Abstract
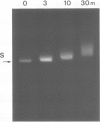
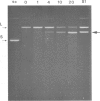
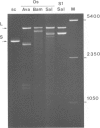
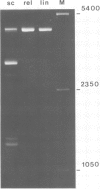
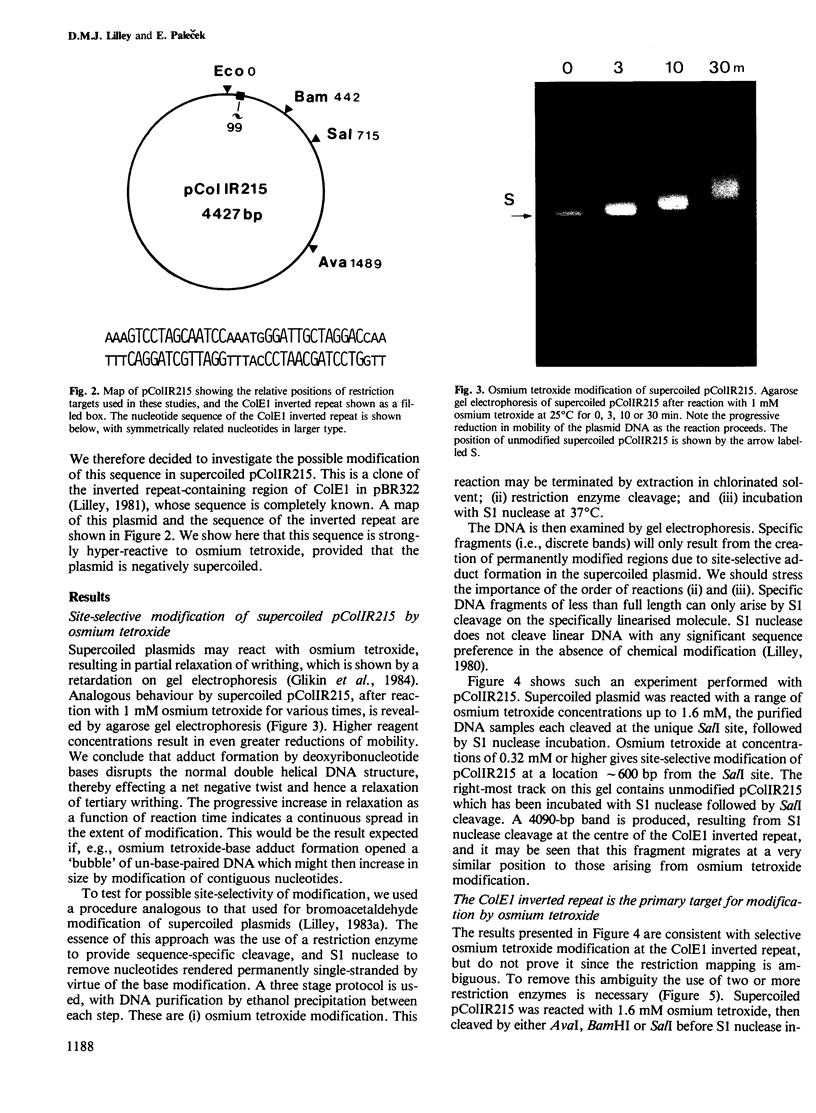
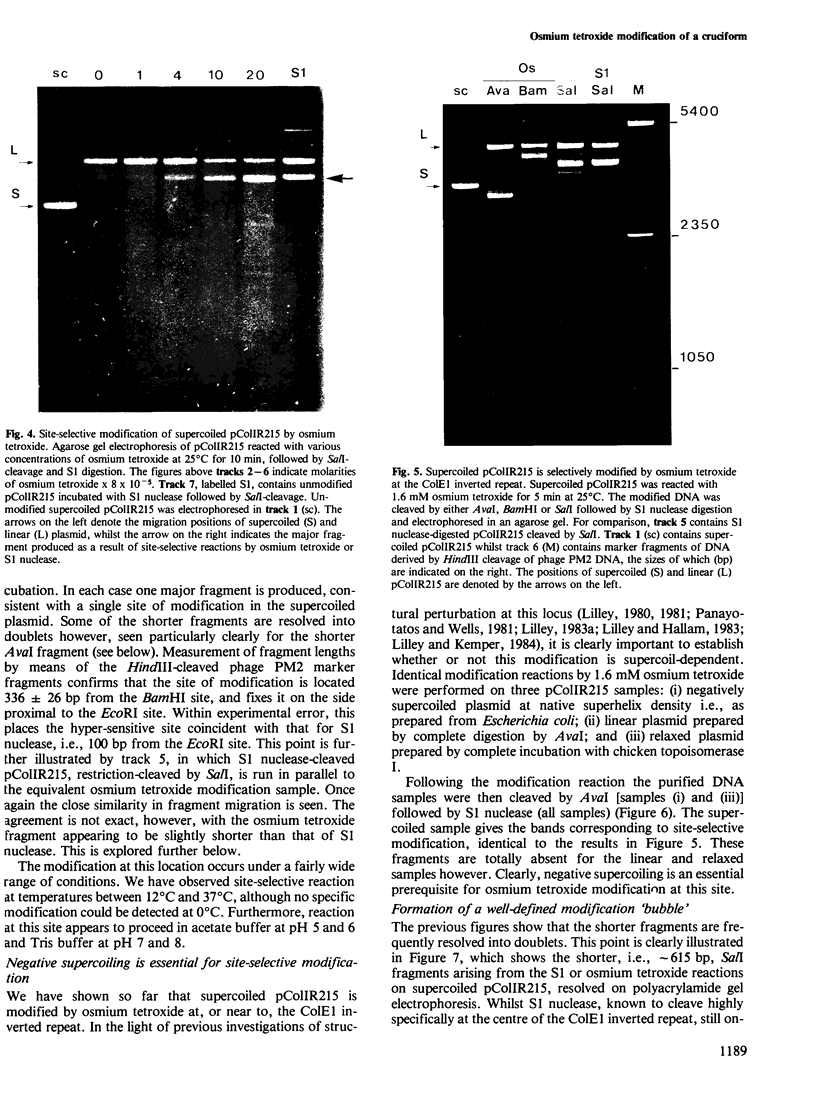
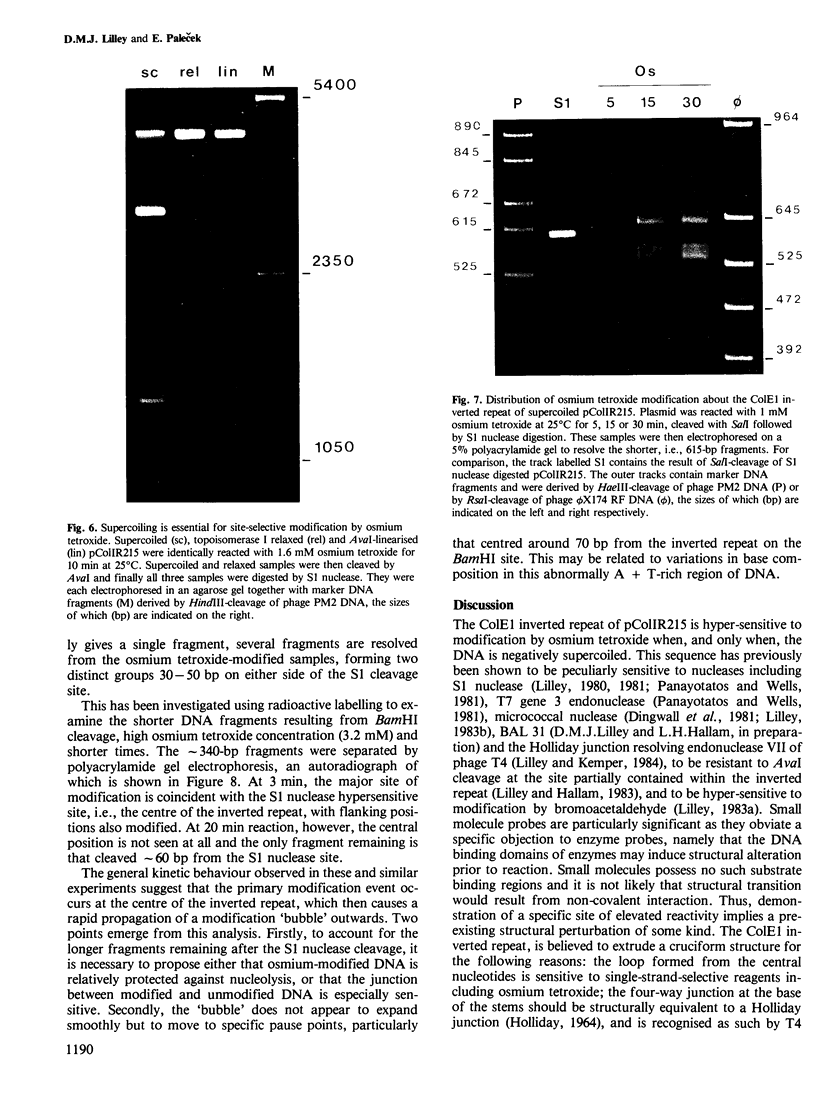
Supercoiled pColIR215 contains a site of pronounced hyper-reactivity towards modification by osmium tetroxide, a reagent known to be single-strand-selective. The site of hypersensitivity has been mapped to the ColE1 inverted repeat, believed to extrude a cruciform in supercoiled DNA. Linear or relaxed plasmids are not modified by the reagent. We conclude that cruciform formation is responsible for the site-selective modification. Fine mapping of the modification site as a function of time has revealed that the initial reaction occurs at the centre of the inverted repeat, i.e., the unpaired loop of the cruciform, but that the modification region rapidly expands outwards from this point.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beer M., Stern S., Carmalt D., Mohlhenrich K. H. Determination of base sequence in nucleic acids with the electron microscope. V. The thymine-specific reactions of osmium tetroxide with deoxyribonucleic acid and its components. Biochemistry. 1966 Jul;5(7):2283–2288. doi: 10.1021/bi00871a017. [DOI] [PubMed] [Google Scholar]
- Benham C. J. Theoretical analysis of transitions between B- and Z-conformations in torsionally stressed DNA. Nature. 1980 Aug 7;286(5773):637–638. doi: 10.1038/286637a0. [DOI] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. Polynucleotide kinase exchange reaction: quantitave assay for restriction endonuclease-generated 5'-phosphoroyl termini in DNA. J Biol Chem. 1977 May 25;252(10):3176–3184. [PubMed] [Google Scholar]
