Abstract
It has been previously observed that the intrinsically weak variant GC donor sites, in order to be recognized by the U2-type spliceosome, possess strong consensus sequences maximized for base pair formation with U1 and U5/U6 snRNAs. However, variability in signal strength is a fundamental mechanism for splice site selection in alternative splicing. Here we report human alternative GC-AG introns (for the first time from any species), and show that while constitutive GC-AG introns do possess strong signals at their donor sites, a large subset of alternative GC-AG introns possess weak consensus sequences at their donor sites. Surprisingly, this subset of alternative isoforms shows strong consensus at acceptor exon positions 1 and 2. The improved consensus at the acceptor exon can facilitate a strong interaction with U5 snRNA, which tethers the two exons for ligation during the second step of splicing. Further, these isoforms nearly always possess alternative acceptor sites and exhibit particularly weak polypyrimidine tracts characteristic of AG-dependent introns. The acceptor exon nucleotides are part of the consensus required for the U2AF35-mediated recognition of AG in such introns. Such improved consensus at acceptor exons is not found in either normal or alternative GT-AG introns having weak donor sites or weak polypyrimidine tracts. The changes probably reflect mechanisms that allow GC-AG alternative intron isoforms to cope with two conflicting requirements, namely an apparent need for differential splice strength to direct the choice of alternative sites and a need for improved donor signals to compensate for the central mismatch base pair (C-A) in the RNA duplex of U1 snRNA and the pre-mRNA. The other important findings include (i) one in every twenty alternative introns is a GC-AG intron, and (ii) three of every five observed GC-AG introns are alternative isoforms.
INTRODUCTION
Two steps of splicing
During RNA splicing, introns are precisely removed and the flanking exons are ligated together. Splicing is carried out by the spliceosome (a large ribonucleoprotein complex containing 5 snRNA molecules and a large number of snRNA-associated proteins as well as other protein factors), which is assembled upon the pre-mRNA. The three important splice signals within the pre-mRNA are the 5′ splice site, the 3′ splice site and the branch point. Splicing involves two catalytic steps (reviewed in 1). During the first step, the 2′ hydroxyl of an adenosine residue at the branch point attacks the phosphate bond at the 5′ splice site to release the 5′ exon and to form an intermediate lariat in which the 5′ end of the intron is attached to the adenosine at the branch point by a 5′-2′ phosphodiester bond. During the second step, the 3′ hydroxyl of the 5′ exon attacks the phosphate at the 3′ splice site resulting in the ligation of the 5′ and 3′ exons and release of the lariat intron.
RNA–RNA interactions
The general splicing process has been elucidated to occur in two steps, both of which are largely RNA-mediated (2). First, U1 snRNA interacts with the pre-mRNA at the 5′ splice site by base pair formation with the nearly invariant GU dinucleotide and the flanking nucleotides (–3 to +8). At a later stage in the first step, the U1 snRNA interaction is replaced by interactions from U5 snRNA (with exon positions –3 to –1) and U6 snRNA (with intron positions +4 to +6). Subsequently the branch point is associated with the U2 snRNA through base pair interactions (3–4). An interaction between the U2AF65 factor and a polypyrimidine tract adjacent to the branch point in the pre-mRNA acts to allow recognition of the branch point (5–10). U2AF65 is a subunit of the U2AF heterodimer in conjunction with U2AF35. In the case where the intron has a strong polypyrimidine tract (named an ‘AG-independent’ intron; 11), recognition of the nearly invariant AG at the acceptor site is needed only during the second step of splicing. In the case where the intron lacks or shows a weak polypyrimidine tract, (named an ‘AG-dependent intron’), U2AF35 can recognize and directly interact with the AG dinucleotide in the first step of splicing (8–10). Such an interaction is critical for the binding of U2AF65 with the weak polypyrimidine tract and the subsequent branch point definition. The U2AF35 binding is sequence-specific and the required sequence on the pre-mRNA includes the acceptor exon positions +1 to +2 in addition to the AG dinucleotide (12). During the second step of splicing, U5 snRNA extends its base pair formation to interact with the same acceptor exon positions and thereby tethers the two exons for ligation (13–15).
Alternative splicing
It is believed that at least one in every three human genes can have alternative exon–intron structure (16–18 and http://www.ebi.ac.uk/~thanaraj/gene_altSplice.html). Choice of alternative splice sites is made early in the assembly of the spliceosome complex. Competition between alternative sites depends on the relative quality of the constitutive splice signals, a balance that presumably can be shifted by protein factors acting on regulatory sequences (19). The splice sites of alternatively spliced exons can deviate from the consensus sequences (20) and variability in the donor site consensus can act as a fundamental regulatory mechanism (21). Further, the strength of the polypyrimidine tract can alter 3′ splice site selection by promoting alternative branch site selection (22). The protein factors SF2/ASF and hnRNP A1 have been implicated in the choice of proximal versus distal 5′ splice sites (of comparable strength) that compete for a common acceptor site (23) as well as in the choice of alternative 3′ splice sites competing for a common donor site (24).
GC-AG introns
It is known that there exist variants to the standard form GT-AG introns (1,25–27). The major splice variant is GC-AG (25,28). Burset and coworkers (29), in the process of creating a database (SpliceDB) of EST-confirmed canonical and non-canonical splice sites, observed that GC splice sites account for 0.5% of annotated donor sites. It has been reported (25,28), based on data sets derived from annotated gene structures, that GC donor sites possess a strong consensus sequence. Since both the GT-AG and GC-AG splice sites are processed by the standard U2-type spliceosome (1,27,30), there is a mismatch base pair in the interaction between the donor site and U1 snRNA (with the C at intron position 2). Thus, it has been proposed that in order to compensate for such a mismatch base pair, the consensus sequence around the GC donor site has evolved to maximize base pair formation with other positions in the U1 snRNA (1,27,28,31).
In this work we carry out spliced-alignments of human genes with human transcripts (EST and mRNA sequences) and identify all potential human GC-AG introns. The GC-AG introns either correspond to the GenBank-annotated introns or represent alternative forms (of both the GT-AG and GC-AG types). Extensive analysis was carried out to ascertain the genuineness of the observed normal and alternative GC-AG intron isoforms, and the splice signals were characterized and compared with those of GT-AG introns. We discuss the implications of our findings in terms of splicing mechanisms.
MATERIALS AND METHODS
Derivation of transcript-confirmed human GC-AG introns
A data set of human genes was obtained by extracting all protein-coding and intron-containing DNA entries in the EMBL/GenBank database (release 117) (32,33). All human mRNA and EST sequences were also extracted. From the start-up data set of genes we removed duplicate entries (multiple copies of the same gene), and any gene with a BLAST (34,35) match to our data set of hypervariable genes (derived from GenBank IgBlast) with Blast expectation <1e–10. Further redundancy is removed in the subsequent steps of the method.
