Abstract
H1 RNA, the RNA component of the human nuclear RNase P, is encoded by a unique gene transcribed by RNA polymerase III (Pol III). In this work, cis-acting elements and trans-acting factors involved in human H1 gene transcription were characterized by transcription assays of mutant templates and DNA binding assays of recombinant proteins. Four elements, lying within 100 bp of 5′-flanking sequences, were defined to be essential for maximal in vitro and in vivo expression, consisting of the octamer, Staf, proximal sequence element (PSE) and TATA motifs. These are also encountered in the promoter elements of vertebrate snRNA genes, where the first two constitute the distal sequence element (DSE). In all the genes examined so far, the DSE is distant from the PSE and TATA box that compose the basal promoter. However, we observed a fundamental difference in the organization of the H1 RNA and snRNA gene promoters with respect to the relative spacing of the DSE and PSE. Indeed, the H1 promoter is unusually compact, with the octamer motif and Staf binding site adjacent to the PSE and TATA motifs. It thus appears that the human RNase P RNA gene has adopted a unique promoter strategy placing the DSE immediately adjacent to the basal promoter.
INTRODUCTION
In higher eukaryotes, RNA polymerase III (Pol III) is responsible for the synthesis of a large variety of small nuclear and cytoplasmic non-coding RNAs. The promoter structures of a large number of genes encoding these RNAs have been determined and it has been found that the nature and localization of the control elements vary between different Pol III transcription units (reviewed in 1). The promoter structures of these units fall into three different types. In types I and II, typified by the 5S RNA and tRNA genes, respectively, the promoter elements are located entirely within the transcribed region. In type III genes, transcription is driven by cis-acting elements found only in the 5′-flanking region. Other Pol III promoters that rely both on internal and upstream sequences for efficient expression include the silkworm tRNAAla (2), human 7SL (3), EBER RNAs (4) and Xenopus tRNASec genes (5). The best characterized type III promoter belongs to the snRNAU6 genes (6–8). A number of other transcription units, such as the 7SK, Y and MRP RNA genes, have similar type III basal promoter elements and can be classified as snRNA-type genes. The sequences required for efficient basal expression of snRNA and snRNA-type genes are a TATA element between –30 and –25, which acts as a major determinant for Pol III specificity, and a proximal sequence element (PSE) between –66 and –47. The PSE recruits a stable protein complex, known as SNAPc or PTF, containing five subunits (9–11). Activated transcription of snRNA and snRNA-type promoters is provided by the distal sequence element (DSE) located between –260 and –190 (12). DSEs are composed of several functional submotifs that can be present either simultaneously or separately. Two of these are often the octamer and the Staf motifs (12–15). The octamer motif binds Oct-1, a homeodomain transcriptional activator (16,17). The transcriptional activator Staf, a seven zinc finger protein originally identified in Xenopus laevis as the transcriptional activator of the tRNASec gene, recognizes the Staf motif (14). ZNF76 and ZNF143 are two human homologs of Staf, ZNF143 being the ortholog whereas ZNF76 is related to Staf and ZNF143 (18).
RNase P is an enzyme that cleaves tRNA precursors to produce the mature 5′-termini. The gene encoding the human nuclear RNase P is transcribed by RNA Pol III (19). In vitro transcription studies previously established that multiple cis-acting elements are necessary for H1 RNA synthesis (20). In this report, various transcription assays on mutant templates, and DNA binding assays with recombinant proteins, led to the discovery that the DNA elements required for transcription of the H1 RNA gene are composed of the octamer, Staf binding site, PSE and TATA motifs, that are typical vertebrate snRNA promoter elements. However, our study shows that the H1 promoter harbors an unusual compact organization within 100 bp of 5′-flanking sequences, with the octamer motif and Staf binding site abutting the PSE and TATA motifs.
MATERIALS AND METHODS
Reporter constructs
Constructs H1 –370/+404, H1 –229/+344, H1 –100/+344 and H1 +1/+344, used in the in vitro transcription assays, were obtained by subcloning into EcoRI–BamH1 digested pBS(–) vector, the –370/+404, –229/+344, –100/+344 and +1/+344 fragments obtained by PCR amplification of the human H1 clone pMBH1 (19). The 5′- and 3′-primers incorporated an EcoRI and a BamHI site, respectively. Nucleotide substitutions described in Figure 3 were introduced into the H1 –229/+344 construct by site-directed mutagenesis. The hybrid H1 constructs used in transfection experiments (Figs 1A and C, and 3C) were obtained by ligating into the KpnI cleaved pU6/Hae/RA.2/EcoRV construct (7), the DNA fragments prepared by PCR amplification of the H1 –370/+404, H1 –229/+344, H1 –100/+344, H1 +1/+344 constructs, and the H1 substitution mutants. The forward primer complementary to positions –370 to –353, –229 to –212 or –100 to –83, and the reverse primers complementary to positions –17 to –1 incorporated a KpnI site. U1wt, U1 ΔDSE, U6wt and U6 ΔDSE used in microinjection assays correspond to the X.laevis U1b2 and U1b2 DDSE (21,22), Xenopus tropicalis U6 (23) and C115 constructs (13), respectively. U1 + AE, U1 + AE +oct, U1 + oct, U6 + AE, U6 + AE + oct, and U6 + oct were obtained by site-directed mutagenesis of U1 ΔDSE and U6 ΔDSE.
Figure 3.