- Chang C. H., Beer M., Marzilli L. G. Osmium-labeled polynucleotides. The reaction of osmium tetroxide with deoxyribonucleic acid and synthetic polynucleotides in the presence of tertiary nitrogen donor ligands. Biochemistry. 1977 Jan 11;16(1):33–38. doi: 10.1021/bi00620a006. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983 Jul;33(3):817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Lomonossoff G. P., Laskey R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glikin G. C., Vojtískova M., Rena-Descalzi L., Palecek E. Osmium tetroxide: a new probe for site-specific distortions in supercoiled DNAs. Nucleic Acids Res. 1984 Feb 10;12(3):1725–1735. doi: 10.1093/nar/12.3.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T. S., Wang J. C. Thermodynamic properties of superhelical DNAs. Biochemistry. 1975 Feb 11;14(3):527–535. doi: 10.1021/bi00674a011. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Quigley G. J., Suddath F. L., McPherson A., Sneden D., Kim J. J., Weinzierl J., Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science. 1973 Jan 19;179(4070):285–288. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- Kłysik J., Stirdivant S. M., Larson J. E., Hart P. A., Wells R. D. Left-handed DNA in restriction fragments and a recombinant plasmid. Nature. 1981 Apr 23;290(5808):672–677. doi: 10.1038/290672a0. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. Dynamic, sequence-dependent DNA structure as exemplified by cruciform extrusion from inverted repeats in negatively supercoiled DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):101–112. doi: 10.1101/sqb.1983.047.01.013. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981 Mar 25;9(6):1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Hallam L. R. The interactions of enzyme and chemical probes with inverted repeats in supercoiled DNA. J Biomol Struct Dyn. 1983 Oct;1(1):169–182. doi: 10.1080/07391102.1983.10507433. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Markham A. F. Dynamics of cruciform extrusion in supercoiled DNA: use of a synthetic inverted repeat to study conformational populations. EMBO J. 1983;2(4):527–533. doi: 10.1002/j.1460-2075.1983.tb01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. Structural perturbation in supercoiled DNA: hypersensitivity to modification by a single-strand-selective chemical reagent conferred by inverted repeat sequences. Nucleic Acids Res. 1983 May 25;11(10):3097–3112. doi: 10.1093/nar/11.