizal responsiveness is seemingly not a general mechanism of plant invasion.
Dominant native plants in the Northern Plains tended to be unresponsive to AMF. Specifically, four of the most prominent plants in the Northern Plains (Artemisia tridentata, Bouteloua gracilis, Hesperostipa comata, and Pascopyrum smithii) routinely associate with AMF (Bethlenfalvay & Dakessian, 1984; Carter, Smith, White, & Serpe, 2014; Reinhart & Anacker, 2014), but we found that only H. comata increased shoot biomass with AMF (Figure 1b). Artemisia tridentata actually had less shoot biomass with AMF than without. Other studies reported no effect of AMF on A. tridentata during mine reclamation (Biondini, Bonham, & Redente, 1985; Stahl, Williams, & Christensen, 1988), restoration postfire (review by Dettweiler‐Robinson et al., 2013), and a controlled mycorrhizal responsiveness test (Busby et al., 2011). Yet, other experiments indicated that A. tridentata seedlings had increased drought tolerance (Stahl, Schuman, Frost, & Williams, 1998), biomass (Stahl et al., 1988), and survival [fall but not spring transplants] with AMF (Davidson, 2015). Prominent congeners responded both positively (A. dracunculus) and negatively (A. frigida) to AMF. Bouteloua gracilis and P. smithii were also used in the Central Plains dataset, and B. gracilis plants were appreciably larger with AMF (Figure 3).
Although not a study aim, we are compelled to mention the apparent differences in AMF responsiveness for the two Plains ecoregions (Figure 1 vs. 3). Specifically, 61% of plants in the Central Plains were larger with AMF than without. Yet, appreciable positive effects of AMF on total plant biomass and shoot biomass were detected for only 28% of grassland plants in the Northern Plains. This apparent difference between Plains regions is intriguing, but we cannot reconcile whether such a difference is due to important methodological differences between studies or important ecological differences between Plains regions. There is a small body of literature suggesting ecological differences are possible—agronomic plants in calcareous soils were often less responsive to AMF (Li, Zhu, Marschner, Smith, & Smith, 2005; Zhu, Smith, & Smith, 2003). Calcium carbonates (which are prevalent in dryland soils like the Northern Plains) are known to reduce the bioavailability of phosphorus (e.g., Belnap, 2011) and may impact mycorrhizal mutualisms. Further testing is necessary to determine whether these regions truly have divergent interactions with AMF and the factors contributing to such differences.
In conclusion, despite substantial differences in functional characteristics, taxa sampled, and growth conditions for the experiments, we found no evidence to support the hypothesis that invasive taxa differ from native taxa in the degree of mycorrhizal responsiveness. Although we considered interspecific differences in the present analysis (i.e., a single genotype represented most plant species), others have noted that plant–AMF interactions vary by cultivars of invasive taxa like A. cristatum (Jun & Allen, 1991; Figure 1). Genotypic and geographical variation in plant–AMF interactions likely complicates formulation of robust generalizations for taxa with broad distributions. For instance, other plant–AMF interactions are conceivable with other pairings of AMF communities (species and genotypes), plant genotypes (e.g., Klironomos, 2003; Stahl & Smith, 1984), and growth conditions. Additional experiments that incorporate population and genotypic variability are necessary to quantify general effects of soil microbes across a plant's range (Callaway, Bedmar, Reinhart, Silvan, & Klironomos, 2011; Reinhart, Royo, van der Putten, & Clay, 2005; Wagner, Antunes, Ristow, Lechner, & Hensen, 2011).
CONFLICT OF INTEREST
None declared.
Supporting information
ACKNOWLEDGMENTS
We thank C. Murphy and B. Garber for assistance in the laboratory. We thank J. Morton and members of the J. Bever's (Indiana University) laboratory for providing training on AMF spore isolation (E. Middleton) and input on production of AMF inocula (K. Vogelsang, W. Kaonongbua). We also appreciate G. Gramig, J. Mangold, L. Vermeire, R. Kilian, and S. Huerd for contributing either input on which plant species to include in the study and/or seed. We also thank J. Muscha and L. Vermeire for comments on an earlier version of the manuscript. This work was funded by USDA appropriated funds (CRIS # 5434‐21630‐003‐00D). YL is grateful to MPG Ranch for funding. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USA Department of Agriculture (USDA). USDA is an equal opportunity provider and employer.
