Abstract
Hepatitis C virus (HCV) is a leading cause of chronic liver disease, cirrhosis, and hepatocellular carcinoma. To address the molecular basis of HCV pathogenesis using tupaias (Tupaia belangeri), we characterized host responses upon HCV infection. Adult tupaias were infected with HCV genotypes 1a, 1b, 2a, or 4a. Viral RNA, alanine aminotransferase, anti-HCV core and anti-nonstructural protein NS3 antibody titres, reactive oxygen species (ROS), and anti-3β-hydroxysterol-Δ24reductase (DHCR24) antibody levels were measured at 2-week intervals from 0 to 41 weeks postinfection. All HCV genotypes established infections and showed intermittent HCV propagation. Moreover, all tupaias produced anti-core and anti-NS3 antibodies. ROS levels in sera and livers were significantly increased, resulting in induction of DHCR24 antibody production. Similarly, lymphocytic infiltration, disturbance of hepatic cords, and initiation of fibrosis were observed in livers from HCV-infected tupaias. Intrahepatic levels of Toll-like receptors 3, 7, and 8 were significantly increased in all HCV-infected tupaias. However, interferon-β was only significantly upregulated in HCV1a- and HCV2a-infected tupaias, accompanied by downregulation of sodium taurocholate cotransporting polypeptide. Thus, our findings showed that humoral and innate immune responses to HCV infection, ROS induction, and subsequent increases in DHCR24 auto-antibody production occurred in our tupaia model, providing novel insights into understanding HCV pathogenesis.
Introduction
Hepatitis C virus (HCV) is a major public health problem that infects approximately 130–170 million people worldwide1, 2. HCV causes chronic hepatitis and is a major cause of liver cirrhosis and hepatocellular carcinoma (HCC)1. The first line of immune defence against HCV relies on cell-intrinsic innate immunity within hepatocytes. The HCV genome is a single-stranded, positive-sense RNA genome. During viral replication, HCV is sensed as non-self by pattern recognition receptors (PRRs) in the host cell, which identify and bind to pathogen-associated molecular patterns (PAMPs) within viral products, leading to activation of innate and adaptive immune responses3. Both effective innate and adaptive immune responses are involved in the control of HCV infections4; however, the role of the humoral immune system in HCV clearance is still unclear5.
Toll-like receptors (TLRs), an important component of innate immunity, play crucial roles in sensing invaders and initiating innate immune responses, thereby limiting the spreading of infections and modulating adaptive immune responses6. Sodium taurocholate cotransporting polypeptide (NTCP), a bile acid transporter expressed at the hepatocyte basolateral membrane7, can act as a regulator of antiviral immunity in HCV infection8. HCV infection can induce reactive oxygen species (ROS) production9, 10 and oxidative stress can lead to the formation of 8-hydroxydeoxyguanosine (8-OHdG), an indicator of oxidative DNA damage11. 3β-Hydroxysterol-Δ24reductase (DHCR24), a cholesterol biosynthetic enzyme12, is an essential host factor that plays a significant role in HCV replication13. Moreover, anti-DHCR24 auto-antibody levels are increased during the progression of HCV infection14. Indeed, the detection rate of HCC by anti-DHCR24 antibodies is higher (70.6%) than that of the standard HCC markers, alpha-fetoprotein (54.8%) or protein induced by vitamin K absence or antagonist-II (42.5%)14.
Recently, HCV can be completely cured by newly approved drugs, direct-acting antivirals (DAAs)15, 16. However, persistent hepatic inflammation, cirrhosis, and HCC have been reported in patients following viral clearance17. To solve these issues and develop an efficient vaccine against HCV, animal models are essential. Lack of small animal models is a great obstacle in the field of HCV research. To date, chimpanzees have been used as infection models for HCV. However, high costs and ethical concerns have restricted the use of chimpanzees in experimental infections. Recently, humanized chimeric mice18 and genetically humanized mice19 have been developed for use in HCV infection models13, 18. However, the use of mice has some disadvantages, including high cost, immunocompromised animal status, donor-to-donor variability, and inability to examine chronic infections. Tupaia belangeri belongs to the Tupaiidae family, which contains four genera and 19 extant species20. The evolutional characterization of 7 S RNA-derived short interspersed elements (SINEs) has shown that tupaias possess specific, chimeric Tu type II SINEs and can be clustered with primates21. Thus, genomic analysis suggested that tupaias are more closely related to humans than to rodents21, 22. Tupaias have been reported to be susceptible to several hepatotropic viruses that also infect humans, including hepatitis B virus23, 24, HCV25, 26, and hepatitis E virus27, and can be developed as an immunocompetent animal infection model. However, the molecular basis of HCV pathogenesis has not been fully characterized in the tupaia model due to a lack of characterization tools (e.g., specific antibodies, quantitative polymerase chain reaction [qPCR] assays, and cDNAs).
Therefore, in this study, we evaluated the susceptibility of tupaias to several viral strains of HCV and characterized the effects of HCV infection on ROS generation and its association with anti-DHCR24 antibody levels. We also characterized humoral immune responses to viral proteins and established a qPCR assay to evaluate TLR, NTCP, and cytokine expression to characterize the innate immune response during HCV infection, which may provide significant insight into HCV pathogenesis.
Results
Alanine aminotransferase (ALT) levels and viral loads in HCV-infected tupaia sera
Tupaias were infected with HCV genotypes 1a (#21), 1b (#22), 4a (#23), and 2a (#24). The level of ALT fluctuated, and intermittent growth of HCV was observed in all tupaias (Figs 1A, 2A, 3A and 4A). The highest ALT level (317.5 IU/L) was observed in tupaia #23 at 29 weeks postinfection (wpi). Virus could be detected in serum at 25 (51 copies/mL) and 31 wpi (43 copies/mL) in tupaia #21; at 13 wpi (21 copies/mL) in tupaia #22; at 7 (4 copies/mL), 29 (13 copies/mL), and 31 wpi (50 copies/mL) in tupaia #23; and at 11 (120 copies/mL), 15 (2 copies/mL), and 23 wpi (75 copies/mL) in tupaia #24 (Figs 1A, 2A, 3A and 4A).
Figure 1.