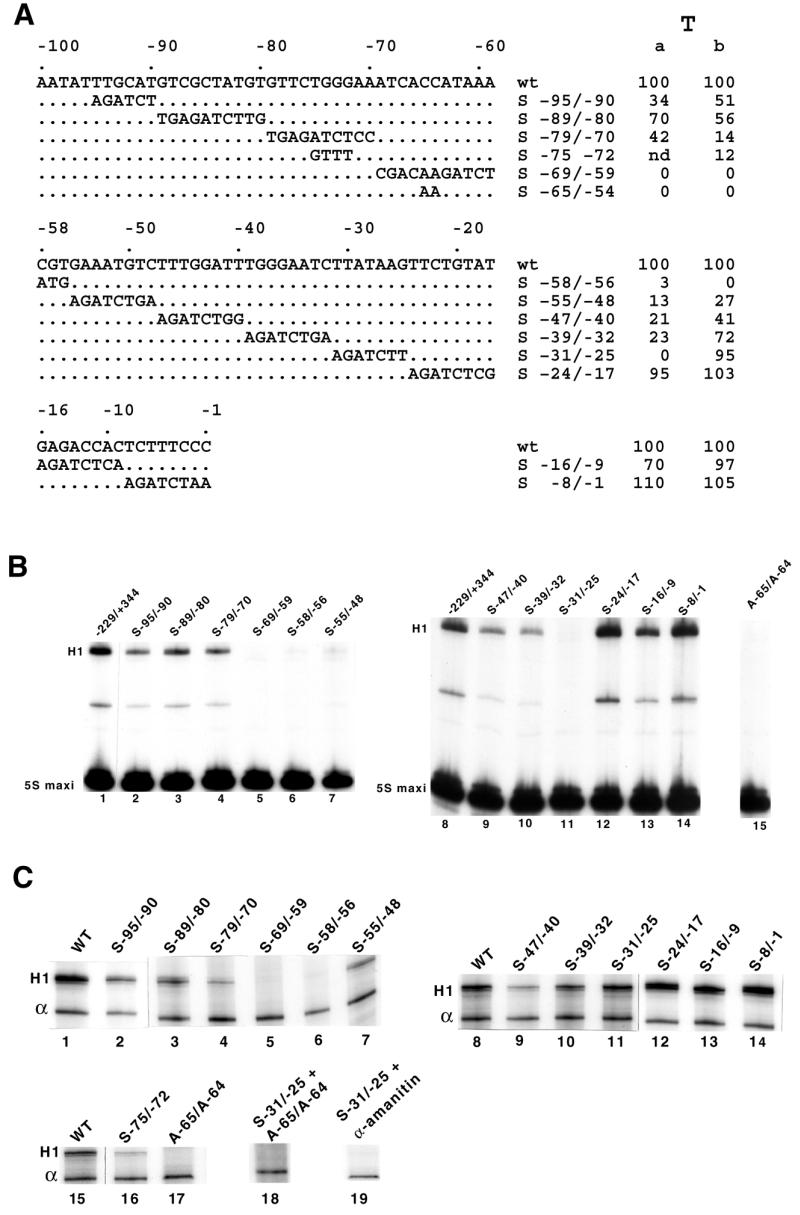
Structure and transcription of different human H1 mutant promoters. (A) Nucleotide sequences of the substitution mutants (S) in the human H1 RNA promoter. The wild-type non-template strand sequence is shown between –100 and –1 (top line). T indicates the in vitro (a) and in vivo (b) transcription levels. (B) Effects of promoter mutations on the H1 RNA gene transcription in vitro. The H1 RNA mutant promoters shown in (A) were used as templates for transcription assays in HeLa whole cell extracts. (C) Analysis of the promoter mutations on H1 RNA gene transcription in vivo. COS-7 cells were transfected with the indicated constructs together with plasmid pα1x72 as the internal control. Recovered RNAs were analyzed as in Figure 1C.
Figure 1.
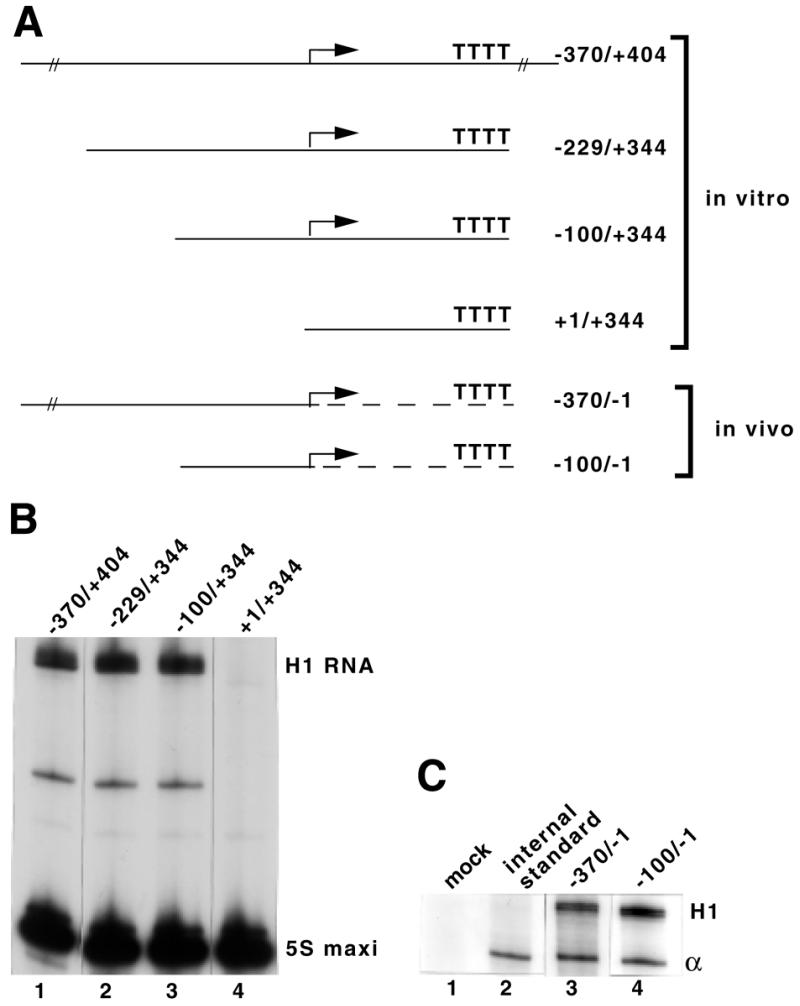
The –100/–1 5′-flanking sequences promote H1 RNA gene transcription. (A) Structures of the various truncated mutants used in the in vitro and in vivo analyses. Drawings are not to scale. An arrow indicates the start of transcription. The part of the hybrid gene derived from the β-globin is indicated by a broken line. (B) The constructs shown in (A) were used as templates for transcription in HeLa whole cell extracts, as described in Materials and Methods. (C) In vivo expression of wild-type and truncated H1 RNA genes. COS-7 cells were transfected with the indicated constructs together with plasmid pα1x72 as the internal control. RNAs were recovered 48 h after transfection and analyzed by the RNase protection assay described in Materials and Methods. Lane 1, mock-transfected cells; lane 2, cells transfected with the internal standard only; H1 and α, protected RNAs derived from the β-globin and internal standard, respectively.
Effector constructs
The pBRN3-Staf-DBD was obtained by cloning into pBRN3 (24) the DNA fragment containing the Staf DBD prepared by PCR amplification of the Staf cDNA, using forward and reverse primers incorporating an EcoRI site. The forward primer complementary to positions 793–810 in the Staf cDNA (14) contains an ATG initiator codon in the Kozak consensus sequence. The reverse primer complementary to positions 1436–1452 carries a TAG stop codon. Construct pBRN3-Staf was as described previously (14).