Reinhart KO, Lekberg Y, Klironomos J, Maherali H. Does responsiveness to arbuscular mycorrhizal fungi depend on plant invasive status?. Ecol Evol. 2017;7:6482–6492. https://doi.org/10.1002/ece3.3226
REFERENCES
- Abbott, L. K. , & Robson, A. D. (1991). Factors influencing the occurrence of vesicular‐arbuscular mycorrhizas. Agriculture, Ecosystems & Environment, 35, 121–150. [Google Scholar]
- Abouheif, E. (1999). A method for testing the assumption of phylogenetic independence in comparative data. Evolutionary Ecology Research, 1, 895–909. [Google Scholar]
- Ackerly, D. (2009). Conservatism and diversification of plant functional traits: Evolutionary rates versus phylogenetic signal. Proceedings of the National Academy of Sciences, 106, 19699–19706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anacker, B. L. , Klironomos, J. N. , Maherali, H. , Reinhart, K. O. , & Strauss, S. Y. (2014). Phylogenetic conservatism in plant‐soil feedback and its implications for plant abundance. Ecology Letters, 17, 1613–1621. [DOI] [PubMed] [Google Scholar]
- ANGIOSPERM PHYLOGENY GROUP (2009). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society, 161, 105–121. [Google Scholar]
- Bell, C. D. , Soltis, D. E. , & Soltis, P. S. (2010). The age and diversification of the angiosperms re‐revisited. American Journal of Botany, 97, 1296–1303. [DOI] [PubMed] [Google Scholar]
- Belnap, J. (2011). Biological phosphorus cycling in dryland regions In Bünemann E., Oberson A., & Frossard E. (Eds.), Phosphorus in action: Biological processes in soil phosphorus cycling (pp. 371–406). Berlin, Heidelberg: Springer Berlin Heidelberg. [Google Scholar]
- Bethlenfalvay, G. J. , & Dakessian, S. (1984). Grazing effects on mycorrhizal colonization and floristic composition of the vegetation on a semiarid range in northern Nevada. Journal of Range Management, 37, 312–316. [Google Scholar]
- Biondini, M. E. , Bonham, C. D. , & Redente, E. F. (1985). Secondary successional patterns in a sagebrush (Artemisia tridentata) community as they relate to soil disturbance and soil biological activity. Vegetatio, 60, 25–36. [Google Scholar]
- Blomberg, S. P. , Garland, T. , & Ives, A. R. (2003). Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution, 57, 717–745. [DOI] [PubMed] [Google Scholar]
- Brundrett, M. C. (2009). Mycorrhizal associations and other means of nutrition of vascular plants: Understanding the global diversity of host plants by resolving conflicting information and developing reliable means of diagnosis. Plant and Soil, 320, 37–77. [Google Scholar]
- Brundrett, M. , Melville, L. , & Peterson, L. (1994). Practical methods in mycorrhiza research. 8727 Lochside Drive, Sidney, BC, V8L 1M8, CANADA: Mycologue Publications. [Google Scholar]
- Bunn, R. A. , Ramsey, P. W. , & Lekberg, Y. (2015). Do native and invasive plants differ in their interactions with arbuscular mycorrhizal fungi? A meta‐analysis. Journal of Ecology, 103, 1547–1556. [Google Scholar]
- Busby, R. R. , Gebhart, D. L. , Stromberger, M. E. , Meiman, P. J. , & Paschke, M. W. (2011). Early seral plant species' interactions with an arbuscular mycorrhizal fungi community are highly variable. Applied Soil Ecology, 48, 257–262. [Google Scholar]
- Busby, R. R. , Stromberger, M. E. , Rodriguez, G. , Gebhart, D. L. , & Paschke, M. W. (2013). Arbuscular mycorrhizal fungal community differs between a coexisting native shrub and introduced annual grass. Mycorrhiza, 23, 129–141. [DOI] [PubMed] [Google Scholar]
- Callaway, R. M. , Bedmar, E. J. , Reinhart, K. O. , Silvan, C. G. , & Klironomos, J. (2011). Effects of soil biota from different ranges on Robinia invasion: Acquiring mutualists and escaping pathogens. Ecology, 92, 1027–1035. [DOI] [PubMed] [Google Scholar]
- Callaway, R. M. , Mahall, B. E. , Wicks, C. , Pankey, J. , & Zabinski, C. (2003). Soil fungi and the effects of an invasive forb on grasses: Neighbor identity matters. Ecology, 84, 129–135. [Google Scholar]
