Abstract
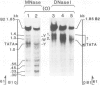
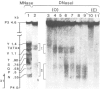
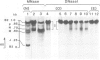
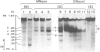
The organization of chromatin was analysed in a segment of the chicken ovalbumin gene extending 6.5 kb upstream from the start site of transcription. Nuclei of chicken oviduct cells and of erythrocytes, and preparations of 'naked' DNA were digested with DNase I and with micrococcal nuclease. The locations of specific nuclease cleavage sites were determined by analyzing the fragments obtained with an indirect end-labeling technique. In oviduct nuclei there are four regions of DNase I hypersensitivity centered at approximately 0.15, 0.80, 3.2 and 6.0 kb upstream from the mRNA cap site. DNase I hypersensitive regions are absent from the 5'-flanking regions of erythrocyte nuclei. Micrococcal nuclease cleavage sites were found that are unique to oviduct nuclei and others that are enhanced in oviduct nuclei, relative to erythrocyte nuclei and to naked DNA. The locations of these micrococcal nuclease cleavage sites are closely associated with the DNase I hypersensitive regions. Nuclease hypersensitivity in the 5'-flanking region of oviduct nuclei reflects alterations in chromatin structure that are specifically correlated with gene expression. Our results suggest the presence at hypersensitive regions of specific proteins which alter the chromatin structure, making the DNA more accessible to nuclease attack.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Bellard M., Dretzen G., Bellard F., Oudet P., Chambon P. Disruption of the typical chromatin structure in a 2500 base-pair region at the 5' end of the actively transcribed ovalbumin gene. EMBO J. 1982;1(2):223–230. doi: 10.1002/j.1460-2075.1982.tb01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Gannon F., Chambon P. Nucleosome structure III: the structure and transcriptional activity of the chromatin containing the ovalbumin and globin genes in chick oviduct nuclei. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):779–791. doi: 10.1101/sqb.1978.042.01.078. [DOI] [PubMed] [Google Scholar]
- Bellard M., Kuo M. T., Dretzen G., Chambon P. Differential nuclease sensitivity of the ovalbumin and beta-globin chromatin regions in erythrocytes and oviduct cells of laying hen. Nucleic Acids Res. 1980 Jun 25;8(12):2737–2750. doi: 10.1093/nar/8.12.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K. S., Anderson J. N. Hormonal regulation of the conformation of the ovalbumin gene in chick oviduct chromatin. J Biol Chem. 1982 Nov 10;257(21):13018–13027. [PubMed] [Google Scholar]
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Buttyan R., Olsson C. A., Sheard B., Kallos J. Steroid receptor-nuclear matrix interactions. The role of DNA. J Biol Chem. 1983 Dec 10;258(23):14366–14370. [PubMed] [Google Scholar]
- Cartwright I. L., Abmayr S. M., Fleischmann G., Lowenhaupt K., Elgin S. C., Keene M. A., Howard G. C. Chromatin structure and gene activity: the role of nonhistone chromosomal proteins. CRC Crit Rev Biochem. 1982;13(1):1–86. doi: 10.3109/10409238209108709. [DOI] [PubMed] [Google Scholar]
- Chambon P. Summary: the molecular biology of the eukaryotic genome is coming of age. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1209–1234. doi: 10.1101/sqb.1978.042.01.122. [DOI] [PubMed] [Google Scholar]
- Chung S. Y., Folsom V., Wooley J. DNase I-hypersensitive sites in the chromatin of immunoglobulin kappa light chain genes. Proc Natl Acad Sci U S A. 1983 May;80(9):2427–2431. doi: 10.1073/pnas.80.9.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciejek E. M., Tsai M. J., O'Malley B. W. Actively transcribed genes are associated with the nuclear matrix. Nature. 1983 Dec 8;306(5943):607–609. doi: 10.1038/306607a0. [DOI] [PubMed] [Google Scholar]