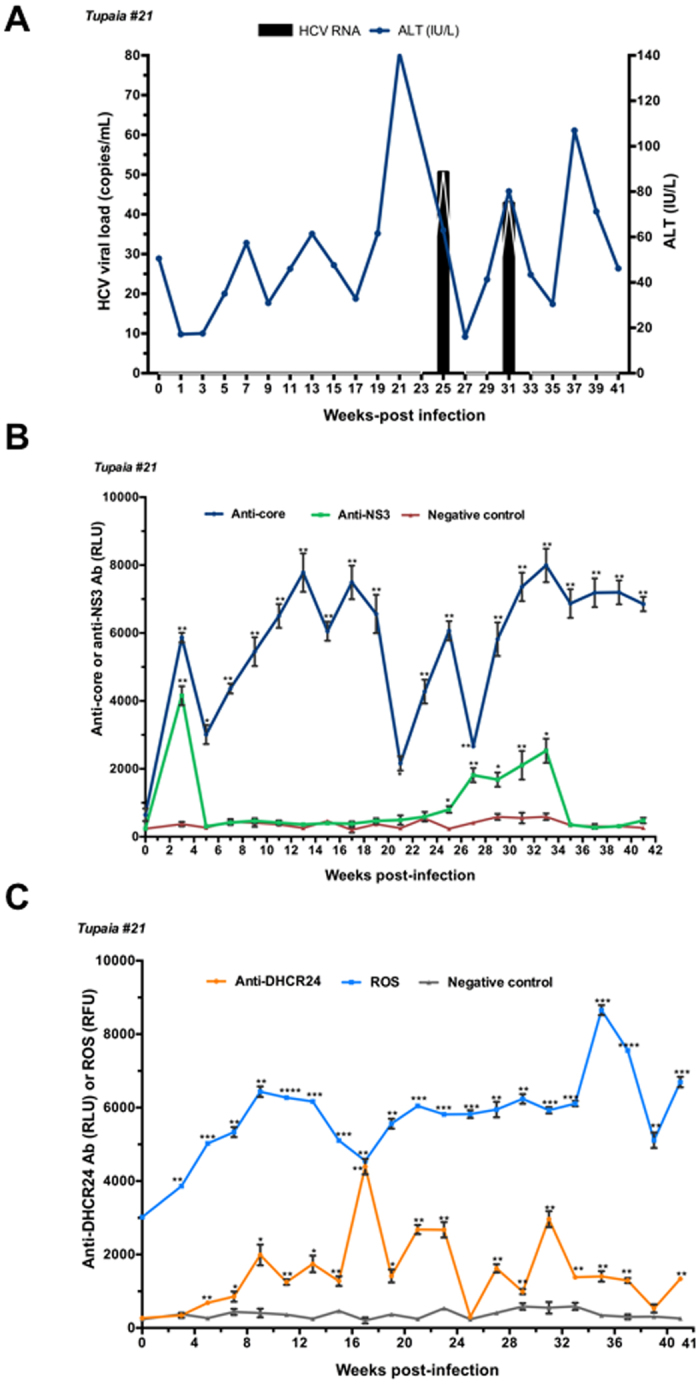
Response of tupaias to HCV1a infection. (A) ALT levels and viral loads in sera from tupaia #21 collected at 2-week intervals from 0 to 41 weeks postinfection (wpi). (B) Anti-HCV core and anti-nonstructural protein NS3 antibody titres in tupaia #21 at 2-week intervals from 0 to 41 wpi. (C) Anti-DHCR24 antibody titres and ROS levels in tupaia #21 at 2-week intervals from 0 to 41 wpi. The empty vector was used as the negative control. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 indicate significant differences in antibody titres or ROS levels, as appropriate, before infection and after infection at different weeks. Data are presented as means ± SDs (n = 2).
Figure 2.
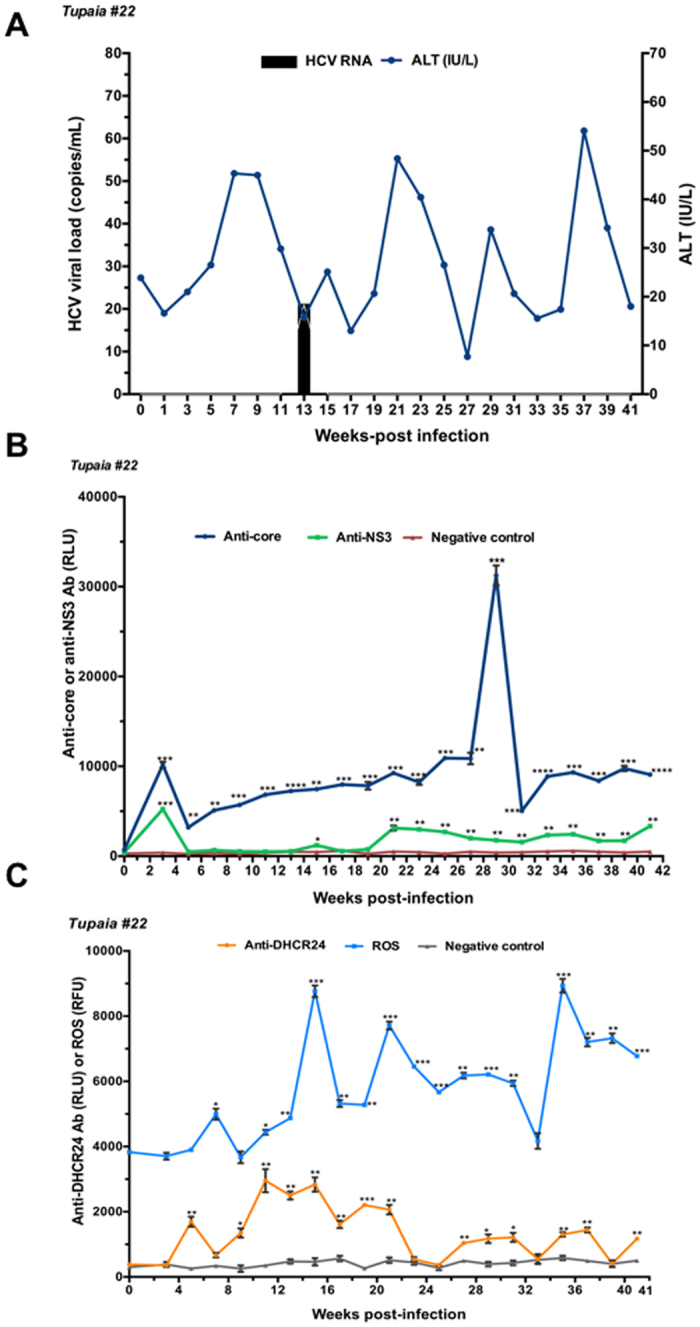
Response of tupaias to HCV1b infection. (A) ALT levels and viral loads in sera from tupaia #22 collected at 2-week intervals from 0 to 41 weeks postinfection (wpi). (B) Anti-HCV core and anti-nonstructural protein NS3 antibody titres in tupaia #22 at 2-week intervals from 0 to 41 wpi. (C) Anti-DHCR24 antibody titres and ROS levels in tupaia #22 at 2-week intervals from 0 to 41 wpi. The empty vector was used as the negative control. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 indicate significant differences in antibody titres or ROS levels, as appropriate, before infection and after infection at different weeks. Data are presented as means ± SDs (n = 2).