- Carter, K. A. , Smith, J. F. , White, M. M. , & Serpe, M. D. (2014). Assessing the diversity of arbuscular mycorrhizal fungi in semiarid shrublands dominated by Artemisia tridentata ssp. wyomingensis . Mycorrhiza, 24, 301–314. [DOI] [PubMed] [Google Scholar]
- Davidson, B. E. (2015). Consequence of pre‐inoculation with native arbuscular mycorrhizae on root colonization and survival of Wyoming big sagebrush (Artemisia tridentata spp. wyomingensis) seedlings after transplanting. M.S., Boise State University. [DOI] [PubMed]
- Dettweiler‐Robinson, E. , Bakker, J. D. , Evans, J. R. , Newsome, H. , Davies, G. M. , Wirth, T. A. , … Dunwiddie, P. W. (2013). Outplanting Wyoming big sagebrush following wildfire: Stock performance and economics. Rangeland Ecology & Management, 66, 657–666. [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 35, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland, T. , Dickerman, A. W. , Janis, C. M. , & Jones, J. A. (1993). Phylogenetic analysis of covariance by computer simulation. Systematic Biology, 42, 265–292. [Google Scholar]
- Gurevitch, J. , Fox, G. A. , Wardle, G. M. , Inderjit, G. , & Taub, D. (2011). Emergent insights from the synthesis of conceptual frameworks for biological invasions. Ecology Letters, 14, 407–418. [DOI] [PubMed] [Google Scholar]
- Hale, A. N. , & Kalisz, S. (2012). Perspectives on allelopathic disruption of plant mutualisms: A framework for individual‐ and population‐level fitness consequences. Plant Ecology, 213, 1991–2006. [Google Scholar]
- Hempel, S. , Götzenberger, L. , Kühn, I. , Michalski, S. G. , Rillig, M. C. , Zobel, M. , & Moora, M. (2013). Mycorrhizas in the Central European flora: Relationships with plant life history traits and ecology. Ecology, 94, 1389–1399. [DOI] [PubMed] [Google Scholar]
- Hierro, J. L. , Khetsuriani, L. , Andonian, K. , Eren, Ö. , Villarreal, D. , Janoian, G. , … Callaway, R. M. (2016). The importance of factors controlling species abundance and distribution varies in native and non‐native ranges. Ecography, https://doi.org/10.1111/ecog.02633. [Google Scholar]
- Huelsenbeck, J. P. , & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogeny. Bioinformatics, 17, 754–755. [DOI] [PubMed] [Google Scholar]
- Janos, D. P. (2007). Plant responsiveness to mycorrhizas differs from dependence upon mycorrhizas. Mycorrhiza, 17, 75–91. [DOI] [PubMed] [Google Scholar]
- Johnson, N. C. , Wilson, G. W. T. , Bowker, M. A. , Wilson, J. A. , & Miller, R. M. (2010). Resource limitation is a driver of local adaptation in mycorrhizal symbioses. Proceedings of the National Academy of Sciences of the United States, 107, 2093–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, N. R. , Aldrich‐Wolfe, L. , Huerd, S. C. , Larson, D. L. , & Muehlbauer, G. (2012). Soil‐occupancy effects of invasive and native grassland plant species on composition and diversity of mycorrhizal associations. Invasive Plant Science and Management, 5, 494–505. [Google Scholar]
- Jordan, N. R. , Larson, D. L. , & Huerd, S. C. (2008). Soil modification by invasive plants: Effects on native and invasive species of mixed‐grass prairies. Biological Invasions, 10, 177–190. [Google Scholar]
- Jun, D. J. , & Allen, E. B. (1991). Physiological responses of 6 wheatgrass cultivars to mycorrhizae. Journal of Range Management, 44, 336–341. [Google Scholar]
- Klironomos, J. N. (2003). Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology, 84, 2292–2301. [Google Scholar]
- Koide, R. T. , & Li, M. (1989). Appropriate controls for vesicular‐arbuscular mycorrhiza research. New Phytologist, 111, 35–44. [Google Scholar]
- Koricheva, J. , Gurevitch, J. , & Mengersen, K. (2013). Handbook of meta‐analysis in ecology and evolution. Princeton, NJ: Princeton University Press. [Google Scholar]
- Lajtha, K. , & Schlesinger, W. H. (1988). The biogeochemistry of phosphorus cycling and phosphorus availability along a desert soil chronosequence. Ecology, 69, 24–39. [Google Scholar]