Using BLAST (2.0.9) (34,35) we compared the EST and mRNA sequences to the gene sequences and identified matches that had a BLAST expectation value of 1e–10 or better and at least 95% similarity. Matches to repeats were defined by two or more individual regions from gene sequences aligning with the same region of a transcript, given an end point tolerance of 20% the length of the match. All such matches to repeats were discarded. This method of repeat removal can identify repeats with low copy number that may otherwise not be identified. Further, after the removal of repeats, any transcript that had matches to more than one gene was removed from the data set. This acts as a specific and local form of redundancy analysis and helps ensure that the matches used in the analysis are between transcripts and the genes from which they derive. Note that if we had undertaken a conventional redundancy purge of the gene data set (say at the 85% level), then one member of a each pair of highly homologous, but distinct, genes would be removed from the data set, and that this may lead to transcripts from the ‘redundant’ gene being incorrectly associated with the ‘non-redundant’ gene. We reiterate that this methodology ensures the avoidance of both introns from redundant genes and from repeated regions within genes.
For each gene, the matches were aligned to determine if they demonstrated the excision of an intron or introns from the gene sequence. Only those alignments unambiguously identifying either a GT-AG or GC-AG intron were further scrutinized (see below). Further, in the case of GC-AG introns, any alignments with a mismatch in a region of 20 bases into either flanking exon were examined on a case by case basis before being removed from, or retained in, the data set. There were six cases, with a single mismatch, that we decided to retain; five of these six showed a mismatch of the type purine–purine or pyrimidine–pyrimidine; the remaining one showed a T–A mismatch at position +17 of the acceptor site.
We classified the identified introns into one of three groups based on the strength of their donor site as determined by the program SpliceProximalCheck (SPC) (36,37, http://www.ebi.ac.uk/~thanaraj/SpliceProximalCheck.html and http://www.ebi.ac.uk/~thanaraj/MZEF-SPC.html). The program is based on a set of six rules that were derived to capture the signals at donor sites. It has been found that in a given population of human donor sites, roughly 80% can be represented by a single major rule, a further 16% can be represented by the remaining five minor rules, and roughly 3–4% of sites cannot be represented by any of these six rules (37). Such a distribution reflects differential strengths of the donor sites. Thus, this tool can categorize splice sites into one of three groups of descending donor strength: SPC Major, SPC Minor and SPC Negative (see Results and Discussion for further details).
Ascertaining that the GC-AG alternative intron isoforms are genuine
We obtained a list of 162 GC-AG introns of which 100 are not annotated as introns in the databases. While 78 of them overlapped with annotated introns, the remaining 22 are ‘cryptic introns’ (they overlapped with annotated exons that split into two alternative exon isoforms with an enclosed cryptic intron). The genuineness of these 100 introns, in particular of the subset with weak donor site (the SPC Minor and SPC Negative groups), is ascertained below.
Is the transcript–gene alignment by chance? Median and average lengths of the alignment to the 5′ and 3′ exons were 133 and 154 bases and 148 and 157 bases, respectively. These high values indicate that the transcript alignments are significant.
Checking the correctness of the DNA sequences and transchromosomal duplications. It is essential to make sure that the observed GC-AG introns, and the consequent findings, are not due to sequence errors in the gene entry. For this purpose, additional copies of the genes were obtained by using BLAST to match a 40 nt region comprising of 20 nt from either side of the donor (or acceptor) site, against all high throughput genome (HTG) sequences and, in the absence of HTG matches, any other DNA/RNA sequences. Such use of HTG sequences for checking the correctness of sequences from annotated gene entries has been suggested previously by Burset et al. (28). In 83 of the 100 alternative GC-AG introns, both the donor and acceptor site sequences could be verified in this manner. Of the 17 introns that could not be double-checked, only three were from the SPC Minor and SPC Negative groups.
It was observed that in 52 of the 100 cases, more than one HTG entry for the gene existed in the database. Interestingly, in 30 of these cases, the corresponding HTG entries were found to correspond to more than one chromosome. This indicates that in at least 30 of the 100 cases, the gene occurs in multiple copies, probably as a result of a transchromosomal duplication event. The set of genes containing the normal GC-AG introns was distinctly different in that only four of the 33 cases (for which HTG entries existed) showed such transchromosomal duplication events. In a handful of instances, copies of the genes showed nucleotide changes. Some of these changes can modulate the splice signals as follows. (i) Donor sites: in three cases, the splice site GC had changed to canonical GT; in at least one case, GC had changed to a non-functional GG. In three cases, changes at –2 (to G/A), at +6 (to T) and at +3 (to A) increased the consensus sequence; in two cases, a change at –5 (to C) and +6 (to A) decreased the splice strength. (ii) Acceptor sites: in two cases, the AG site had changed to either GG or AT, thereby probably aborting splicing at this site; in three other cases changes at either +1 (G→A) or at +2 (C→T) occurred, thereby modulating splice consensus.
Common subsequences between donor and acceptor site regions can point to more than one possible spliced-alignment. It can be difficult to exactly assign positions to splice sites in cases where the spliced alignments overlap due to a common subsequence adjacent to the 5′ and 3′ splice sites. An illustrative example of such cases is shown in Figure 1. In such situations, a number of rearrangements are possible for the gene–transcript alignment without changing the order of coding nucleotides. It is important to examine all such possibilities and ascertain that the observed GC-AG splice sites are genuine.
Figure 1.

Illustration of common subsequences, at the donor and acceptor sites, that allow for different interpretations of the gene–transcript alignment.
First, it was ascertained that rearrangements do not suggest the possibility of a canonical or standard non-canonical splice site. In none of the 100 cases did rearrangement give rise to the possibility of a GT-AG, AT-AC or even another GC-AG splice site. Secondly, it was checked whether rearrangements could suggest the possibility of any of the unusual non-canonical splice sites: GX-AG, XT-AG, GT-AX, GT-XG, AX-AC, XA-AC, AT-AX or AT-XC. These types were chosen for the following reasons. (i) Observed non-canonical splice sites in the literature, such as GG-AG, CT-AG (25); GT-CG, GT-TG (31); AT-AG, GA-AG (28); and GT-GG, TT-AG, GT-AC (as suggested in 28) are of the above types. (ii) It is generally believed that mismatching can be tolerated in splice sites at either 5′ or 3′ cleavage site but not at both. Examination of these possibilities revealed nine cases (from nine genes) that involved four types of non-canonical sites (GT-AT, GT-AA, GA-AG, AT-GC). We further examined each of these nine cases for its potential to be a splice site. In the five cases that are variants of GT-AG, the donor sites were tested for validation by our SPC program (36,37). All but one site was predicted to be false. As SPC validated the corresponding GC donor site as true, these other possibilities were disregarded. The single positive case was ignored for the reason that SPC also validated the corresponding GC donor site as true. In the four cases that are variants of AT AC, we checked the donor sites against the U12 intron consensus sequence of ATCCTTT at the donor intron positions +3 to +9 (as suggested in 1). None of the four sequences showed the U12-type donor consensus sequence. Thus, we consider the originally determined GC-AG sites to be genuine.