Figure 3.
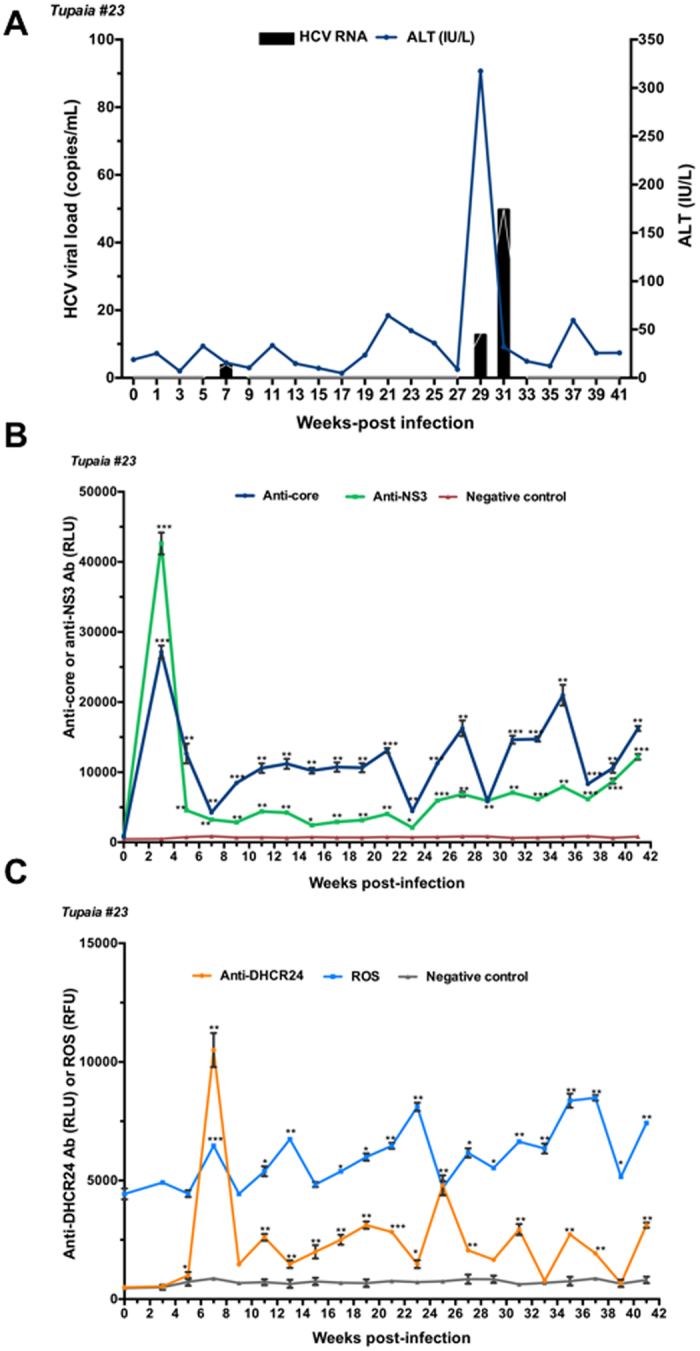
Response of tupaias to HCV4a infection. (A) ALT levels and viral loads in sera from tupaia #23 collected at 2-week intervals from 0 to 41 weeks postinfection (wpi). (B) Anti-HCV core and anti-nonstructural protein NS3 antibody titres in tupaia #23 at 2-week intervals from 0 to 41 wpi. (C) Anti-DHCR24 antibody titres and ROS levels in tupaia #23 at 2-week intervals from 0 to 41 wpi. The empty vector was used as the negative control. *p < 0.05, **p < 0.01, and ***p < 0.001 indicate significant differences in antibody titres or ROS levels, as appropriate, before infection and after infection at different weeks. Data are presented as means ± SDs (n = 2).
Figure 4.
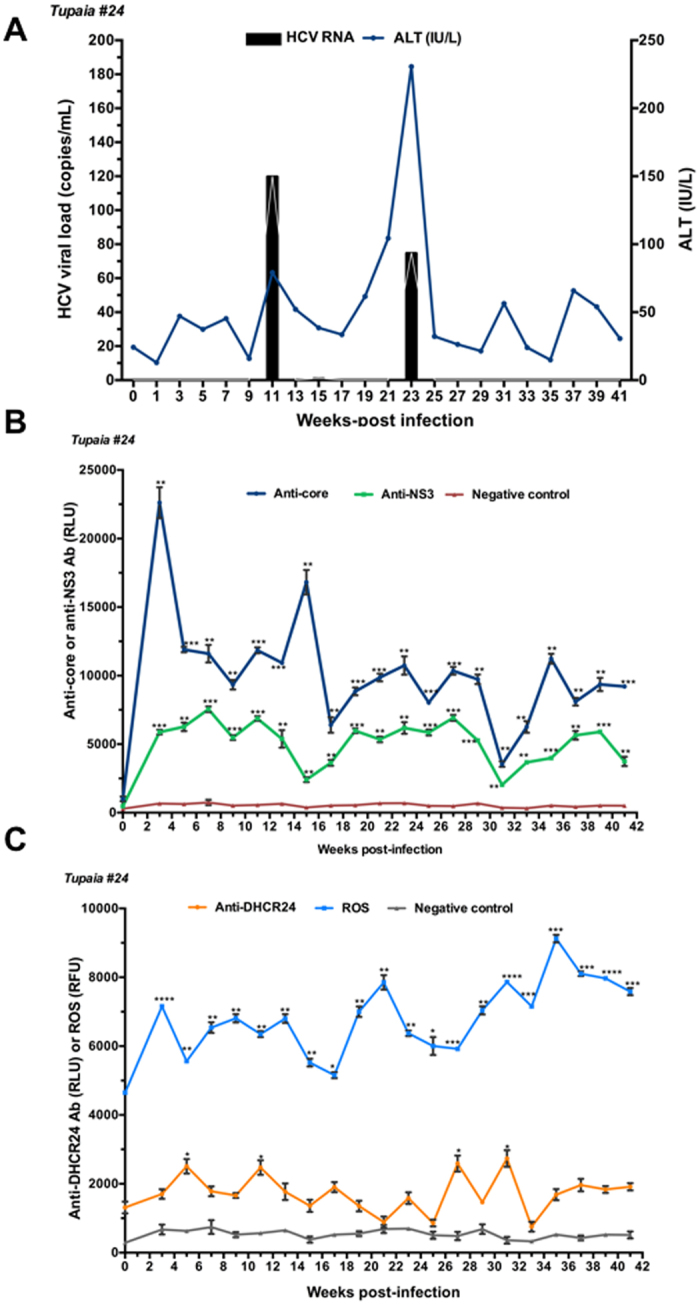
Response of tupaias to HCV2a infection. (A) ALT levels and viral loads in sera from tupaia #24 collected at 2-week intervals from 0 to 41 weeks postinfection (wpi). (B) Anti-HCV core and anti-nonstructural protein NS3 antibody titres in tupaia #24 at 2-week intervals from 0 to 41 wpi. (C) Anti-DHCR24 antibody titres and ROS levels in tupaia #24 at 2-week intervals from 0 to 41 wpi. The empty vector was used as the negative control. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 indicate significant differences in antibody titres or ROS levels, as appropriate, before infection and after infection at different weeks. Data are presented as means ± SDs (n = 2).