In vitro transcription assays
Transcription reactions were carried out in a final volume of 25 µl, essentially as described previously (19), in the presence of 15 µl of HeLa whole cell extract (9 µg/µl), 250 ng of H1 template and 25 ng of 5S RNA maxigene.
DNA binding assays
Gel retardation and DNase I footprinting assays were performed essentially as described in Myslinski et al. (13) and Schuster et al. (14). The template strand of the human H1 RNA gene (positions –279 to –1) was 5′-end-labeled by PCR amplification of the H1 –370/+404 construct using the proximal 32P-labeled primer.
Transfection and RNA analysis
COS-7 cells were cotransfected by the calcium phosphate coprecipitation procedure with 13 µg of reporter constructs, 3 µg of the reference plasmid pα1x72 (7) and pBS carrier DNA to adjust the total amount of DNA to 20 µg. An aliquot of 10 µg of RNA collected 48 h after transfection was used for RNase mapping, as described by Hernandez and Lucito (25). In the α-amanitin experiment shown in Figure 3C, the medium was removed 43 h after transfection and fresh medium containing 50µg/ml of α-amanitin was added. RNA was recovered 5 h later. Transcription efficiencies were quantitated with a Fuji Bioimage Analyzer Bas 2000 and normalized relative to the α-globin transcription level.
Oocyte microinjections
In the experiments shown in Figure 5B and C, X.laevis oocytes were coinjected with 4 ng of wild-type or mutant templates, 0.2 µCi [α-32P]GTP (800 Ci/mmol) and 5S RNA maxigene (75 pg for U1 and 100 pg U6) as an internal control for nuclear injection and RNA recovery. In the experiments shown in Figure 6, capped mRNAs were synthesized in vitro according to the manufacturer (Ambion) and injected (20 nl; 0.1, 1 or 5 ng) into the cytoplasm of X.laevis oocytes 20 h before the nuclear injection of 20 nl containing the U6 reporter DNA (200 µg/ml), the 5S RNA maxigene (5 µg/ml) and [α-32P]GTP (800 Ci/mmol, 0.2 µCi/oocyte). Oocytes were incubated at 19°C for 8 h (U6 constructs) or 16 h (U1 constructs). RNAs were extracted from batches of 10 oocytes and analyzed as previously described (13). Transcription efficiencies were quantitated with a Fuji Bioimage Bas 2000 Analyzer and normalized relative to the transcription level of the 5S maxi RNA.
Figure 5.
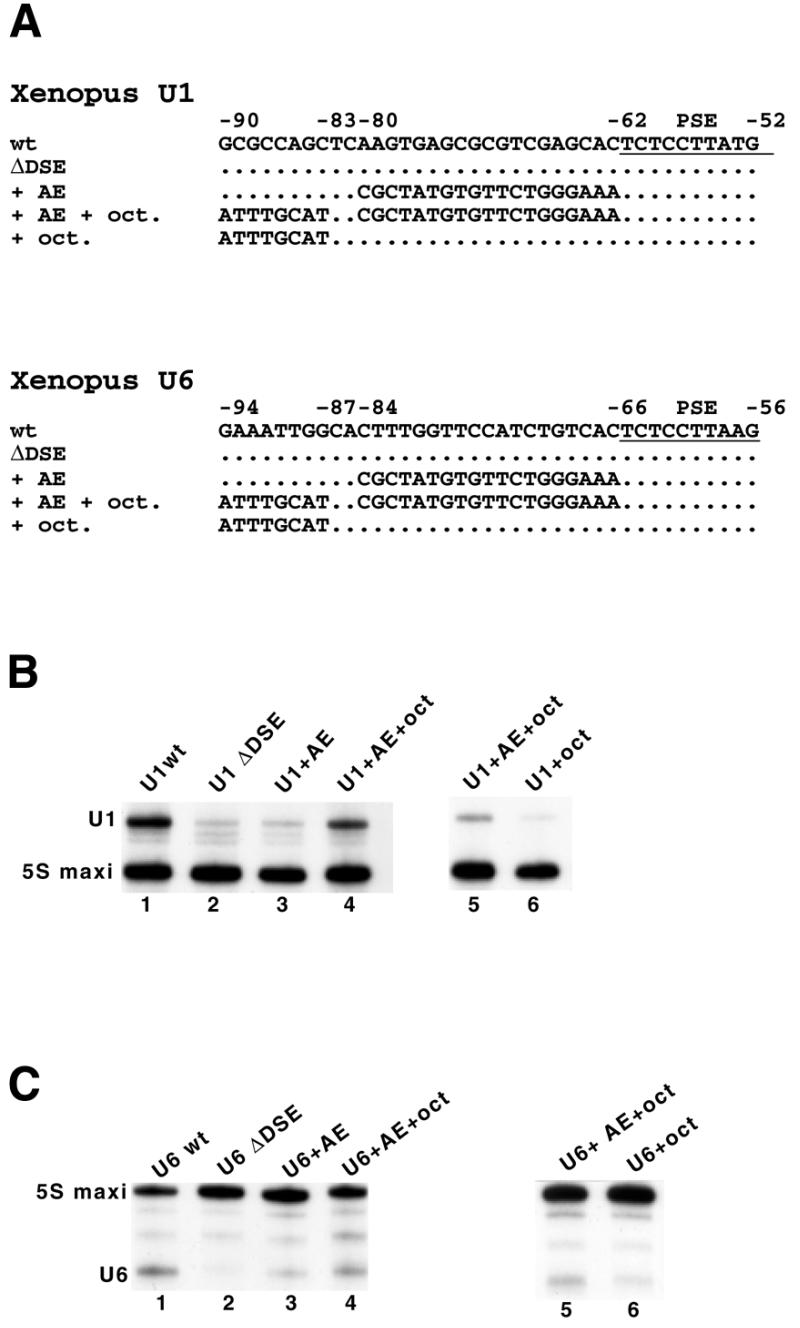
Octamer and Staf motifs engineered at proximal positions can stimulate transcription from snRNA Pol II and Pol III promoters, in X.laevis oocytes. (A) Structure of the Xenopus U1 and U6 constructs used in the analysis. The wild-type (wt) non-template strand sequences of the Xenopus U1 and U6 snRNA genes are shown between –90/–52 and –94/–56, respectively (top line). The positions of the PSE, octamer motif (oct) and Staf binding site (AE) are indicated. The PSE is underlined. U1 ΔDSE and U6 ΔDSE are U1 and U6 genes truncated at positions –177 and –190, respectively. (B and C) Xenopus laevis oocyte nuclei injected with the wild-type (lane 1) or mutants (lanes 2–6) U1 and U6 templates described in (A). Positions of the 5S maxi RNA, U1 and U6 are indicated. Results from lanes 1–4, and lanes 5 and 6 arose from separate experiments.