It was noted that only one of the above nine cases occurred in the SPC Minor or SPC Negative groups. Thus, these two groups have no ambiguity in the assignment of splice sites. In order to be doubly sure that the introns in these two groups are real, we checked their possible alignment rearrangements for extreme types of non-canonical sites such as GT-XX, XX-AG, GC-XX, AT-XX and XX-AC. There were seven cases from these two groups, which showed the possibilities of CG-AG, AG-AG, AA-AG, AC-AG, TG-AG, GT-CA and GT-CT. All these possible donor sites were predicted to be false by the SPC program.
Ascertaining that there are no insertions/deletions in the transcripts due to sequencing errors. U2-type donor sites often contain a GT dinucleotide at positions +5 and +6. Since GT dinucleotides were also observed at other positions, we scrutinized whether they could represent valid donor sequences. We examined every such occurrence in a 20 nt region around each GC donor site. It was interesting to note that none of the proximal GT sequences could be validated as true by the SPC program. This exercise ascertained that the observed GC donor sites are not due to small insertion or deletion errors in the transcripts.
Are the alternative GC-AG intron isoforms aberrant splicing rather than genuine alternative splicing? Aberrant splicing can take place in the following two situations: (i) mutations occurring at the normal splice site (or surrounding nucleotides) destroy the consensus sequence and thereby abort the normal splicing; (ii) mutations that occur elsewhere create consensus sequences at cryptic splice sites (38). Aberrant splicing can further be characterized by the observation that the supporting transcripts are solely derived from clone libraries that correspond to ‘diseased’ tissues.
We ascertained that the 100 observed alternative GC-AG intron isoforms were not aberrant events caused by mutations to, or near, the splice signals. Such mutations, if they existed, would have been observed either in the transcript–gene alignments, or during the double-check against HTG and additional EMBL entries. In addition, our method has allowed only those alignments with ≥95% base identity. The six cases that did show single base mismatches within 20 nt of the splice site, as discussed previously, were examined and considered harmless (such as purine–purine or pyrimidine–pyrimidine). There were only 14 cases where verifying sequences could not be found. As it is the case that mutations at intron positions do not show up in the alignments, we determined, for the donor sites, that none of the nearby GT dinucleotides possessed any level of splice consensus (although we did not perform this check for AG dinucleotides near the alternative acceptor sites). These observations, taken in their entirety, leave little reason to suppose that these introns are an aberrant consequence of mutation events.
We examined the type of tissue from which the supporting transcripts were derived for each of the 100 alternate GC-AG introns. Twenty-two cases had multiple transcripts from different tissue types. The remaining 78 cases were supported either by a single transcript or by multiple transcripts of same tissue type; 24 of these were from ‘diseased tissues’, 24 corresponded to ‘developmental stages’ and 30 were from ‘normal tissues’. This does not constitute a particular bias towards ‘diseased tissues’, and given that identification of a transcript in a ‘diseased’ library does not necessarily mean that the transcript is due to an aberrant splicing, we chose to retain this data. It should further be pointed out that even if the intron has arisen as the product of aberrant splicing, this does not in all likelihood detract from its use in the current study.
Data on transcript coverage and minor isoforms. Examination of the transcript coverage data indicated that while only 16% of the annotated GC-AG introns (10 of 62) showed single-transcript coverage, 61% of the alternative intron isoforms (61 of 100) were supported by a single transcript. A similar situation existed for the GT-AG introns where 26% of normal GT-AG introns (3651 of 14 157) had single-transcript support compared to 61% for the alternative GT-AG introns (1174 of 1941). It is to be expected that transcript coverage data reflect the expression level of genes rather than the relative quality of the data sets. Thus these alternative introns are minor isoforms that have not been observed so far by conventional experimental approaches. It is important to identify these minor isoforms as they might be biologically important. Examination of the nature of the genes containing GC-AG alternative introns, using bibliography information and a compilation of diseased genes (GeneCards at http://www.cgal.icnet.uk/genecards; 39), revealed that 46% of these genes are disease-associated.
Branch point signals. We assessed whether reasonable branch point signals could be identified for the alternative GC-AG introns. The branch point has a consensus sequence 5′-YTRAY-3′ and is located in a region upstream of the acceptor site. It is involved in an interaction with a region on the U2 snRNA (namely 3′-GAUG-3′) (3–4,25) where the unpaired branch point adenosine bulges out of the RNA duplex and attacks the 5′ splice site. We considered an extended region from the U2 snRNA, namely 3′-AU GAUG UGAA-3′ and searched for complementary sequences in a region of 70 bases upstream of the acceptor site. We used the criteria that such an RNA duplex contained base pairs involving at least three bases around the bulge adenosine and additional bases from the extended region. Such reasonable branch point signals could be identified in 65% of alternative GC-AG intron isoforms (the corresponding value for GT-AG introns was 71%).
RESULTS AND DISCUSSION
Scrutiny of high quality spliced alignments of human genes with human transcripts resulted in 162 transcript-confirmed GC-AG introns derived from 145 genes. These introns were categorized into two groups, namely GC-AG Normal, containing 62 cases corresponding to introns annotated in GenBank (and are thus normal isoforms), and GC-AG Alternative, containing 100 alternative introns. Of these 100 alternative introns, 22 overlapped with annotated exons and thus are ‘cryptic introns’. In 58 out of 78 alternative non-cryptic intron isoforms, transcript confirmation could be obtained for their normal intron isoforms. Similarly, transcript-confirmed GT-AG introns were obtained giving 14 157 GT-AG Normal introns and 1941 GT-AG Alternative introns.
It should be mentioned that (i) great care has been exercised in the methodology to obtain only non-redundant introns, and (ii) such a data set of around 16 000 non-redundant introns from around 2800 non-redundant genes is large enough to be representative of the human genome, which is predicted to have around 26 000–40 000 genes. Thus, the reported estimates and analysis of such introns presented here are as sound and realistic as possible.
Significance of GC-AG introns
Comparison of the above data for GT-AG and GC-AG introns led to the following findings. (i) The occurrence of GC-AG introns can be estimated at 1%, which is twice the earlier estimates based on only the annotated introns (27,28,31). If only the annotated introns are considered, this work also indicates that roughly 0.5% of the annotated introns are GC-AG, in agreement with the estimates of the earlier workers. (ii) One in 20 observed alternative introns is a GC-AG intron. This is significant considering that only one in 200 introns is a GC-AG intron. (iii) 62% of the observed GC-AG introns are alternative introns.
It was further observed that 72 of the 78 GC-AG Alternative non-cryptic introns are isoforms of GT-AG introns. The preferential use of GC-AG introns in alternative splicing is probably because both GC-AG and GT-AG introns are processed by the same U2-type spliceosome albeit with different efficiencies (40), and that variability of this type is often used as a regulatory mechanism in cellular processes.