Measurement of anti-core and anti-NS3 antibody titres
To investigate the course of HCV-specific humoral immune responses in HCV-infected tupaias, we measured the levels of anti-core and anti-NS3 antibody titres in serum samples before infection (0 wpi) and every 2 weeks from 3 wpi to until the tupaias were sacrificed at 41 wpi. Despite fluctuations, significant increases in anti-core antibody levels were observed in all tupaias from 3 to 41 wpi (Figs 1B, 2B, 3B and 4B). In tupaia #21, significant increases in anti-NS3 antibody titres were observed at 3 and 25–35 wpi (Fig. 1B). In tupaia #22, significant increases in anti-NS3 antibody titres were observed at 3, 15, and 21–41 wpi (Fig. 2B). Significant increases in anti-NS3 antibodies were observed in tupaias #23 and #24 from 3 to 41 wpi (Figs 3B and 4B).
At 3 wpi, though virus could not be detected in sera, a significant increase in anti-core and anti-NS3 antibody production was observed in all HCV-infected tupaias. Anti-core antibody production was decreased after the initial increase at 3 wpi, which further increased during or around peak viral propagation and fluctuated (Figs 1B, 2B, 3B and 4B). The highest anti-core antibody production was observed in tupaia #22 at 29 wpi (Fig. 2B). Additionally, anti-NS3 antibody production sharply decreased after 3 wpi and fluctuated. The highest anti-NS3 antibody production was observed in tupaia #23 at 3 wpi (Fig. 3B). Overall, peak NS3 antibody production was observed during or after peak viral propagation. Anti-core and anti-NS3 antibody titres in uninfected controls and positive controls were measured to ensure the specificity of the assay (Fig. S1A,B).
Histological analysis of liver tissues from HCV-infected tupaias
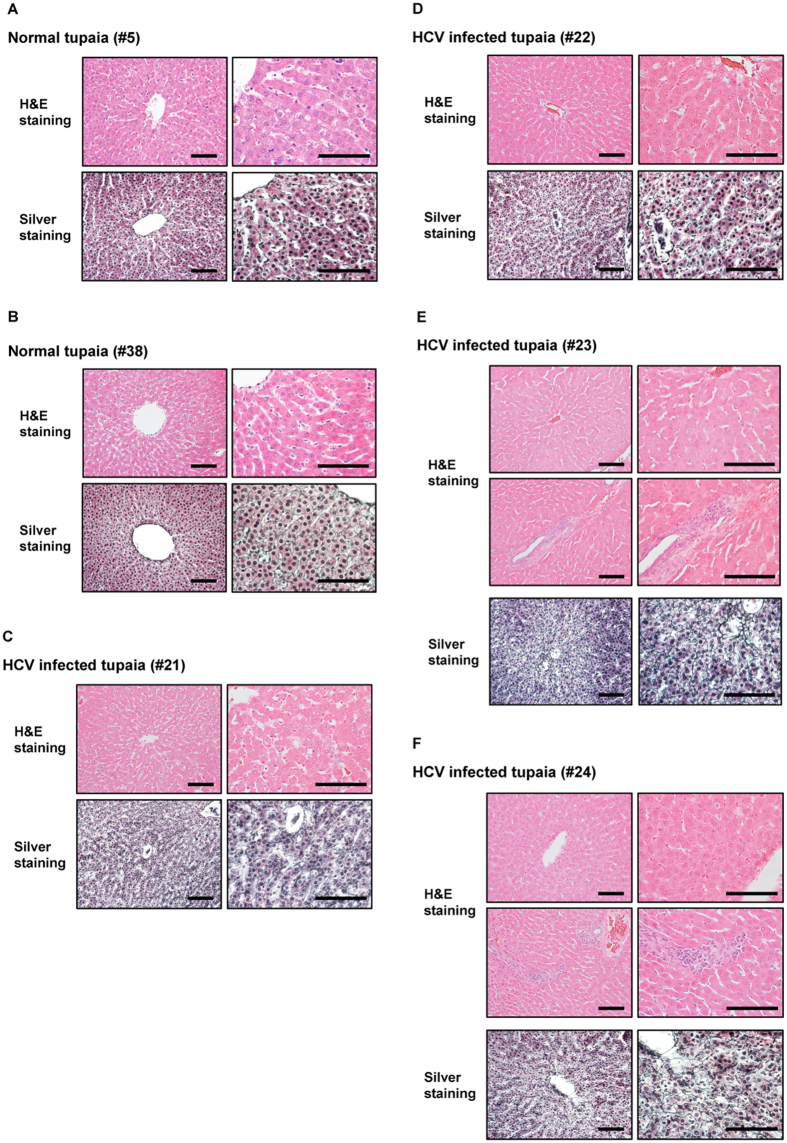
Histological analysis showed chronic hepatitis including abnormal architecture of liver cell cords, piecemeal necrosis, hepatocyte swelling, and lymphocytic infiltration into liver tissues of HCV-infected tupaias compared with that of normal tupaia liver tissues (Fig. 5). Silver staining showed increased fibres among hepatocytes (tupaias #21 and #22) (Fig. 5C, D) and thickened fibres (tupaias #23 and #24) (Fig. 5E, F), indicating the progression of fibrosis in HCV-infected tupaia livers.
Figure 5.