Figure 6.
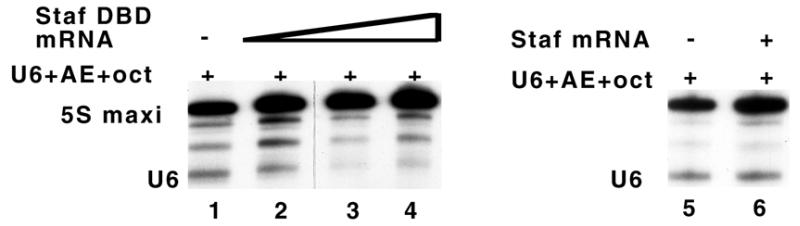
The Staf DNA binding domain can repress transcriptional activation of a U6 gene carrying a compact promoter. Identities of the effector proteins and reporter genes are indicated above each lane. Lanes 1 and 5, no effector was expressed. Lanes 2–4 and 6, cytoplasmic expression of variable amounts of mRNA effectors: lane 2, 0.1 ng; lane 3, 1 ng; lanes 4 and 6, 5 ng. Lanes 1–6, oocyte nuclei microinjected with 0.5 ng of the indicated reporter.
RESULTS
Boundaries of the human H1 RNA gene
To investigate the boundaries of the gene, mutant H1 DNA templates were constructed that carry various lengths of 5′- and 3′-flanking regions (Fig. 1A). The transcription efficiencies of the different constructs were analyzed in vitro by incubation in HeLa whole cell extracts under conditions optimal for RNA Pol III transcription. The transcription signals were normalized to the expression of the 5S RNA maxigene included in the assays as an internal standard. The H1 transcript shown in Figure 1B corresponds to the H1 RNA described by Baer et al. (19). The shortened construct H1 +1/+344, which retains the region to be transcribed and a run of Ts, was unable to drive transcription (Fig. 1B, lane 4). Constructs H1 –100/+344 (Fig. 1B, lane 3) and H1 –229/+344 (Fig. 1B, lane 2), which lack the 3′-flanking sequence but retain only 100 or 229 bp of 5′-flanking sequences, showed no difference in transcription levels with construct H1 –370/+404 containing 60 bp of 3′-flanking and 370 bp of 5′-flanking regions (Fig. 1B, lane 1). We concluded from these preliminary experiments that the H1 coding region alone is transcriptionally inert, and that elements required for maximal in vitro expression of the H1 RNA gene reside between –100 and the start site of transcription.
Prompted by the in vitro results, we used transfection of hybrid human H1 RNA genes into COS-7 cells to investigate the requirements of 5′-flanking sequences for transcription in vivo. The hybrid genes consist of wild-type or shortened H1 5′-flanking sequences, followed by a 137 bp spacer derived from the β-globin gene (Fig. 1A) (7). Thus, no H1 coding sequence is present in these constructs. The β-globin fragment is followed by different 3′-end formation signals: one acts on the termination of RNA polymerase II (Pol II) transcription from snRNA promoters; the second one consists of a run of Ts, constituting an efficient RNA Pol III termination site. RNAs were analyzed by RNase protection assay of an antisense RNA probe and normalization to the expression of an α-globin mRNA included as an internal standard. The construct which retained only 100 bp of 5′-flanking sequences showed no difference in transcription level with that containing 300 bp of 5′-flanking region (Fig. 1C, compare lanes 3 and 4). The above data clearly established that the elements required for H1 RNA gene expression reside between –100 and the start site of transcription.
A compact promoter for the human and mouse H1 RNA gene
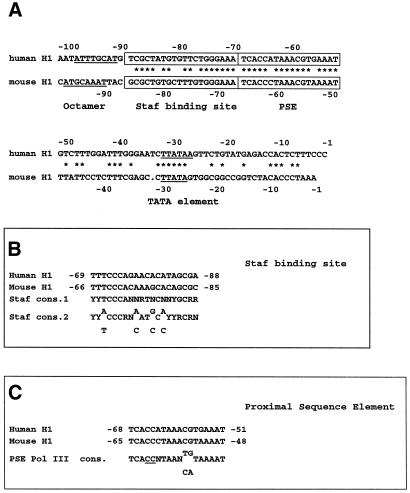
Sequence comparisons of the 5′-flanking region between –100 and –1 in the human and mouse H1 RNA genes (26) revealed the presence of four conserved motifs displaying striking similarities with promoter elements in other RNA Pol III transcription units (Fig. 2). Among these was the octamer element ATTTGCAT (in the reverse orientation) at positions –97/–90 in the human gene. This sequence is also present at essentially the same location (positions –96/–89) in the mouse H1 RNA gene, but in the ATGCAAAT orientation. The 19 bp region centered to –79 and –77 bp in the human and mouse H1 RNA genes, respectively, and harbors 85% sequence identity with the consensus sequences of the Staf binding site (Fig. 2A and B) (15). Figure 2A and C indicates that an element, lying –68/–51 bp upstream of the transcription start site in the human H1 gene, and –65/–48 bp in the mouse H1 counterpart, shows 100% sequence identity to the consensus sequence of the human PSE in other Pol III snRNA genes (12). Finally, both the human and mouse H1 genes contain a TATA element centered at –29 and –28 bp, respectively (Fig. 2).
Figure 2.
The promoter of the human and mouse H1 RNA genes contains four common putative control elements. (A) Sequence comparison of the –100/–1 5′-flanking regions of the human and mouse H1 RNA genes. The wild-type non-template strands of the human and mouse genes are shown between –100/–1 and –97/–1, respectively. Nucleotide identity is indicated by an asterisk. Octamer and TATA elements are underlined. Presumed PSE and Staf binding sites are boxed. (B) Sequence of the putative Staf binding sites in the human and mouse H1 RNA genes compared with the consensus Staf binding site. Staf cons.1, consensus sequence determined by footprint assay of native gene promoters (15); Staf cons. 2, consensus sequence determined by PCR-mediated binding site selection (15). (C) Sequence of the putative H1 RNA gene PSE compared with the human Pol III PSE consensus. The invariant CC at positions 4 and 5 of the consensus is underlined.