Types of alternative intron isoforms
The alternative isoforms can be categorized depending on whether an alternative site is used at the donor and/or the acceptor site. The distribution is shown in Table 1. It can be seen that in the case of GC-AG isoforms, there is a significant bias towards alternative forms in which both the donor and acceptor sites are alternative (68% for GC-AG alternative isoforms compared to 12% for GT-AG alternative isoforms). This might indicate that use of an alternative donor site in GC-AG introns is significantly coupled with use of an alternative acceptor site.
Table 1. Percentage distribution of the alternative intron isoforms as per the use of Normal or Alternative donor and acceptor splice sites.
| Type of sites used at donor and
acceptor sitesa |
GC-AG Alternative introns |
GT-AG Alternative introns |
| Nor, Nor | 3 | 18 |
| Nor, Alt | 3 | 38 |
| Alt, Nor | 26 | 33 |
| Alt, Alt | 68 | 12 |
Note that an exon-skipping event will generate an alternative intron that may use two normal splice sites.
aNor indicates that the site is the same as that used in a normal intron isoform; Alt indicates that the site is alternative to the one used in any normal intron isoform.
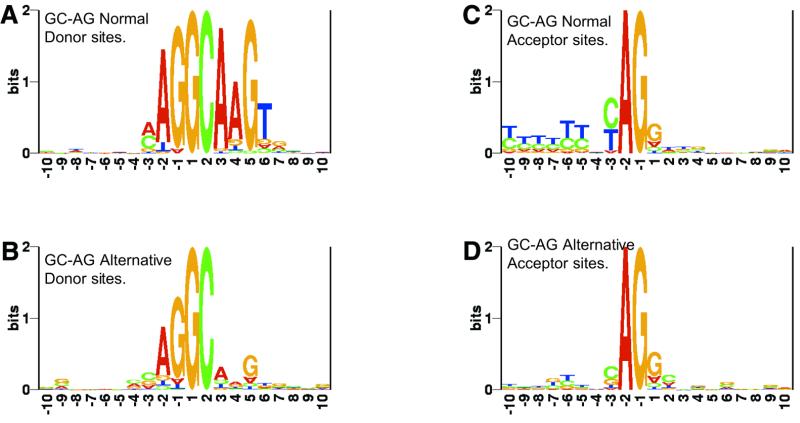
GC donor sites do not always show strong consensus
Sequence logos representing information content (41) at the splice sites of GC-AG introns were derived using the ‘RNA Structure Logo’ program (42) and are presented in Figure 2A–D. GC-AG alternative intron isoforms differ from GC-AG normal isoforms in the level of consensus around the splice sites. In agreement with the literature (25,28), the donor sites of normal GC-AG introns show strong consensus at positions –2 to +6 (Fig. 2A). However, the donor sites of the alternative GC-AG introns (Fig. 2B) differ significantly at intron positions (+3 to +6) and considerably at the exon positions (–2 to –1).
Figure 2.
Sequence logos at splice sites from normal and alternative GC-AG intron isoforms. The ‘RNA structure logo’ program (42) was used to derive the logos.
The extent of the consensus at acceptor sites also differs between the two sets. While the normal introns show a strong signal along the polypyrimidine tract and at the –3 position (Fig. 2C), the alternative introns display a weak polypyrimidine tract and less Y at the –3 position (Fig. 2D). Though such differences in the polypyrimidine tract can arise due to the use of an alternative site at the acceptor (22), we show in the subsequent sections that the polypyrimidine tract becomes significantly weaker when the pairing donor is a weak alternative GC site.
Variability in donor site signals and alternative splicing
As it is simpler to characterize the signals at donor sites than at acceptor sites, we decided to classify introns in terms of the donor strength. For this purpose we chose to use our SPC program (36,37). This tool implements a set of rules that identify true splice sites from proximal false positive sites. The rules utilize signals from mononucleotides as well as from dinucleotides involving positions from the upstream and downstream region of the splice sites. SPC can categorize donor sites into three groups: (i) SPC Major, the sites that could be validated by a single major rule; (ii) SPC Minor, the sites that could still be validated as true but not by the major rule; and (iii) SPC Negative, the sites that could not be validated by any of the rules.
The percentage distribution of donor sites, as per the above categorization, for GC-AG and GT-AG intron isoforms is given in Table 2. It can be seen that, while all the normal GC donor sites possess strong signals (compared to 81% for GT donor sites), only 56% of the alternative GC donor sites possess strong signals (with 63% for GT alternative donor sites). It is important to note that, contrary to the conventional understanding of GC-AG donor splice sites, nearly half of the alternative GC-AG intron isoforms show weak or no detectable donor signals.
Table 2. Percentage distribution of the intron isoforms as per the signal strength at the donor sites.
| Categories |
SPC Major |
SPC Minor |
SPC Negative |
| GC-AG introns | |||
| Normal isoforms | 100 | 0 | 0 |
| Alternative isoformsa | 56 | 16 | 28 |
| GT-AG introns | |||
| Normal isoforms | 81 | 13 | 6 |
| Alternative isoformsb | 63 | 17 | 20 |
The SPC program was used to assess the strength of donor site signals. SPC Major sites possess strong signals, SPC Minor sites possess weak signals, and SPC Negative sites possess signals that are too weak to be detected by programs that are based on splice signals alone.
aOf the 100 GC-AG alternative intron isoforms, 94 use alternative donor sites and 71 use alternative acceptor sites (see Table 1).
bIn the case of GT-AG alternative introns, only those that use alternative donor sites were included in the calculation. The percentage distribution in the case of alternative isoforms using a normal donor site followed the same pattern as for normal GT-AG isoforms.
These observations, as well as showing a definite correlation between alternative splicing and differential splice strength, raise an important question with regard to the processing of GC donor sites. The GC-AG isoforms have the following two conflicting requirements: (i) the GC donor sites need a strong consensus sequence in order to compensate for the mismatch in the central base pair of the RNA duplex involving the donor site and the U1 snRNA; (ii) alternative splicing is facilitated by weak splice signals. How do these GC-AG alternative isoforms with weak donor sites cope with these conflicting requirements? Examination of the alternative GC-AG introns with weak donor sites (the SPC Minor and SPC Negative categories) revealed that almost all of these isoforms use alternative acceptor sites as well. In contrast, this observation was found to hold for only 27% of the GT-AG alternative isoforms with weak donor sites. Thus, as pointed out earlier, a strong dependence may exist between the donor and acceptor sites for GC-AG alternative intron isoforms with weak donor sites. We characterize this dependence in the following sections.