- Lekberg, Y. , Gibbons, S. M. , Rosendahl, S. , & Ramsey, P. W. (2013). Severe plant invasions can increase mycorrhizal fungal abundance and diversity. The ISME Journal, 7, 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekberg, Y. , & Waller, L. P. (2016). What drives differences in arbuscular mycorrhizal fungal communities among plant species?. Fungal Ecology Part B, 24, 135–138. [Google Scholar]
- Li, H. Y. , Zhu, Y. G. , Marschner, P. , Smith, F. A. , & Smith, S. E. (2005). Wheat responses to arbuscular mycorrhizal fungi in a highly calcareous soil differ from those of clover, and change with plant development and P supply. Plant and Soil, 277, 221–232. [Google Scholar]
- Lim, J. , Crawley, M. J. , de Vere, N. , Rich, T. , & Savolainen, V. (2014). A phylogenetic analysis of the British flora sheds light on the evolutionary and ecological factors driving plant invasions. Ecology and Evolution, 4, 4258–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack, R. N. , Simberloff, D. , Lonsdale, W. M. , Evans, H. , Clout, M. , & Bazzaz, F. A. (2000). Biotic invasions: Causes, epidemiology, global consequences, and control. Ecological Applications, 10, 689–710. [Google Scholar]
- Marler, M. J. , Zabinski, C. A. , & Callaway, R. M. (1999). Mycorrhizae indirectly enhance competitive effects of an invasive forb on a native bunchgrass. Ecology, 80, 1180–1186. [Google Scholar]
- Maron, J. L. , Waller, L. P. , Hahn, M. A. , Diaconu, A. , Pal, R. W. , Müller‐Schärer, H. , … Callaway, R. M. (2013). Effects of soil fungi, disturbance and propagule pressure on exotic plant recruitment and establishment at home and abroad. Journal of Ecology, 101, 924–932. [Google Scholar]
- McGonigle, T. P. , Miller, M. H. , Evans, D. G. , Fairchild, G. L. , & Swan, J. A. (1990). A new method which gives an objective measure of colonization of roots by vesicular‐arbuscular mycorrhizal fungi. New Phytologist, 115, 495–501. [DOI] [PubMed] [Google Scholar]
- Mooney, H. A. , & Hobbs, R. J. (2000). Invasive species in a changing world, CSIRO.
- Müller, G. , Horstmeyer, L. , Rönneburg, T. , van Kleunen, M. , & Dawson, W. (2016). Alien and native plant establishment in grassland communities is more strongly affected by disturbance than above‐ and below‐ground enemies. Journal of Ecology, 104, 1233–1242. [Google Scholar]
- Nylander, J. A. (2004). MrModeltest v2. 2.13 ed. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Öpik, M. , Vanatoa, A. , Vanatoa, E. , Moora, M. , Davison, J. , Kalwij, J. M. , … Zobel, M. (2010). The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytologist, 188, 223–241. [DOI] [PubMed] [Google Scholar]
- Pakpour, S. , & Klironomos, J. (2015). The invasive plant, Brassica nigra, degrades local mycorrhizas across a wide geographical landscape. Royal Society Open Science, DOI: 10.1098/rsos.150300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, L. B. , & Nowak, R. S. (2012). Soil conditioning and plant‐soil feedbacks affect competitive relationships between native and invasive grasses. Plant Ecology, 213, 1337–1344. [Google Scholar]
- Perkins, L. B. , & Nowak, R. S. (2013). Native and non‐native grasses generate common types of plant‐soil feedbacks by altering soil nutrients and microbial communities. Oikos, 122, 199–208. [Google Scholar]
- Pimentel, D. , Lach, L. , Zuniga, R. , & Morrison, D. (2000). Environmental and economic costs of nonindigenous species in the United States. BioScience, 50, 53–65. [Google Scholar]
- Pringle, A. , Bever, J. D. , Gardes, M. , Parrent, J. L. , Rillig, M. C. , & Klironomos, J. N. (2009). Mycorrhizal symbioses and plant invasions. Annual Review of Ecology, Evolution, and Systematics, 40, 699–715. [Google Scholar]
- van der Putten, W. H. , Klironomos, J. N. , & Wardle, D. A. (2007). Microbial ecology of biological invasions. ISME Journal, 1, 28–37. [DOI] [PubMed] [Google Scholar]