- Compton J. G., Schrader W. T., O'Malley B. W. DNA sequence preference of the progesterone receptor. Proc Natl Acad Sci U S A. 1983 Jan;80(1):16–20. doi: 10.1073/pnas.80.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D. C., Knoll B. J., Riser M. E., O'Malley B. W. A 5'-flanking sequence essential for progesterone regulation of an ovalbumin fusion gene. Nature. 1983 Oct 6;305(5934):551–554. doi: 10.1038/305551a0. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Lomonossoff G. P., Laskey R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvas A., Price P. A. Some effects of calcium and magnesium ions on the activity of bovine pancreatic deoxyribonuclease A. Biochim Biophys Acta. 1975 Jun 16;395(2):201–212. doi: 10.1016/0005-2787(75)90159-8. [DOI] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Fisher L. M., Mizuuchi K., O'Dea M. H., Ohmori H., Gellert M. Site-specific interaction of DNA gyrase with DNA. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4165–4169. doi: 10.1073/pnas.78.7.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritton H. P., Sippel A. E., Igo-Kemenes T. Nuclease-hypersensitive sites in the chromatin domain of the chicken lysozyme gene. Nucleic Acids Res. 1983 Jun 11;11(11):3467–3485. doi: 10.1093/nar/11.11.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Berg P. Simian virus 40 early- and late-region promoter functions are enhanced by the 72-base-pair repeat inserted at distant locations and inverted orientations. Mol Cell Biol. 1983 Jun;3(6):991–999. doi: 10.1128/mcb.3.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Heilig R., Perrin F., Gannon F., Mandel J. L., Chambon P. The ovalbumin gene family: structure of the X gene and evolution of duplicated split genes. Cell. 1980 Jul;20(3):625–637. doi: 10.1016/0092-8674(80)90309-8. [DOI] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongstra J., Reudelhuber T. L., Oudet P., Benoist C., Chae C. B., Jeltsch J. M., Mathis D. J., Chambon P. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984 Feb 23;307(5953):708–714. doi: 10.1038/307708a0. [DOI] [PubMed] [Google Scholar]
- Keene M. A., Elgin S. C. Micrococcal nuclease as a probe of DNA sequence organization and chromatin structure. Cell. 1981 Nov;27(1 Pt 2):57–64. doi: 10.1016/0092-8674(81)90360-3. [DOI] [PubMed] [Google Scholar]
- Knoll B. J., Zarucki-Schulz T., Dean D. C., O'Malley B. W. Definition of the ovalbumin gene promoter by transfer of an ovalglobin fusion gene into cultured cells. Nucleic Acids Res. 1983 Oct 11;11(19):6733–6754. doi: 10.1093/nar/11.19.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Gelinas R., Weintraub H. Detection of an altered DNA conformation at specific sites in chromatin and supercoiled DNA. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4389–4393. doi: 10.1073/pnas.80.14.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lawson G. M., Knoll B. J., March C. J., Woo S. L., Tsai M. J., O'Malley B. W. Definition of 5' and 3' structural boundaries of the chromatin domain containing the ovalbumin multigene family. J Biol Chem. 1982 Feb 10;257(3):1501–1507. [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Shermoen A. W., Beckendorf S. K. A transposable element inserted just 5' to a Drosophila glue protein gene alters gene expression and chromatin structure. Cell. 1983 Aug;34(1):75–84. doi: 10.1016/0092-8674(83)90137-x. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Shermoen A. W., Heemskerk J., Beckendorf S. K. DNA sequence changes in an upstream DNase I-hypersensitive region are correlated with reduced gene expression. Proc Natl Acad Sci U S A. 1983 Feb;80(4):1063–1067. doi: 10.1073/pnas.80.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Mills F. C., Fisher L. M., Kuroda R., Ford A. M., Gould H. J. DNase I hypersensitive sites in the chromatin of human mu immunoglobulin heavy-chain genes. Nature. 