Variability in donor signals for alternative GC-AG isoforms is accompanied by changes at the acceptor sites
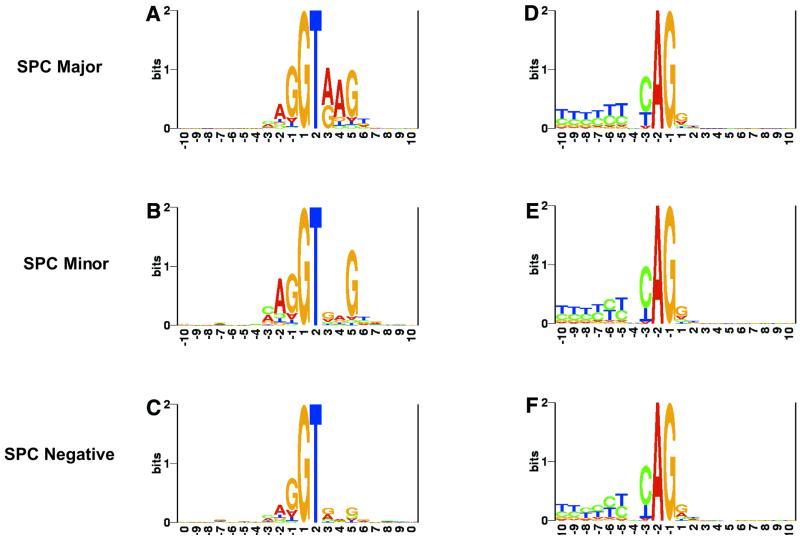
The sequence logos for the splice sites of normal and alternative GT-AG and GC-AG isoforms for the three categorizations are shown in Figures 3–5. Note that there are no introns, and hence no logos, for GC-AG normal introns in the SPC Minor and SPC Negative categories.
Figure 3.
Sequence logos at splice sites for the three categories of GT-AG normal intron isoforms. Categorization is in terms of donor site strength as assessed by our SPC program (see text). SPC Major introns are those in which the donor sites could be validated by a single major rule of SPC. SPC Minor donor sites could still be validated by SPC but not by the single major rule. SPC Negative donor sites could not be validated by any of the SPC rules.
Figure 5.
Sequence logos at splice sites for the three categories of GC-AG alternative intron isoforms. In some of the SPC Major introns, only the donor site is alternative. Almost all of the SPC Minor and SPC Negative introns use alternative donor and acceptor sites.
Logos for donor sites. While the SPC Major donor sites show strong signals at both the intron and exon positions, the SPC Minor and SPC Negative sites show weak signals in either the intron positions or both intron and exon positions. The logos indicate that the GC-AG alternative intron isoforms show considerable variability in donor signals across the three SPC categories (Fig. 5A–C), with this variability following a similar pattern to that for the GT-AG normal or alternative isoforms (Figs 3A–C and 4A–C). Note the normal GC-AG intron isoforms do not show any variability of this type (Fig. 2A) as all of them are categorized into the SPC Major group.
Figure 4.
Sequence logos at splice sites for the three categories of GT-AG alternative intron isoforms. Used are only those isoforms that use alternative donor and acceptor sites [corresponding to (Alt, Alt) in Table 1].
Logos for acceptor sites. The logos for the acceptor sites highlight a distinct feature in the case of alternative GC-AG introns with weak (SPC Minor and SPC Negative) donor sites (Fig. 5E–F); they possess an increased consensus sequence at the acceptor exon positions and almost lack the polypyrimidine tract. Such behavior is not seen in the case of normal or alternative GT-AG introns (Figs 3E–F and 4E–F); in these cases, categorization in terms of donor strength has not significantly influenced the strength at the acceptor site (except that the polypyrimidine tract becomes comparatively weak in the case of alternative GT-AG isoforms of weak donor sites).
The information contents of splice signals relevant to the above observations for GC-AG introns is shown in Table 3. It can be seen that the information content at the donor sites of GC-AG introns, irrespective of whether they are normal or alternative isoforms, tends to be higher than that of their GT-AG counterparts (this is believed to balance the mismatch base pair at the GC dinucleotide with U1 snRNA). When the information content reduces at the donor site (either within the intron position or both the exon and intron positions) it is seen that the information content at the acceptor exon increases substantially. Further, the acceptor site shows a drastically reduced content of pyrimidines.
Table 3. Variable donor strength and the accompanying change at the acceptor sites for the alternative GC-AG intron isoforms.
| Category of intronsa |
Information content
(bits)d |
|
Percentage occurrence of pyrimidine |
||
| |
Donor exon |
Donor intron |
Acceptor exon |
Polypyrimidine tracte |
Y at YAGf |
| GC-AG introns | |||||
| I. GC-AG Normal | |||||
| SPC Major | 3.2 | 5.3 | 0.5 | 79 | 95 |
| II. GC-AG Alternativeb | |||||
| SPC Major | 3.0 | 3.3 | 0.3 | 70 | 78 |
| SPC Minor | 3.7 | 1.6 | 1.0 | 51 | 56 |
| SPC Negative | 0.8 | 0.8 | 2.3 | 54 | 46 |
| Control experiments | |||||
| III. GC-AG Alternative (Alt, Alt) | |||||
| SPC Major (Alt, Alt)c | 3.1 | 2.6 | 0.5 | 65 | 60 |
| IV.GT-AG Alternative (Alt, Alt) | |||||
| SPC Major | 1.6 | 2.6 | 0.3 | 81 | 95 |
| SPC Minor | 1.9 | 1.3 | 0.3 | 70 | 78 |
| SPC Negative | 1.3 | 0.3 | 0.5 | 69 | 76 |
| V. GT-AG Alternative (Alt, Nor) | |||||
| SPC Major | 1.5 | 2.4 | 0.4 | 83 | 95 |
| SPC Minor | 1.7 | 1.2 | 0.4 | 82 | 97 |
| SPC Negative | 1.2 | 0.4 | 0.4 | 81 | 96 |
| VI. GT-AG Alternative (Nor, Alt) | |||||
| SPC Major | 1.6 | 3.2 | 0.2 | 75 | 86 |
| SPC Minor | 2.0 | 1.9 | 0.2 | 74 | 90 |
| SPC Negative | 1.1 | 0.8 | 0.5 | 75 | 86 |
| VII. GT-AG Normal | |||||
| SPC Major | 1.5 | 3.1 | 0.3 | 81 | 95 |
| SPC Minor | 1.7 | 1.7 | 0.3 | 81 | 95 |
| SPC Negative | 1.1 | 0.6 | 0.3 | 80 | 95 |
a(Alt, Alt) indicates that both donor and acceptor are alternative sites; (Alt, Nor) indicates that donor is alternative and acceptor is normal; (Nor, Alt) indicates that donor is normal and acceptor is alternative.
bSome of the SPC Major introns from the GC-AG Alternative group used alternative sites only at the donor. Except in a couple of cases, the introns from SPC Minor and SPC Negative groups used alternative donor and acceptor sites.
cThis SPC Major (Alt, Alt) group is a subset of the SPC Major introns from GC-AG Alternative group (II); the introns from this subset use alternative donor and acceptor sites.
dDonor exon corresponds to positions –2 and –1 of the donor site; donor intron corresponds to positions +3 to +6 of the donor site; and acceptor exon corresponds to +1 and +2 of the acceptor site.
eCorresponds to average percentage occurrence of a pyrimidine in the region –5 to –15 of the acceptor site. The corresponding value for a similar region on the exon side is consistently between 45 and 49%.
fCorresponds to the percentage occurrence of a pyrimidine at –3 position of the acceptor site.