To identify in more detail the H1 promoter elements, we introduced a series of clustered point mutations between positions –95 and –1 in the human 5′-flanking region (Fig. 3A). Figure 3B (lanes 1–14) shows the in vitro transcription activities of the different constructs, the efficiencies normalized to the expression of the 5S RNA maxigene being displayed in Figure 3A. Transcription was abolished by the mutant templates S–69/–59, S–58/–56, S–31/–25 (Fig. 3B, lanes 5, 6 and 11) and reduced by S–95/–90 (Fig. 3B, lane 2), S–79/–70 (Fig. 3B, lane 4), S–55/–48 (Fig. 3B, lane 7; correct with the low level of 5S RNA maxi), S–47/–40 (Fig. 3B, lane 9) and S–39/–32 (Fig. 3B, lane 10).
Analysis of the in vivo effects of those same clustered point mutations (Fig. 3C; see Fig. 3A for quantifications) revealed an overall similar pattern of H1 RNA expression, except that the construct carrying point mutations at positions –31/–25 in the TATA element generated transcripts in vivo but not in vitro (compare lanes 11 in Fig. 3B and C). Remarkably, it appears that the in vitro and in vivo down mutations map essentially to the PSE motif. Sequence comparisons of the human and mouse PSEs underscored the occurrence of two positionally invariant C residues at positions 4 and 5 in the consensus sequence (Fig. 2C). Double point mutations were introduced at these positions in the wild-type H1 gene to generate the H1 template with A–65/A–64. Transcriptional analysis of this mutant template showed that its ability to drive transcription in vitro (Fig. 3B, lane 15) and in vivo (Fig. 3C, lane 17) was entirely lost. These results demonstrate that the two C residues in the H1 RNA gene PSE play a crucial role for transcription, as in other PSEs (3,27).
In the above transfection experiments, we found that mutant S–31/–25, which changes the sequence of the TATA element, was deleterious in vitro while leaving a transcription level in vivo amounting to 95% of the wild-type level. This was reminiscent of the effects obtained by substituting the TATA element in different Pol III snRNA and snRNA-type genes (3,6,8). A template carrying both the A–65/A–64 substitution destroying the PSE activity and S–31/–25 led to complete abolition of H1 RNA transcription in vivo (Fig. 3C, lane 18). Thus, it appears that the transcriptional activity observed in Figure 3C, lane 11 in the absence of the TATA element was PSE-dependent, suggesting that the lack of TATA sequence rendered the H1 RNA promoter RNA Pol II dependent. We tested this possibility by examining the in vivo transcription products 5 h after addition of α-amanitin under conditions inhibiting Pol II transcription. The in vivo transcription product observed in S–31/–25 disappeared in the presence of α-amanitin (Fig. 3C, compare lanes 11 and 19). In this experiment, in the absence of transcription, the RNA derived from the H1 hybrid construct was unstable and disappeared within 5 h, whereas the α-globin transcript remained stable for the same period of time (7). Thus, the α-globin RNA served as an internal reference for RNA recovery in the α-amanitin experiment. Primer extension analysis revealed that transcription directed by the S–31/–25 substitution mutant initiated predominantly 7 bp upstream of the wild-type transcription start site (data not shown). Altogether, our findings established that the human H1 promoter harbors a remarkable unusual and unique architecture, with the classical four promoter elements of the snRNA genes that are here located within 100 bp of 5′-flanking sequence.
A Staf binding site abuts the PSE in the human H1 RNA gene
To determine whether the functional site identified at positions –88/–69 of the human gene is a bona fide Staf binding site, its binding abilities were assessed by electrophoretic mobility shift assays. Labeled DNA fragments encompassing positions –279/–1 of the gene were incubated with Staf-containing bacterial extracts. Two major complexes, C1 and C2, were obtained (Fig. 4A, lane 2). The C2 complex arose from proteolytic cleavages of Staf that could not be prevented by inclusion of any inhibitor (14,15). The two complexes are specific because they were competed by an excess of unlabeled Staf binding site (AE) of the Xenopus tRNASec gene (Fig. 4A, lane 4), but not by an excess of mutant AE (Fig. 4A, lane 3). To localize the Staf binding site, DNase I footprint analysis was carried out with an H1 DNA probe labeled on the template strand. Figure 4B shows that Staf protected a sequence of 25 bp from –91 to –67 on the template strand. We next examined the binding capacities of Staf to H1 mutant DNAs carrying alterations in the mapped Staf binding site. The binding activities were assayed by electrophoresis on non-denaturating gels and the relative efficiency with which Staf bound to the different mutant sites was estimated by comparing the amounts of the shifted complexes formed with the different labeled sites. Figure 4C shows the capacity of Staf to bind to three mutant versions of the H1 Staf binding site. S–89/–80 and S–79/–70 correspond to substitutions in the 5′- and 3′-parts of the site, respectively (Fig. 3A); in S–75/–72, the highly conserved CCCA sequence in the characterized Staf binding sites (15) was replaced by AAAC (Fig. 3A). With S–89/–80, S–79/–70 and S–75/–72, the binding efficiencies of Staf decreased to 14, 8 and 7% of the wild-type levels, respectively (Fig. 4C, compare lane 5 with lanes 6–8). More importantly, the relative affinities of Staf for the different mutant DNAs could be correlated with the in vivo transcriptional activity of the H1 RNA gene harboring the same mutations (Fig. 3A and C).
Figure 4.