1983 Dec 22;306(5945):809–812. doi: 10.1038/306809a0. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill E. R., LePennec J. P., Chambon P. Chicken oviduct progesterone receptor: location of specific regions of high-affinity binding in cloned DNA fragments of hormone-responsive genes. Cell. 1982 Mar;28(3):621–632. doi: 10.1016/0092-8674(82)90217-3. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D., Mulvihill E. R., Shepherd J. H., McKnight G. S. Steroid hormone regulation of ovalbumin and conalbumin gene transcription. A model based upon multiple regulatory sites and intermediary proteins. J Biol Chem. 1981 Aug 10;256(15):7910–7916. [PubMed] [Google Scholar]
- Palmiter R. D. Quantitation of parameters that determine the rate of ovalbumin synthesis. Cell. 1975 Mar;4(3):189–189. doi: 10.1016/0092-8674(75)90167-1. [DOI] [PubMed] [Google Scholar]
- Parslow T. G., Granner D. K. Chromatin changes accompany immunoglobulin kappa gene activation: a potential control region within the gene. Nature. 1982 Sep 30;299(5882):449–451. doi: 10.1038/299449a0. [DOI] [PubMed] [Google Scholar]
- Picard D., Schaffner W. A lymphocyte-specific enhancer in the mouse immunoglobulin kappa gene. Nature. 1984 Jan 5;307(5946):80–82. doi: 10.1038/307080a0. [DOI] [PubMed] [Google Scholar]
- Robinson S. I., Small D., Idzerda R., McKnight G. S., Vogelstein B. The association of transcriptionally active genes with the nuclear matrix of the chicken oviduct. Nucleic Acids Res. 1983 Aug 11;11(15):5113–5130. doi: 10.1093/nar/11.15.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royal A., Garapin A., Cami B., Perrin F., Mandel J. L., LeMeur M., Brégégègre F., Gannon F., LePennec J. P., Chambon P. The ovalbumin gene region: common features in the organisation of three genes expressed in chicken oviduct under hormonal control. Nature. 1979 May 10;279(5709):125–132. doi: 10.1038/279125a0. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Cereghini S., Yaniv M. Fine structure of the regulatory region of simian virus 40 minichromosomes revealed by DNAase I digestion. J Mol Biol. 1982 Sep 15;160(2):133–146. doi: 10.1016/0022-2836(82)90171-1. [DOI] [PubMed] [Google Scholar]
- Shermoen A. W., Beckendorf S. K. A complex of interacting DNAase I-hypersensitive sites near the Drosophila glue protein gene, Sgs4. Cell. 1982 Jun;29(2):601–607. doi: 10.1016/0092-8674(82)90176-3. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Stumph W. E., Baez M., Beattie W. G., Tsai M. J., O'Malley B. W. Characterization of deoxyribonucleic acid sequences at the 5' and 3' borders of the 100 kilobase pair ovalbumin gene domain. Biochemistry. 1983 Jan 18;22(2):306–315. doi: 10.1021/bi00271a012. [DOI] [PubMed] [Google Scholar]
- Sweet R. W., Chao M. V., Axel R. The structure of the thymidine kinase gene promoter: nuclease hypersensitivity correlates with expression. Cell. 1982 Dec;31(2 Pt 1):347–353. doi: 10.1016/0092-8674(82)90128-3. [DOI] [PubMed] [Google Scholar]
- VONHIPPEL P. H., FELSENFELD G. MICROCOCCAL NUCLEASE AS A PROBE OF DNA CONFORMATION. Biochemistry. 1964 Jan;3:27–39. doi: 10.1021/bi00889a006. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Augereau P., Chambon P. The SV40 72 bp repeat preferentially potentiates transcription starting from proximal natural or substitute promoter elements. Cell. 1983 Feb;32(2):503–514. doi: 10.1016/0092-8674(83)90470-1. [DOI] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Glotov B. O., Schnell H., Zachau H. G. Differences in the nuclease sensitivity between the two alleles of the immunoglobulin kappa light chain genes in mouse liver and myeloma nuclei. Nucleic Acids Res. 1982 Jun 25;10(12):3627–3645. doi: 10.1093/nar/10.12.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]