In order to further substantiate that the above observations are specific to alternative GC-AG isoforms with weak donor sites, we carried out the following control experiments. (i) As noted earlier, almost all the alternative GC-AG introns with weak donor sites (SPC Minor and SPC Negative) also used alternative acceptor sites. We scrutinized a subset of the GC-AG alternative introns from the SPC Major group, which used alternative sites at both the donor and acceptor but possessed strong donor sites. These introns did not show changes at the acceptor sites (Table 3, III). (ii) In a similar manner, alternative GT-AG introns with weak donor sites (either as normal or alternative) did not show the accompanying changes at the acceptor sites (irrespective of whether normal or alternative; Table 3, IV–VII). (iii) GT-AG introns with weak or no polypyrimidine tract (such introns formed only 6% of the total) were scrutinized to see whether they showed weak donor sites and/or increased consensus at the acceptor exon. Similarly, GT-AG introns with increased consensus at the acceptor exon (such introns formed only 10% of the total) were scrutinized to see whether they showed weak donor sites and/or weak/no polypyrimidine tracts. In both cases no correlation was observed among the three structural elements (data not shown).
Thus, the observation made for alternative GC-AG intron isoforms with weak donor sites is significant, and is specific to the processing of GC-AG alternative introns.
Appropriateness of the SPC categories
The appropriateness of using the SPC program to categorize the introns becomes apparent in the light of the following biological interpretation of the above observations. As discussed above, U1 snRNA interacts with the donor site (at both exon and intron positions), initiating assembly of the spliceosome. At a later stage in the first step, this interaction is replaced by those of U5 snRNA with the exon nucleotides and U6 snRNA with intron nucleotides. Thus, while the SPC Major sites can have good interactions with both U5 and U6 snRNA (as well as U1 snRNA), the SPC Minor sites will have a poor interaction with U6 snRNA, and the SPC Negative sites would have poor interactions with both U5 and U6 snRNA molecules. The population sizes of the SPC Minor and SPC Negative categories are much higher when alternative intron isoforms are considered. In the case of GC-AG alternative introns, such gradual changes (as we go from category SPC Major to Minor and Negative) at donor sites showed gradual compensatory changes at the acceptor sites.
Implications of the observed changes at the acceptor sites
The GC-AG alternative introns possessing weak donor sites have, so far, been shown to have dramatically improved consensus at the acceptor site, and to have very weak polypyrimidine tracts.
The frequencies of the bases within the pre-mRNA that can form Watson–Crick base pairs with the snRNAs are shown in Table 4. It can be seen that SPC Minor and SPC Negative groups, irrespective of the type of intron, showed low occurrences of such bases within the intron and/or exon at the donor site. However, increased occurrences of such bases at the corresponding acceptor exon positions can be seen in (only) the case of GC-AG alternative introns. The SPC Minor group for these introns showed a guanosine at +1 position in 88% of cases; SPC Negative group showed a guanosine at +1 in 89% of cases and a cytosine at +2 position in 75% of cases.
Table 4. Frequencies (in %) of bases that can form Watson–Crick base pair with the interacting bases from the snRNAs.
| Category of intronsa | Nucleotide frequency (in %)d at positions | |||||
| Donor exon | Donor intron | Acceptor exon | ||||
| |
–2 = A |
–1 = G |
+4 = A |
+5 = G |
+1 = G |
+2 = C (C/U) |
| GC-AG introns | ||||||
| I. GC-AG Normal | ||||||
| SPC Major | 90 | 97 | 81 | 98 | 59 | 16 (54) |
| II.GC-AG Alternativeb | ||||||
| SPC Major | 85 | 100 | 56 | 80 | 44 | 38 (56) |
| SPC Minor | 94 | 100 | 6 | 63 | 88 | 31 (56) |
| SPC Negative | 50 | 64 | 18 | 29 | 89 | 75 (76) |
| Control experiments | ||||||
| III. GC-AG Alternativec | ||||||
| SPC Major (Alt, Alt)c | 83 | 97 | 53 | 67 | 50 | 47 (70) |
| IV. GT-AG Alternative (Alt, Alt) | ||||||
| SPC Major | 66 | 83 | 70 | 72 | 46 | 20 (52) |
| SPC Minor | 72 | 81 | 33 | 75 | 47 | 25 (56) |
| SPC Negative | 62 | 66 | 32 | 36 | 54 | 22 (70) |
| V. GT-AG Alternative (Alt, Nor) | ||||||
| SPC Major | 59 | 83 | 65 | 74 | 53 | 19 (58) |
| SPC Minor | 69 | 82 | 42 | 77 | 53 | 16 (56) |
| SPC Negative | 55 | 76 | 31 | 48 | 47 | 22 (65) |
| VI. GT-AG Alternative (Nor, Alt) | ||||||
| SPC Major | 59 | 86 | 78 | 83 | 42 | 20 (49) |
| SPC Minor | 81 | 79 | 44 | 88 | 47 | 21 (56) |
| SPC Negative | 49 | 73 | 38 | 42 | 49 | 9 (20) |
| VII. GT-AG Normal | ||||||
| SPC Major | 61 | 82 | 76 | 81 | 51 | 20 (56) |
| SPC Minor | 75 | 76 | 43 | 88 | 54 | 20 (56) |
| SPC Negative | 55 | 72 | 37 | 53 | 51 | 21 (59) |
| Interacting bases from snRNAs | ||||||
| U1 snRNA | U | C | U | C | ||
| U6 snRNA | A | C | ||||
| U5 snRNA | U | C | C | G |
a(Alt, Alt) indicates that both donor and acceptor sites are alternative; (Alt, Nor) indicates that the donor is alternative and acceptor is normal; (Nor, Alt) indicates that donor is normal and the acceptor is alternative.
bSome of the SPC Major introns from the GC-AG Alternative group used alternative sites only at the donors. Except in a couple of cases, the introns from the SPC Minor and SPC Negative groups used both alternative donor and acceptor sites.
cThis SPC Major (Alt, Alt) group is a subset of the SPC Major introns that are Alternative GC-AG (II); the introns from this subset use alternative donor and acceptor sites.
dValues ≤50% at the donor positions are shown in italic. Significant and high values at the acceptor exon positions are shown in bold.
The implications of the accompanying change at acceptor sites are at least 2-fold, as discussed below.