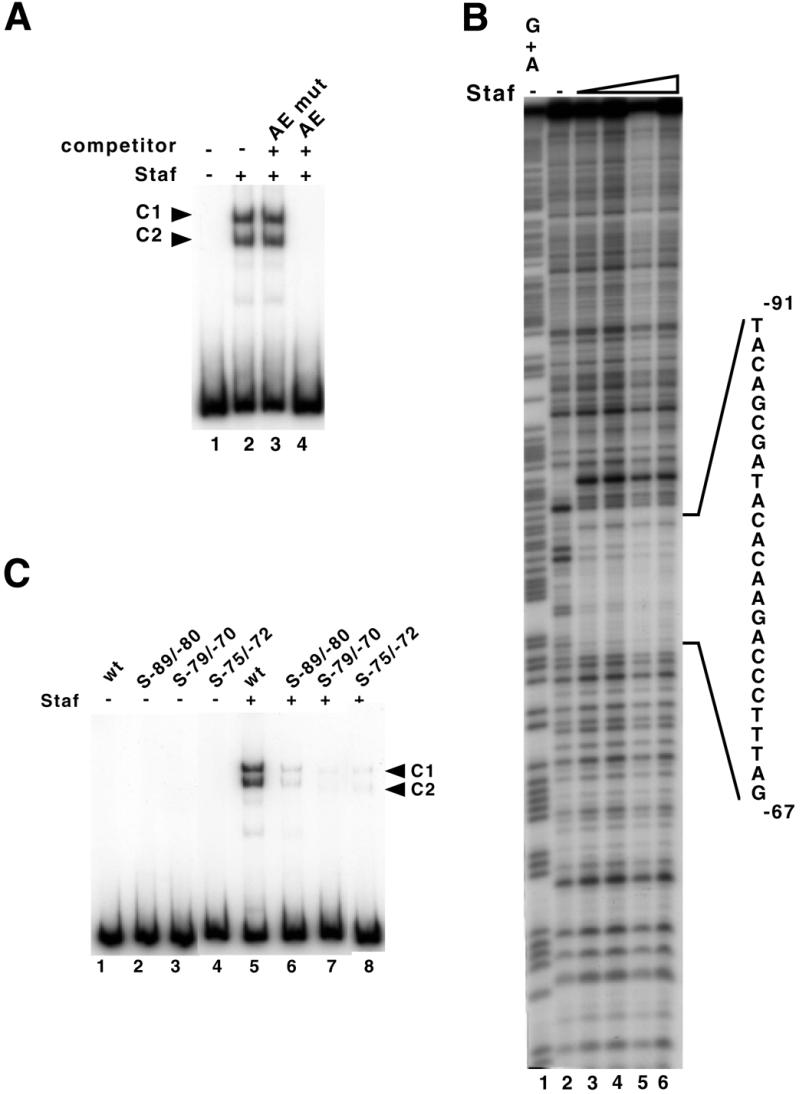
Identification of the Staf binding site in the human H1 RNA gene. (A) Gel retardation assay with a labeled DNA fragment encompassing positions –279/–1 upstream of the transcription start site. The 32P-end-labeled DNA probe was incubated in the absence (lane 1) or presence (lanes 2–4) of Staf-containing bacterial extracts. Reactions in lanes 3 and 4 contained a 1000-fold excess of unlabeled unspecific (AE mut) and specific (AE) competitor, respectively. (B) Footprint analysis of Staf–DNA complexes formed on the H1 RNA gene. DNase I digestion of the human H1 probe, labeled on the template strand, in the absence (lane 2) or presence (lanes 3–6) of increasing amounts of Staf. Probes submitted to G + A chemical cleavage were used as markers (lane 1). The protected region is indicated. (C) Staf binding assays on wild-type and mutant versions of the human H1 RNA gene. A 223 bp end-labeled fragment containing wild-type or mutant versions of the Staf binding site was used in the binding studies. Lanes 1–4, no protein added. Lanes 5–8 contained 1 µl of 20-fold diluted bacterial extracts containing the recombinant Staf. Probes are indicated above the lanes.
Collectively, these results demonstrate unambiguously that a functional Staf binding site is located adjacent to the PSE in the human H1 RNA gene.
Engineered compact Pol II or Pol III snRNA gene promoters are functional in Xenopus oocytes
To investigate whether compact snRNA-type promoters are still functional in Xenopus oocytes, the mutant Xenopus U1 (Pol II) and U6 (Pol III) snRNA templates shown in Figure 5A were constructed from the U1 ΔDSE and U6 ΔDSE templates. The U1 and U6 ΔDSE correspond to U1 and U6 templates truncated at positions –177 and –190, respectively. In the constructs + AE, the wild-type sequences from positions –80/–62 in U1 ΔDSE and –84/–66 in U6 ΔDSE were replaced by the 19 bp sequence of the Staf binding site identified in the human H1 RNA (Fig. 5A). In constructs + AE + oct, the octamer sequence ATGCAAAT was introduced in a reverse orientation, as it is found in the human H1 RNA gene, 2 bp upstream of the Staf binding site (positions –90/–83 in U1 and –94/–87 in U6, Fig. 5A). The transcriptional capacities of the various constructs were assayed by microinjection into X.laevis oocytes. The normalized values for the U1 ΔDSE and U6 ΔDSE templates dropped to 21 and 4% of the wild-type levels, respectively (compare lanes 1 and 2 in Fig. 5B and C). Introduction of a Staf binding site adjacent to the PSE resulted in a slight increase in template activity to 14% for U6 (Fig. 5C, lane 3). No significant transcription enhancement could be observed for the U1 template (Fig. 5B, lane 3). Remarkably, the simultaneous presence of the Staf binding site and octamer sequence produced a functional promoter with transcriptional efficiencies of 58 and 45% of the wild-type level for U1 and U6, respectively (Fig. 5B and C, compare lanes 1 and 4). The octamer sequence alone is unable to enhance transcription to a level equivalent to that observed in the presence of both the octamer and Staf binding site (compare lanes 5 and 6 in Fig. 5B and C). These results show that the unusual compact structure identified in the human and mouse H1 gene promoters can also be functional in other snRNA genes.
The DNA binding domain of Staf can repress transcription from an engineered compact snRNA promoter
To determine whether Staf could recognize in vivo a Staf binding site abutting a PSE, we used a microinjection assay in Xenopus oocytes to test whether the sole presence of the Staf DNA binding domain (Staf-DBD) can repress transcription from a compact snRNA promoter. The reporter construct used in the assay was U6 + AE + oct, which contains, in addition to the PSE and TATA elements, the Staf binding site immediately adjacent to the PSE and the octamer motif 2 bp upstream of the Staf binding site (Fig. 5A). The Staf-DBD mRNA was produced by T7 transcription in vitro and then microinjected into the oocyte cytoplasm. After 20 h of incubation, the U6 reporter gene was injected into oocyte nuclei with [α-32P]GTP and the 5S RNA maxigene as the internal standard. After a second incubation, the labeled RNAs were extracted and the level of transcribed U6, normalized relative to the 5S RNA maxigene expression, was used to determine the transcriptional capacities of the effector proteins. Injecting increasing amounts of Staf-DBD mRNAs resulted in a progressive reduction of U6 gene expression (Fig. 6, compare lane 1, without effector, to lanes 2–4, for Staf-DBD). At higher mRNA concentrations (Fig. 6, lanes 3 and 4), the U6 level decreased dramatically to become similar to that of a U6 gene devoid of DSE. A control experiment, performed with the mRNA encoding the full-length Staf, confirmed that the observed effects were effectively caused by the unproductive binding of the Staf-DBD to the target DNA sequences, since expression of the U6 reporter was unaffected by the presence of full-length Staf (Fig. 6, compare lanes 5 and 6). These data established that the DNA binding domain of Staf alone is able to efficiently compete for DNA binding sites with the full-length endogenous Staf, therefore reducing the level of transcriptional activation. This observation strongly suggests that Staf could efficiently recognize in vivo a Staf binding site abutting a PSE.