- Reinhart, K. O. , & Anacker, B. L. (2014). More closely related plants have more distinct mycorrhizal communities. AoB Plants, 2014, https://doi.org/10.1093/aobpla/plu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, K. O. , Royo, A. A. , van der Putten, W. H. , & Clay, K. (2005). Soil feedback and pathogen activity in Prunus serotina throughout its native range. Journal of Ecology, 93, 890–898. [Google Scholar]
- Reinhart, K. O. , Wilson, G. W. T. , & Rinella, M. J. (2012). Predicting plant responses to mycorrhizae: Integrating evolutionary history and plant traits. Ecology Letters, 15, 689–695. [DOI] [PubMed] [Google Scholar]
- Revell, L. J. (2012). phytools: An R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution, 3, 217–223. [Google Scholar]
- Richardson, D. M. , Allsopp, N. , D'Antonio, C. M. , Milton, S. J. , & Rejmanek, M. (2000). Plant invasions—The role of mutualisms. Biological Reviews of the Cambridge Philosophical Society, 75, 65–93. [DOI] [PubMed] [Google Scholar]
- Seifert, E. K. , Bever, J. D. , & Maron, J. L. (2009). Evidence for the evolution of reduced mycorrhizal dependence during plant invasion. Ecology, 90, 1055–1062. [DOI] [PubMed] [Google Scholar]
- Smith, S. E. , & Read, D. J. (2008). Mycorrhizal symbiosis. New York, NY: Academic Press. [Google Scholar]
- Stahl, P. D. , Schuman, G. E. , Frost, S. M. , & Williams, S. E. (1998). Arbuscular mycorrhizae and water stress tolerance of Wyoming big sagebrush seedlings. Soil Science Society of America Journal, 62, 1309–1313. [Google Scholar]
- Stahl, P. D. , & Smith, W. K. (1984). Effects of different geographic isolates of Glomus on the water relations of Agropyron smithii . Mycologia, 76, 261–267. [Google Scholar]
- Stahl, P. D. , Williams, S. E. , & Christensen, M. J. (1988). Efficacy of native vesicular‐arbuscular mycorrhizal fungi after severe soil disturbance. New Phytologist, 110, 347–354. [Google Scholar]
- Stinson, K. A. , Campbell, S. A. , Powell, J. R. , Wolfe, B. E. , Callaway, R. M. , Thelen, G. C. , … Klironomos, J. N. (2006). Invasive plant suppresses the growth of native tree seedlings by disrupting belowground mutualisms. PLoS Biology, 4, 727–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintjer, T. , & Rudgers, J. A. (2006). Grass–herbivore interactions altered by strains of a native endophyte. New Phytologist, 170, 513–521. [DOI] [PubMed] [Google Scholar]
- Veresoglou, S. D. , & Rillig, M. C. (2014). Do closely related plants host similar arbuscular mycorrhizal fungal communities? A meta‐analysis. Plant and Soil, 377, 395–406. [Google Scholar]
- Vogelsang, K. M. , & Bever, J. D. (2009). Mycorrhizal densities decline in association with nonnative plants and contribute to plant invasion. Ecology, 90, 399–407. [DOI] [PubMed] [Google Scholar]
- Wagner, V. , Antunes, P. M. , Ristow, M. , Lechner, U. , & Hensen, I. (2011). Prevailing negative soil biota effect and no evidence for local adaptation in a widespread Eurasian grass. PLoS One, 6, e17580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller, L. P. , Callaway, R. M. , Klironomos, J. N. , Ortega, Y. K. , & Maron, J. L. (2016). Reduced mycorrhizal responsiveness leads to increased competitive tolerance in an invasive exotic plant. Journal of Ecology, 104, 1599–1607. [Google Scholar]
- Webb, C. O. , & Donoghue, M. J. (2004). Phylomatic: Tree assembly for applied phylogenetics. Molecular Ecology Notes, 5, 181–183. [Google Scholar]
- Wilson, G. W. T. , & Hartnett, D. C. (1998). Interspecific variation in plant responses to mycorrhizal colonization in tallgrass prairie. American Journal of Botany, 85, 1732–1738. [PubMed] [Google Scholar]
- Zhang, Q. , Yang, R. , Tang, J. , Yang, H. , Hu, S. , & Chen, X. (2010). Positive feedback between mycorrhizal fungi and plants influences plant invasion success and resistance to invasion. PLoS One, 5, e12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y.‐G. , Smith, A. F. , & Smith, S. E. (2003). Phosphorus efficiencies and responses of barley (Hordeum vulgare L.) to arbuscular mycorrhizal fungi grown in highly calcareous soil. Mycorrhiza, 13, 93–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