Improved interaction for acceptor exon with U5 snRNA during the second step of splicing. It is known that U5 snRNA interacts with the exon sequence at the 5′ end of U2-type introns during the first step of splicing. While this interaction continues during the second step, an additional interaction forms with the exon sequence at the 3′ splice site (13–15). These RNA–RNA interactions are stabilized by the PRP8 auxiliary protein (43,44). Thus, the U5 snRNA acts to tether the exons in the correct orientation for the second catalytic step of splicing. Such an interaction is illustrated in Figure 6. The observed high frequency of G at +1 position and C at +2 position (of the acceptor exon) allows the formation of strong Watson–Crick base pairs with U5 snRNA and thus can lead to a much more stable ‘tethering’ of exons. Such strong interactions are possible only when there is a GC in the first two positions of the acceptor exon, and this only tends to occur in cases of GC-AG alternative introns with weak donor sites. Interestingly, in two of the categories of GT-AG alternative introns, both involving weak alternative donor sites, an increased occurrence of C/U (predominantly U) was observed at the +2 acceptor position (see Table 4). These bases can form either a Watson–Crick C-G base pair (as above), or a non-Watson–Crick U.G base pair, respectively, with the U5 snRNA.
Figure 6.

Interaction of the Loop 1 of U5 snRNA with regions from the 5′ and 3′ exons. Bases that occurred in ≥70% of the cases in a group are shown in upper case; those that occurred in 35–70% of cases are shown in lower case (see Table 4 for actual values). Watson–Crick base pairs are indicated by a vertical line; non-Watson–Crick base pairs are indicated by a center dot.
GC-AG alternative intron isoforms with weak donor sites are probably ‘AG-dependent introns’. Introns have been categorized into two groups based on the strength of the polypyrimidine tract. Introns with a strong polypyrimidine tract are called ‘AG-independent’ and those that lack or possess a weak polypyrimidine tract are called ‘AG-dependent’ (9,11). In the case of AG-independent introns, the binding of U2AF65 with the polypyrimidine tract is sufficient to define the branch point, with recognition of the AG dinucleotide needed only for the second step of splicing. In the case of AG-dependent introns, the AG dinucleotide is recognized in the first step and bound by U2AF35 (8–10), such an interaction being critical for the binding of U2AF with the weak polypyrimidine tract and the subsequent branch point definition. The U2AF35 binding is sequence-specific and the consensus sequence at the acceptor site includes the acceptor exon positions +1 and +2 (12).
Our observation that the GC-AG Alternative introns with weak donor sites possess an extremely weak polypyrimidine tract indicates that they are probably ‘AG-dependent’ introns. The observed strong consensus sequence (GC) at their acceptor exon positions can help in the sequence-specific binding of U2AF35 to the AG site during the first step of splicing. Such an early recognition of the AG site during spliceosome assembly may even facilitate some sort of cooperative interaction between the 5′ and 3′ splice sites (45) and help in the identification of the weak donor sites.
Ascertaining that the GC-AG alternative intron isoforms are genuine
As discussed so far, the GC-AG alternative intron isoforms with weak donor sites have shown uniform and distinct features that fit the current models for RNA splicing. Systematic analysis, as detailed in the Methods and Materials, was carried out to ascertain that these isoforms are not artifacts of the analysis, but are indeed genuine observations. The quality of the gene–transcript alignments, the sequences and the assignment of splice sites were assessed. In a significant fraction of cases (30 of the 52 cases for which HTG coverage was obtained) the genes containing the alternative GC-AG intron isoforms were found to occur in multiple copies and on more than one chromosome; and in certain copies nucleotide changes modulating splice site strength could be observed. Transcript coverage was characterized and found to indicate that the alternative isoforms represent a class of ‘minor isoforms’. The possibility that the observed alternative GC-AG introns were the result of aberrant splicing was assessed and it was concluded that this was not the case. Sensible branch point signals could be identified in 65% of alternative GC-AG intron isoforms. This indicates that variability in donor signals may not particularly influence changes at the branch point signals. However, the corresponding value for normal GC-AG isoforms is 79%, indicating that GC-AG normal isoforms possess strong branch point signals as well.
Ascertaining that the observed GC-AG alternative intron isoforms are genuine U2-type introns
It is now known that some introns with GT and AG boundary sequences are spliced by the minor U12-type spliceosome (which splices AT-AC introns) rather than the major U2-type spliceosome (1,46). Such U12-type introns show highly conserved consensus sequences at the donor site (ATCCTTT at intron positions +3 to +9) and at the branch point (TTCCRACTC located at 11–20 positions upstream of AG). Since many of the alternative GC-AG introns showed poor U2-type consensus sequence at the donor site, we checked whether any of the reported GC-AG introns displayed the U12-type consensus sequences. It was observed that none of the GC-AG introns (alternative or normal) showed either of the above two U12-type consensus sequences. Thus the reported GC-AG introns are genuine U2-type introns and the observations that are made in this work pertain to U2-type spliceosomes.
CONCLUSION
The mammalian GC donor sites are intrinsically weak because of a mismatch base pair in the RNA duplex that is formed with the U1 snRNA. In order to compensate for this, constitutive GC-AG intron isoforms possess a strong consensus in the surrounding nucleotides of the donor site. However, in accordance with a need for differential splice strength in alternatively spliced isoforms, a large subset of GC-AG alternative intron isoforms from human show fewer consensus nucleotides at their donor sites. These introns use alternative donor and acceptor splice sites, and lack a reasonable polypyrimidine tract. However, these introns show a compensatory effect in terms of a dramatic increase in consensus at the acceptor exon positions. These acceptor exon nucleotides serve the following two purposes: (i) they serve as a strong consensus sequence for the U2AF35-mediated recognition of AG, which can happen in the first step of splicing, and (ii) they promote a strong interaction with the U5 snRNA, which tethers the two exons for ligation during the second step of splicing. These enhanced interactions may ensure an overall splice accuracy for introns with weak donor sites and polypyrimidine tracts. In addition other cis-acting elements such as exonic splicing enhancers and silencers can be present in and around these GC-AG alternative introns and might also have a role in determining splicing efficiency and accuracy.
Availability of the data set
The reported data set of GC-AG introns is of use in at least two ways. (i) It would be useful in the development of appropriate computational tools to predict normal and alternatively spliced exons involving GC-AG splice sites. This is important since one in every 20 alternative isoforms may be a GC-AG intron and as many as three of every five GC-AG introns may be alternative isoforms. (ii) We provide here a list of genes that can serve as experimental model systems to characterize the correlation among variant donor sites, differential splice strength, alternative splicing, the compensatory effects at the acceptor exon positions and the possible presence of exon splicing enhancers/silencers.
A value-added data set of these GC-AG introns is available from our web sites http://www.ebi.ac.uk/~thanaraj/gcag/ and http://www.bit.uq.edu.au/gcag/.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Alan Robinson for his support and encouragement. We gratefully acknowledge Millennium Pharmaceuticals Inc., USA for a small donation that enabled the visit of F.C. to EBI.
References
- 1.Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World—The Nature Of Modern RNA Suggests a Prebiotic RNA. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- 2.Madhani H.D. and Guthrie,C. (1994) Dynamic RNA–RNA interactions in the spliceosome. Annu. Rev. Genet., 28, 1–26. [DOI] [PubMed] [Google Scholar]
- 3.Query C.C., Moore,M.J. and Sharp,P.A. (1994) Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev., 8, 587–597. [DOI] [PubMed] [Google Scholar]
- 4.Valcarcel J., Gaur,R.K., Singh,R. and Green,M.R. (1996) Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science, 273, 1706–1709. [DOI] [PubMed] [Google Scholar]
- 5.Kramer A. (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- 6.Caceres J.F. and Krainer,A.R. (1997) Mammalian pre-mRNA splicing factors. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. Oxford University Press, Oxford, pp. 174–212.