DISCUSSION
The present study has provided characterization of the cis- and trans-acting elements involved in the RNA Pol III transcription of the human H1 RNA gene. Using in vitro and in vivo transcription studies, we have demonstrated that the sequence elements that are required for transcription of the H1 RNA lie entirely within 100 bp of 5′-flanking region. The H1 promoter elements are comprised of four cis-acting elements: the TATA motif, the PSE and the DSE containing an octamer motif, and a Staf binding site. These elements are characteristic features of snRNA and snRNA-type genes transcribed by RNA Pol II or Pol III. However, their peculiar arrangement with, in particular, the adjacency of the Staf and PSE sequences, makes the H1 RNA gene unique. The compact structure of the H1 promoter arises from juxtaposition of the two functional submotifs of the DSE to the PSE. The octamer sequence is distant by only 21 bp from the 5′-part of the PSE, the Staf binding site being perfectly adjacent to the PSE. We have shown that the PSE and TATA motifs are absolutely required for in vitro and in vivo Pol III synthesis of the H1 RNA. The transcription level observed in vivo with templates carrying TATA mutations was Pol II dependent. As in the case of the human and Xenopus U6 genes for which the TATA box is a major determinant specifying RNA Pol III transcription (7,8), the lack of TATA sequence rendered the H1 RNA gene promoter RNA Pol II-dependent. Surprisingly, the sequence substitution in the region between the PSE and the TATA motif (S–47/–40) resulted in an important reduction in RNA synthesis, both in vitro and in vivo. The sequence at these positions neither resembles previously identified transcriptional control elements nor is conserved in the mouse H1 RNA gene (26). This observation suggests that this sequence can influence transcription per se, perhaps by inducing a particular conformation of the DNA.
Our band-shift experiment showed that the mapped Staf binding site can actually be recognized by Staf. Site-directed mutagenesis demonstrated that both motifs of the DSE, the octamer and the Staf binding site, were required for full activity of the H1 promoter. However, their individual contribution toward the global promoter activity was not equivalent. Mutation in the octamer sequence reduced the activity by 50% in vivo, whereas mutations in the TGGG sequence of the Staf binding site resulted in ∼90% reduction of the promoter activity. These results suggest the putative existence of cooperative interactions between octamer and/or Staf sites-bound factors, and protein(s) of the basal transcription complex. Although we originally identified Staf in X.laevis as a trans-acting factor controlling the expression of the tRNASec, we could also show that it more generally controls snRNA and snRNA-type gene transcription (14,15). In humans, the two proteins ZNF76 and ZNF143 are functional homologs to the Xenopus Staf (18). The present study extends the roles of ZNF143 and/or ZNF76 to the transcription of the H1 RNA gene. Indeed, our results previously established that Xenopus Staf could stimulate transcription from Pol II and Pol III promoters harboring a Staf binding site in a distal (classical) position. Here, we showed that this transcriptional activator can also stimulate transcription from promoters containing an engineered Staf binding site in a proximal position. As ZNF143 constitutes the human ortholog of the Xenopus Staf, it is conceivable that ZNF143 functions in human H1 RNA transcription.
The observation that a Staf binding site is juxtaposed to the PSE is intriguing and raises questions with regard to the simultaneous presence of proteins bound to the two elements, especially in terms of steric hindrance. The classical Pol III snRNA and snRNA-type gene promoters also contain the DSE, PSE and TATA motifs, but the DSE/PSE distance is highly conserved to ∼150 bp. Several lines of evidence suggested that in this case, the DNA may be bound to a histone octamer to form a nucleosome in vivo (28–32). First, in a reconstituted chromatin system, the human U6 gene positions a nucleosome between the DSE and the PSE, and the DSE can participate to transcriptional activation in vitro, in contrast to the situation with naked DNA (28). Secondly, the pattern of DNase I protection in vivo suggests that a nucleosome lies between the DSE and the PSE of the 7SK gene (29). Finally, potentiation by Oct-1 of SNAPc/PTF binding to the PSE in vitro, as well as transcription of the human U6 and 7SK RNA genes in vitro, are highly inefficient unless an octamer element is positioned close to the PSE. This suggests that the DSE- and PSE-bound factors contact each other in vivo (9,30–32). It is tempting to speculate that the H1 compact promoter enables the DSE- and PSE-bound factors to directly interact, while this interaction is made possible by the presence of a nucleosome in classical snRNA and snRNA-type genes.
Another interesting feature is the presence of the poly(ADP–ribose) polymerase 2 (PARP-2) gene in an opposite orientation relative to the H1 RNA gene (26,33). The transcription start sites of the human and mouse PARP-2 genes are separated by only 152 and 113 bp from that of H1 RNA gene, respectively (26). The functional significance of the compact character of the H1 RNA gene is unclear at the present time, but the presence of the two genes in opposite orientation can require a particular chromatin configuration incompatible with a DSE distant from the PSE. This will be addressed in future investigations.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to S.Altman for the gift of human H1 RNA clone. We also thank N.Hernandez for the pU6/Hae/Ra.2/EcoRV and pα1x72 constructs, D.Fagegaltier for the HeLa whole cell extracts and C.Loegler for excellent technical assistance.