Histopathological analysis of liver tissues from normal and HCV-infected tupaias at 41 wpi. Histopathological analysis was performed with H&E or silver staining. H&E staining images (upper panel) and silver staining images (lower panel) of liver tissues from (A) Normal tupaia #5, (B) Normal tupaia #38, (C) HCV genotype 1a-infected tupaia #21, (D) HCV genotype 1b-infected tupaia #22, (E) HCV genotype 4a -infected tupaia #23, and (F) HCV genotype 2a -infected tupaia #24 have been shown. Black bars indicate 100 μm.
Measurement of ROS levels and anti-DHCR24 antibody titres
To investigate the effects of HCV infection on ROS generation, we measured ROS levels in sera from HCV-infected tupaias at 2-week intervals from 0 to 41 wpi. ROS levels were increased in all HCV-infected tupaias compared to ROS levels before infection (Figs 1C, 2C, 3C and 4C). ROS levels were also evaluated in mock-infected controls and with different concentrations of H2O2 as a positive control to ensure the appropriateness of the assay conditions (Fig. S1C).
To investigate whether increased ROS production induced anti-DHCR24 auto-antibody levels in HCV-infected tupaias, we measured the levels of serum anti-DHCR24 antibodies at 2-week intervals from 0 to 41 wpi. High levels of serum anti-DHCR24 antibodies were observed in all HCV-infected tupaias following ROS induction (Figs 1 C, 2 C, 3C and 4C). To ensure the specificity of the assay, anti-DHCR24 antibody titres in normal and positive controls were also evaluated (Fig. S1D).
Measurement of oxidative stress in the liver
To determine intrahepatic oxidative stress, we measured ROS levels in liver tissues at 41 wpi in normal and HCV-infected tupaias and found significantly higher ROS levels in HCV-infected tupaia liver tissues compared to normal controls (Fig. 6A). Additionally, at 41 wpi, we measured 8-OHdG levels in genomic DNA extracted from HCV-infected tupaia liver tissues to evaluate oxidative DNA damage. A significant increase in 8-OHdG levels was observed in all HCV-infected tupaias compared to normal controls (Fig. 6B).
Figure 6.
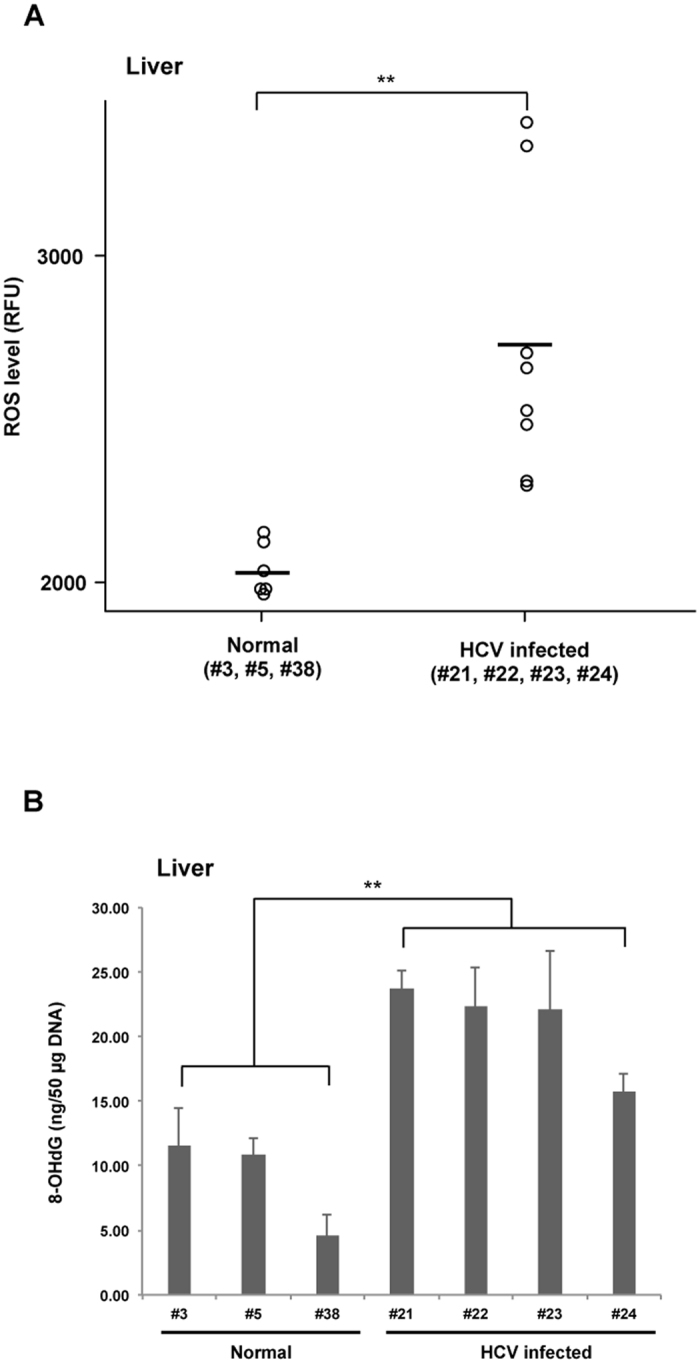
Intrahepatic ROS and 8-OHdG levels in normal and HCV-infected tupaias at 41 wpi. (A) ROS in liver lysates and (B) 8-OHdG levels in genomic DNA from HCV-infected tupaias were compared to those of normal controls. Horizontal bars in (A) indicate the mean value. **p < 0.01 indicates significant differences compared to normal controls.
Changes in TLR, NTCP, and cytokine expression in liver tissues
To investigate the mRNA expression patterns of TLRs, NTCP, and cytokines in liver tissues from HCV-infected tupaias, we isolated RNA from the livers of HCV-infected tupaias and measured the level of TLR3, TLR7, TLR8, NTCP, and cytokines (interferon [IFN]-β and interleukin [IL]-6) mRNAs at 41 wpi. Upregulation of TLR3, TLR7, and TLR8 was observed in the liver tissues of all HCV-infected tupaias (#21, #22, #23, and #24) compared to uninfected normal tupaias (#3, #5, and #38; Fig. 7). NTCP was significantly suppressed in tupaias #21 and #24 and significantly upregulated in tupaias #22 and #23 (Fig. 8A). Moreover, significant upregulation of IFN-β was observed in all tupaias, except tupaia #22 (Fig. 8B). IL-6 levels were significantly increased in tupaias #22 and #24 (Fig. 8C).
Figure 7.