- 7.Guth S., Martinez,C., Gaur,R.K. and Valcarcel,J. (1999) Evidence for substrate-specific requirement of the splicing factor U2AF(35) and for its function after polypyrimidine tract recognition by U2AF(65). Mol. Cell. Biol., 19, 8263–8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merendino L., Guth,S., Bilbao,D., Martinez,C. and Valcarcel,J. (1999) Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site. Nature, 402, 838–841. [DOI] [PubMed] [Google Scholar]
- 9.Wu S., Romfo,C.M., Nilsen,T.W. and Green,M.R. (1999) Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature, 402, 832–835. [DOI] [PubMed] [Google Scholar]
- 10.Zorio D.A.R. and Blumenthal,T. (1999) Both subunits of U2AF recognize 3′ splice site in Caenorhabditis elegans. Nature, 402, 835–838. [DOI] [PubMed] [Google Scholar]
- 11.Reed R. (1989) The organization of 3′ splice-site sequences in mammalian introns. Genes Dev., 3, 2113–2123. [DOI] [PubMed] [Google Scholar]
- 12.Moore M.J. (2000) Intron recognition comes of AGe. Nat. Struct. Biol., 7, 14–16. [DOI] [PubMed] [Google Scholar]
- 13.Newman A.J. and Norman,C. (1992) U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell, 65, 115–123. [DOI] [PubMed] [Google Scholar]
- 14.Newman A.J. (1997) The role of U5 snRNP in pre-mRNA splicing. EMBO J., 16, 5797–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Keefe R.T. and Newman,A.J. (1998) Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J., 17, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mironov A.A., Fickett,J.W. and Gelfand,M.S. (1999) Frequent alternative splicing of human genes. Genome Res., 9, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thanaraj T.A. (1999) A clean data set of EST-confirmed splice sites from Homo sapiens and standards for clean-up procedures. Nucleic Acids Res., 27, 2627–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croft L., Schandorff,S., Clark,F., Burrage,K., Arctander,P. and Mattick,J.S. (2000) ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nat. Genet., 24, 340–341. [DOI] [PubMed] [Google Scholar]
- 19.Hedjran F., Yeakley,J.M., Huh,G.S., Hynes,R.O. and Rosenfeld,M.G. (1997) Control of alternative pre-mRNA splicing by distributed pentameric repeats. Proc. Natl Acad. Sci. USA, 94, 12343–12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamm S., Zhang,M.Q., Marr,T.G. and Helfman,D.M. (1994) A sequence compilation and comparison of exons that are alternatively spliced in neurons. Nucleic Acids Res., 9, 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob M. and Gallinaro,H. (1989) The 5′ splice site: phylogenetic evolution and variable geometry of association with U1RNA. Nucleic Acids Res., 17, 2159–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coolidge C.J., Seely,R.J. and Patton,J.G. (1997) Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res., 25, 888–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caceres J.F., Stamm,S., Helfman,D.M. and Krainer,A.R. (1994) Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science, 265, 1706–1709. [DOI] [PubMed] [Google Scholar]
- 24.Bai Y., Lee,D., Yu,T. and Chasin,L.A. (1999) Control of 3′ splice site choice in vivo by ASF/SF2 and hnRNP A1. Nucleic Acids Res., 27, 1126–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senapathy P., Sharpiro,M.B. and Harris,N.L. (1990) Splice junctions, branch point sites and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol., 183, 252–278. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M.Q. (1998) Statistical features of human exons and their flanking regions. Hum. Mol. Genet., 7, 919–932. [DOI] [PubMed] [Google Scholar]
- 27.Mount S.M. (2000) Genome sequence, splicing, and gene annotation. Am. J. Hum. Genet., 67, 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burset M., Seledtsov,I.A. and Solovyev,V.V. (2000) Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res., 28, 4364–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burset M., Seledtsov,I.A. and Solovyev,V.V. (2001) SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res., 29, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aebi M., Hornig,H. and Weissmann,C. (1987) 5′ cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5′ splice region, not by the conserved 5′ GU. Cell, 50, 237–246. [DOI] [PubMed] [Google Scholar]
- 31.Jackson I.J. (1991) A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res., 19, 3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoesser G., Baker,W., van den Broek,A., Camon,E., Garcia-Pastor,M., Kanz,C., Kulikova,T., Lombard,V., Lopez,R., Parkinson,H., Redaschi,N., Sterk,P., Stoehr,P. and Tuli,M.A. (2001) The EMBL nucleotide sequence database. Nucleic Acids Res., 29, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) GenBank. Nucleic Acids Res., 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 35.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thanaraj T.A. (2000) Positional characterisation of false positives from computational prediction of human splice sites. Nucleic Acids Res., 28, 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thanaraj T.A. and Robinson,A.J. (2000) Prediction of exact boundaries of exons. Briefings in Bioinformatics, 1, 343–356. [DOI] [PubMed]
- 38.Nakai K. and Sakamoto,H. (1994) Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene, 141, 171–177. [DOI] [PubMed] [Google Scholar]
- 39.Rebhan M., Chalifa-Caspi,V., Prilusky,J. and Lancet,D. (1997) GeneCards: encyclopedia for genes, proteins and diseases. Weizmann Institute of Science, Bioinformatics Unit and Genome Center, Rehovot, Israel.
- 40.Aebi M., Hornig,H., Padgett,R.A., Reiser,J. and Weissman,C (1986) Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell, 47, 555–565. [DOI] [PubMed] [Google Scholar]
- 41.Schneider T.D. and Stephens,R.M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res., 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorodkin J., Heyer,L.J., Brunak,S. and Stormo,G.D. (1997) Displaying the information contents of structural RNA alignments: the structure logos. Comput. Appl. Biosci., 13, 583–586. [DOI] [PubMed] [Google Scholar]
- 43.Beggs J.D., Teigelkamp,S. and Newman,A.J. (1995) The role of PRP8 protein in nuclear pre-mRNA splicing in yeast. J. Cell Sci. [Suppl.], 19, 101–105. [DOI] [PubMed] [Google Scholar]
- 44.Dix I., Russel,C.S., O’Keefe,R.T., Newman,A,J. and Beggs,J.D. (1998) Protein–RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA, 4, 1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker R. and Siliciano,P.G. (1993) Evidence for an essential non-Watson–Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature, 361, 660–662. [DOI] [PubMed] [Google Scholar]
- 46.Sharp P.A. and Burge,C.B. (1997) Classification of introns: U2-type or U12-type. Cell, 91, 875–879. [DOI] [PubMed] [Google Scholar]