References
- 1.Paule R.P. and White,R.J. (2000) Transcription by RNA polymerases I and III. Nucleic Acids Res., 28, 1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sprague K.U., Larson,D. and Morton,D. (1980) 5′ flanking sequences are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell, 22, 171–172. [DOI] [PubMed] [Google Scholar]
- 3.Ullu E. and Weiner,A.M. (1985) Upstream sequences modulate the internal promoter of the human 7SL RNA gene. Nature, 318, 371–374. [DOI] [PubMed] [Google Scholar]
- 4.Howe J.G. and Shu,M.-D. (1989) Epstein–Barr virus small RNA (EBER) genes: unique transcription units that combine RNA polymerase II and III promoters elements. Cell, 57, 825–834. [DOI] [PubMed] [Google Scholar]
- 5.Carbon P. and Krol,A. (1991) Transcription of the Xenopus laevis selenocysteine tRNASec gene: a system that combines an internal B box and upstream elements also found in U6 snRNA genes. EMBO J., 10, 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carbon P., Murgo,S., Ebel,J.P., Krol,A., Tebb,G. and Mattaj,J. (1987) A common octamer motif binding protein is involved in the transcription of U6 snRNA by RNA polymerase III and U2 snRNA by RNA polymerase II. Cell, 51, 71–79. [DOI] [PubMed] [Google Scholar]
- 7.Lobo S.M. and Hernandez,N. (1989) A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell, 58, 55–67. [DOI] [PubMed] [Google Scholar]
- 8.Mattaj I., Dathan,N.A., Parry,H.D., Carbon,P. and Krol,A. (1988) Changing the RNA polymerase specificity of U snRNA gene promoters. Cell, 55, 435–442. [DOI] [PubMed] [Google Scholar]
- 9.Murphy S., Yoon,J.-B., Gerster,T. and Roeder,R.G. (1992) Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol. Cell. Biol., 12, 3247–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadowski C.L., Henry,R.W., Lobo,S.M. and Hernandez,N. (1993) Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev., 7, 1535–1548. [DOI] [PubMed] [Google Scholar]
- 11.Henry R.W., Sadowski,C.L., Kobayashi,R. and Hernandez,N. (1995) A TBP–TAF complex required for transcription of human snRNA genes by RNA polymerases II and III. Nature, 374, 653–657. [DOI] [PubMed] [Google Scholar]
- 12.Lobo S.M. and Hernandez,N. (1994) Trancription of snRNA genes by RNA polymerases II and III. In Conaway,R.C. and Conaway,J.W. (eds), Transcription, Mechanisms and Regulation. Raven Press, Ltd, New York, NY, pp. 127–159.
- 13.Myslinski E., Krol,A. and Carbon,P. (1992) Optimal tRNASec gene activity requires an upstream SPH motif. Nucleic Acids Res., 20, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuster C., Myslinski,E., Krol,A. and Carbon,P. (1995) Staf, a novel zinc finger protein that activates the RNA polymerase III promoter of the selenocysteine tRNA gene. EMBO J., 14, 3777–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaub M., Myslinski,E., Schuster,C., Krol,A. and Carbon,P. (1997) Staf, a promiscuous activator for enhanced transcription by RNA polymerases II and III. EMBO J., 16, 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herr W. (1992) Oct-1 and Oct-2: differential transcriptional regulation by proteins that bind to the same DNA sequence. In MacKnight,S.L. and Yamamoto,K.R. (eds), Transcriptional Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 1, pp. 1103–1135.
- 17.Herr W. and Cleary,M.A. (1995) The POU domain : versatility in transcriptional regulation by a flexible two-in one DNA-binding domain. Genes Dev., 9, 1679–1693. [DOI] [PubMed] [Google Scholar]
- 18.Myslinski E., Krol,A. and Carbon,P. (1998) ZNF76 and ZNF143 are two human homologs of the transcriptional activator Staf. J. Biol. Chem., 273, 21998–22006. [DOI] [PubMed] [Google Scholar]
- 19.Baer M., Nilsen,T.W., Costigan,C. and Altman,S. (1990) Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res., 18, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannon G.J., Chubb,A., Maroney,P.A., Hannon,G., Altman,S. and Nilsen,T.W. (1991) Multiple cis-acting elements are required for RNA polymerase III transcription of the gene encoding H1 RNA, the RNA component of human RNase P. J. Biol. Chem., 266, 22796–22799. [PubMed] [Google Scholar]
- 21.Krol A., Lund,E. and Dahlberg,J.E. (1985) The two embryonic U1 RNA genes of Xenopus laevis have both common and gene-specific transcription signals. EMBO J., 4, 1529–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murgo S., Krol,A. and Carbon,P. (1991) Sequence, organization and transcriptional analysis of a gene encoding a U1 snRNA from the axolotl, Ambystoma mexicanum. Gene, 99, 163–170. [DOI] [PubMed] [Google Scholar]
- 23.Krol A., Carbon,P., Ebel,J.P. and Appel,B. (1987) Xenopus tropicalis U6 snRNA genes transcribed by RNA polymerase III contain the upstream promoter elements used by pol II dependent U-snRNA genes. Nucleic Acids Res., 15, 2463–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemaire P., Garett,N. and Gurdon,J.B (1995) Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell, 81, 85–94. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez N. and Lucito,R. (1988) Elements required for transcription initiation of the human U2 snRNA gene coincide with elements required for 3′ end formation. EMBO J., 7, 3125–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amé J.-C., Schreiber,V., Fraulob,V., Dollé,P., de Murcia,G. and Niedergang,C. (2001) A bidirectional promoter connects the PARP-2 gene to the gene for RNase P RNA: structure and expression of the mouse PARP-2 gene. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 27.Lescure A., Carbon,P. and Krol,A. (1991) The different positioning of the proximal sequence element in the Xenopus RNA polymerase II and III snRNA promoters is a key determinant which confers RNA polymerase III specificity. Nucleic Acids Res., 19, 435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stunkel W., Kober,I. and Seifart,K.H. (1997) A nucleosome positioned in the distal promoter region activates transcription of the human U6 gene. Mol. Cell. Biol., 17, 4397–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyd D.C., Greger,I.H. and Murphy,S. (2000) In vivo footprinting studies suggest a role for chromatin in transcription of the human 7SK gene. Gene, 247, 33–44. [DOI] [PubMed] [Google Scholar]
- 30.Murphy S., Pierani,A., Scheidereit,C., Melli,M. and Roeder,R.G. (1989) Purified octamer binding transcription factors stimulate RNA polymerase III-mediated transcription of the 7SK RNA gene. Cell, 59, 1071–1080. [DOI] [PubMed] [Google Scholar]
- 31.Danzeiser D.A., Urso,O. and Kunkel,G.R. (1993) Functional characterization of elements in a human U6 small nuclear RNA gene distal control region. Mol. Cell. Biol., 13, 4670–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mittal V., Cleary,M.A., Herr,W. and Hernandez,N. (1996) Oct-1 POU-specific domain can stimulate small nuclear RNA gene transcription by stabilizing the basal transcription complex SNAPc. Mol. Cell. Biol., 16, 1955–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amé J.C., Rolli,V., Schreiber,V., Niedergang,C., Apiou,F., Decker,P., Muller,S., Höger,T., Ménissier-de Murcia,J. and de Murcia,G. (1999) PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem., 274, 17860–17868. [DOI] [PubMed] [Google Scholar]