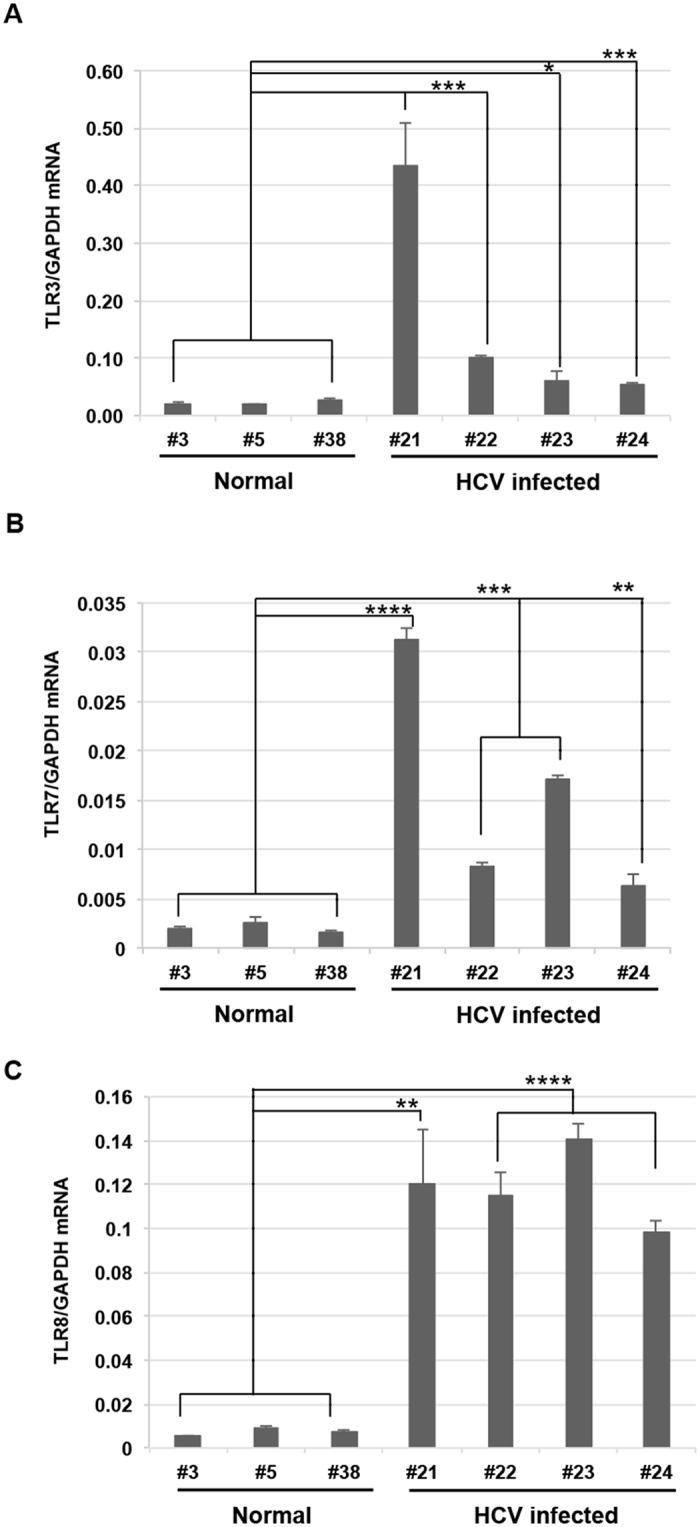
Changes in the expression of TLR mRNAs in HCV-infected tupaias at 41 wpi. (A) TLR3, (B) TLR7, and (C) TLR8 mRNA expression in livers of HCV-infected tupaias was measured by one-step qRT-PCR. Gene expression levels were normalized to the expression level of GAPDH mRNA. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 indicate significant differences compared to normal controls. Data are presented as means ± SDs (n = 3).
Figure 8.
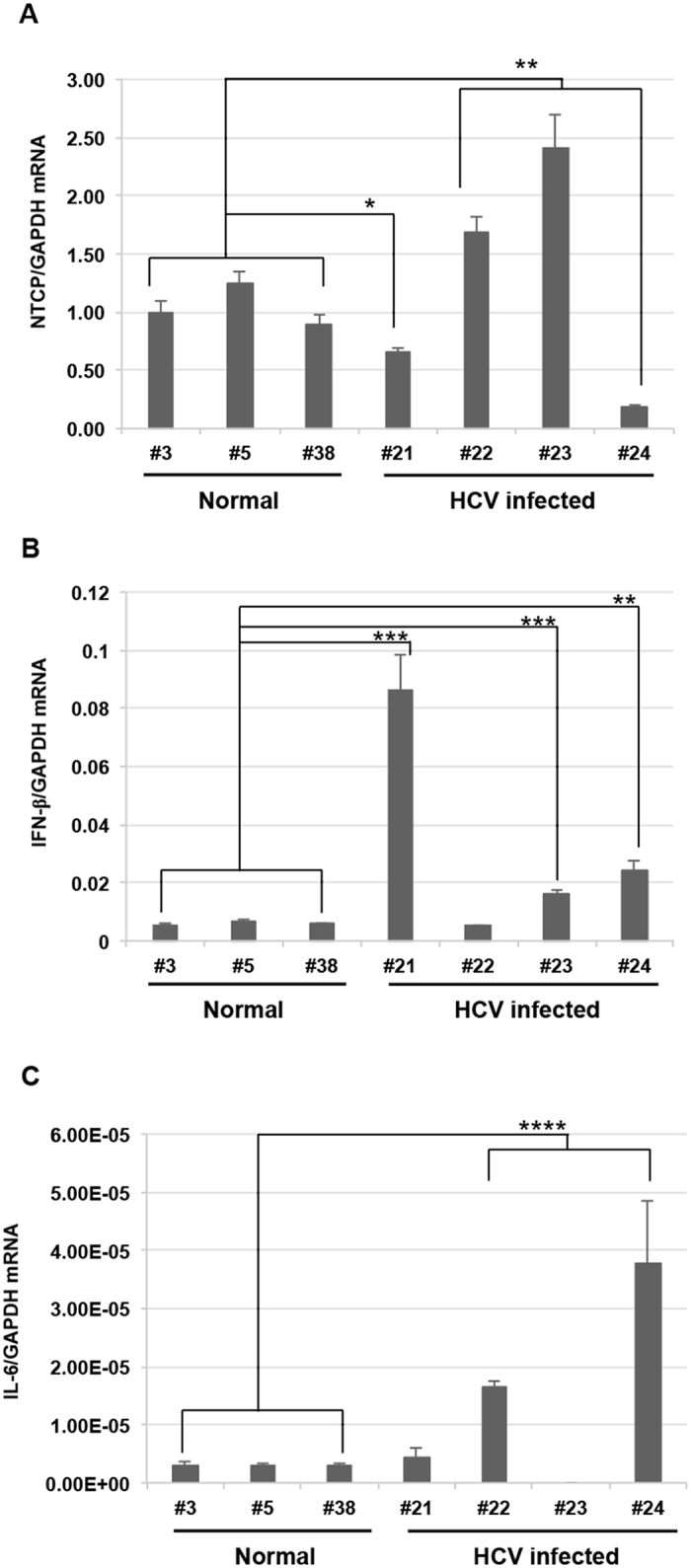
Changes in the expression of NTCP and cytokine mRNAs in HCV-infected tupaias at 41 wpi. (A) NTCP, (B) IFN-β, and (C) IL-6 mRNA expression in livers of HCV-infected tupaias was measured by one-step qRT-PCR. Gene expression levels were normalized to the expression level of GAPDH mRNA. * p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 indicate significant differences compared to normal controls. Data are presented as means ± SDs (n = 3).