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Panyutin I. G., Frank-Kamenetskii M. D. Evidence of cruciform structures in superhelical DNA provided by two-dimensional gel electrophoresis. FEBS Lett. 1983 Mar 21;153(2):298–302. doi: 10.1016/0014-5793(83)80628-0. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Neidle S., Stuart D. I. The crystal and molecular structure of an osmium bispyridine adduct of thymine. Biochim Biophys Acta. 1976 Jan 19;418(2):226–231. doi: 10.1016/0005-2787(76)90072-1. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Panyutin I. G., Lyamichev V. I., Lyubchenko YuL A sharp structural transition in pA03 plasmid DNA caused by increased superhelix density. FEBS Lett. 1982 Nov 8;148(2):297–301. doi: 10.1016/0014-5793(82)80828-4. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Nordheim A., Rich A., Wang J. C. Flipping of cloned d(pCpG)n.d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa J. J., Sigler P. B. The site of covalent attachment in the crystalline osmium-tRNA-fMet isomorphous derivative. Biochemistry. 1974 Dec 3;13(25):5102–5110. doi: 10.1021/bi00722a008. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Wells R. D. Relationship between superhelical density and cruciform formation in plasmid pVH51. J Biol Chem. 1982 Jun 10;257(11):6292–6295. [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J. Physical and topological properties of circular DNA. J Gen Physiol. 1966 Jul;49(6):103–125. doi: 10.1085/jgp.49.6.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Watson R. Early and late helix-coil transitions in closed circular DNA. The number of superhelical turns in polyoma DNA. J Mol Biol. 1968 Apr 14;33(1):173–197. doi: 10.1016/0022-2836(68)90287-8. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Frank-Kamenetskii M. D. Premelting of superhelical DNA: an expression for superhelical energy. FEBS Lett. 1981 Aug 17;131(1):178–180. doi: 10.1016/0014-5793(81)80914-3. [DOI] [PubMed] [Google Scholar]