Discussion
In this study, we demonstrated, for the first time, that ROS levels were higher in sera and liver tissues from HCV-infected tupaias than in that from uninfected tupaias. These data were consistent with previous findings of higher ROS levels in sera28 and liver tissues29, 30 of HCV-infected patients. We also observed increased 8-OHdG levels in livers from HCV-infected tupaias. These data were consistent with previous findings of higher 8-OHdG levels in livers from patients with chronic HCV infections31, 32. Previous studies have found that ROS induces the upregulation of DHCR2410, 33. Consistent with these results, we showed that anti-DHCR24 antibody production was increased with ROS generation during the course of HCV infection in tupaias. Thus, the tupaia HCV infection model supported the possibility that anti-DHCR24 antibodies could be a valuable marker to monitor HCV infection in vivo and should be consistent with previous evidence demonstrating that anti-DHCR24 auto-antibodies could be a useful biomarker for hepatitis C progression14.
In this study, we also characterized humoral and intrahepatic innate immune responses in tupaias infected with different HCV strains. Anti-NS3 and anti-core antibodies have been reported to be predominant in chronic HCV infections34. In fact, at 3 wpi, we found that all infected tupaias produced anti-core and anti-NS3 antibodies but were negative for serum HCV RNA. In our previous study, we detected HCV RNA only in the liver after 172 wpi26; therefore, HCV may replicate in the liver but not be released into the serum via an unknown mechanism. A longitudinal study in humans, with a median follow-up of 7 years, also reported cases in which core antibody was positive but HCV RNA was negative35. Additionally, the highest anti-core antibody levels in tupaia #22 were observed at 29 wpi, indicating robust viral replication occurred somewhere in the tupaia body. At 41 wpi, upon histological analysis of liver tissues of HCV-infected tupaias, lymphocytic infiltration was observed, indicating the occurrence of inflammation in the liver.
TLRs are important initiators of cytokine production, and the TLR signalling pathway serves as a link between innate and acquired immunity36, 37. TLRs act as PRRs for recognizing HCV PAMPs38. In our study, we found significant increases in TLR3, TLR7, and TLR8 mRNA levels in liver tissues in chronically HCV-infected tupaias with different genotypes. Our results highlighted that HCV could trigger innate immune responses in livers of chronically infected tupaias. Thus, our data confirmed previous studies reporting that HCV could be recognized by TLR3, TLR7, and TLR8 in cell cultures39, 40.
HCV mainly infects hepatocytes, and two PRRs (retinoic acid-inducible genes I [RIG-I] and TLR3) recognize HCV RNA to trigger production of multiple cytokines, including type I IFN. HCV develops strategies to evade these immune responses through several mechanisms; for example, the cleavage or relocalisation of IFN-β promoter stimulator 1 by HCV NS3/4 A protease inhibits RIG-I signalling41–43. Furthermore, NS3/4 A disrupts the TLR3 pathway by degradation of TIR-domain-containing adapter-inducing IFN-β43. HCV core inhibits IFN signalling by interfering with the Janus kinase/signal transducer and activator of transcription pathway44, and HCV NS5A blocks 2′5′ oligoadenylate synthetase and induces IL-845, 46. In livers from HCV-infected tupaias, TLR3 was induced in all four tupaias, whereas downstream IFN-β induction was observed in three tupaias (#21, #23, and #24).
Silencing of NTCP could inhibit HCV infection, whereas overexpression of NTCP could enhance HCV infection in cell culture8. NTCP can act as a regulator of antiviral immune responses in the liver. Moreover, NTCP is associated with the IFN response, and increased NTCP expression could suppress interferon-induced transmembrane protein (IFITM)-2 and IFITM-3 expression, and vice versa8. In our study, we found downregulation of NTCP but upregulation of IFN-β in tupaias #21 and #24. Significant upregulation of NTCP was observed in tupaias #22 and #23, and upregulation of IFN-β was only observed in tupaia #23. Thus, further studies are needed to explore the detailed mechanisms involved in NTCP-IFN interactions in HCV infection.
In conclusion, the tupaia infection model developed in this study was an effective approach to analyse the pathogenesis of HCV infection. ROS generation induced by HCV infection may be a trigger for the generation of anti-DHCR24 antibodies. Production of anti-core and anti-NS3 antibodies and intrahepatic innate immune responses upon HCV infection by alterations of TLR, NTCP, and cytokine expression highlights the potential applications of the tupaia infection model for the evaluation of HCV pathogenesis. Furthermore, tupaias could be a suitable small animal model for the evaluation of vaccines. The findings of this study provide novel insights into HCV pathogenesis and virus-host interactions.
Materials and Methods
Animals
The tupaias used in this study were obtained from the Laboratory Animal Center at the Kunming Institute of Zoology, Chinese Academy of Sciences (Kunming, China). This study was carried out following the Guidelines for Animal Experimentation of the Japanese Association for Laboratory Animal Science and the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All experimental protocols were approved by the institutional review boards of the regional ethics committees of Kagoshima University (VM15051 and VM13044).
Animals were individually housed in cages and fed a daily regimen of eggs, fruit, water, and dry mouse food. The animals were humanely handled in accordance with the Institutional Animal Care and Use Committee for Laboratory Animals.
Virus infection
All animals were found to be negative for HCV by quantitative real-time detection (RTD)-PCR before viral infection. Four adult tupaias (#21, #22, #23, and #24) were infected with HCV (genotypes 1a, 1b, 4a, and 2a [JFH1]), as described previously26, and three normal tupaias (#3, #5, and #38), which were not infected with HCV, were used as controls. Briefly, tupaias (#21, #22, #23, and #24) were infected at 12 months of age under anaesthesia induced by intramuscular injection of ketamine hydrochloride and atropine at 50 mg/kg body weight prior to virus inoculation and bleeding. Inocula derived from chimeric mice were introduced twice intraperitoneally at 1.5 × 108 copies/mL for genotype 1a, 107 copies/mL for genotypes 1b and 4a, and 1.2 × 108 copies/mL for genotype 2a (JFH1) for tupaias #21, #22, #23, and #24, respectively.
The infected animals were bled (0.5 mL) biweekly, and sera were separated, aliquoted, and immediately used or stored at −80 °C for further analysis. At 41 wpi, infected and control animals were sacrificed, and liver tissues were extracted. RNA from liver tissues was isolated using the acid guanidinium-phenol-chloroform extraction method and purified with an RNeasy Mini Kit (Qiagen, Valencia, CA, USA). For histological analysis, liver tissues were characterized by haematoxylin and eosin (H&E) or silver staining, as described previously47.
Measurement of ALT levels and viral load
Serum ALT level was determined using a Transnase Nissui kit (Nissui Pharmaceutical Co. Ltd., Tokyo, Japan); data were standardized and represented in IU/L. RNA was isolated from sera (50 µL) using SepaGene RV-R (Sanko Junyaku Co., Ltd., Tokyo, Japan). HCV RNA levels were quantified using qRT-PCR, as reported previously48.
Gaussia luciferase immunoprecipitation system (GLIPS) assays
To evaluate antibody levels, GLIPS assays were performed as reported previously49, with some modifications, using SureBeads Protein G magnetic beads (Bio-Rad, Hercules, CA, USA). Briefly, the HCV core gene (1b, nucleotides 341–759), HCV NS3 gene (1b, nucleotides 3991–4753), and tupaia DHCR24 gene (nucleotides 31–1599) were subcloned into the Gaussia luciferase vector (pGLIP vector)49 and expressed in HEK293 cells with Lipofectamine LTX and Plus Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. At 24–48 h after transfection, cells were lysed with Renilla Luciferase Assay Lysis Buffer (1×) and centrifuged at 20,380 × g for 5 min at 4 °C, and supernatants were collected and used immediately or stored at −80 °C. The luminescence of the crude extract was measured for 10 s using the Renilla luciferase assay system (Promega, Madison, WI, USA) on the GloMax-Multi + Detection System (Promega).
Immunoprecipitation assays were performed in duplicate in 96-well plates, and 100-fold diluted serum (1 µL equivalent) was used. To prepare beads, 25 μL of the Protein G magnetic bead suspension was added to each well and washed with 200 μL phosphate-buffered saline (PBS) + 0.1% Tween 20 (PBS-T) three times using a DynaMag-96 Side Skirted plate (Invitrogen). Supernatants were removed using an aspirator. Next, 100 μL of diluted serum from HCV-infected or mock-infected tupaias or known antibodies (used as a positive control) of different concentrations (10, 100, or 1000 ng) was added to the beads and incubated for 30 min at room temperature on a rotator. The beads were then washed three times with PBS-T, and lysates containing corresponding antigen or negative control antigens (empty vector) of 107 light units were added to each well. After 1-h incubation at room temperature on a rotator, beads were washed with PBS-T at least three times. Finally, the beads were transferred with 50 μL of PBS into plates to be read, and 50 μL of Renilla substrate-buffer mixture (1×) was added to each well using an injector system during luminescence measurement, as stated above. The antibody titre was expressed as relative light units (RLU).
Quantification of ROS levels
Total free radicals in serum or liver samples from HCV-infected and uninfected control tupaias were measured using an OxiSelect In Vitro ROS/RNS Assay Kit (Cell Biolabs, Inc., San Diego, CA, USA) according to the manufacturer’s protocol. Each reaction was performed in duplicate. Different concentrations of H2O2 were used in each reaction plate as positive controls to confirm the test conditions. Sera were diluted in PBS (1:200), and 50 μL of the diluted samples was collected into each well for the assay. For tissue lysates, 30 mg of liver tissues was homogenised using TissueLyser LT (Qiagen) in 1 mL PBS and centrifuged at 10,000 × g for 5 min. The supernatant was diluted in PBS (1:100), and 50 μL of the diluted samples was used in each well for the assay. The stabilized, highly reactive 2′,7′-dichlorodihydrofluorescin (DCFH) form was oxidized by ROS in the samples, and a catalyst was added to accelerate the reaction. The fluorescence of the oxidized DCFH to 2′,7′-dichlorofluorescein (DCF) was proportional to the concentration of ROS in the samples. Green fluorescence was monitored at 525 nm excitation/580–640 nm emission using the GloMax-Multi + Detection System (Promega). The ROS level was expressed as relative fluorescence units (RFU).
Determination of 8-OHdG levels in genomic DNA from liver tissues
DNA was extracted from frozen liver tissues using the phenol-chloroform extraction method. The level of 8-OHdG in extracted DNA was determined using the OxiSelect Oxidative DNA Damage ELISA Kit (Cell Biolabs, Inc.) according to the manufacturer’s protocol with some modifications. Briefly, 100 µg DNA was digested with 6 units of nuclease P1 for 2 h at 37 °C in a final concentration of 200 nM sodium acetate (pH 5.5). Then, the DNA reaction mixture was subjected to further digestion with 2 units of alkaline phosphatase for 1 h at 37 °C in a final concentration of 1M Tris (pH 7.5). Finally, the reaction mixture was centrifuged for 5 min at 6,000 × g and the supernatant was used for 8-OHdG ELISA assay. Each reaction was performed in duplicate. The 8-OHdG content in unknown samples was determined by comparison with predetermined 8-OHdG standard curves.
Gene expression analysis by qRT-PCR
Tupaia TLR and cytokine mRNA expression levels were measured by qRT-PCR, as described previously50. Primers used to detect tupaia NTCP gene [GenBank accession number: JQ608471] were as follows: forward primer (5′-TGTGGGCAAGAGCATCATGT-3′) and reverse primer (5′-CACTGTGCATTGAGGCGAAA-3′), and the reaction conditions were similar to that used for TLR8 50. Tupaia GAPDH was used as an endogenous control for normalization of the results.
Statistical analysis
To analyse the statistical significance of antibody titres and ROS levels in animals before and after infection and to determine gene expression differences between uninfected controls and virus-infected individuals, unpaired t-tests were conducted using GraphPad software (http://graphpad.com/quickcalcs/ttest1/). Differences with P values of less than 0.05 were considered significant. All tests were two-sided.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the Japan Agency for Medical Research and Development (AMED, J15000952, 16fk0210108s0201), the Tokyo Metropolitan Government; the Ministry of Health, Labour and Welfare of Japan; and the Ministry of Education, Culture, Sports, Science and Technology of Japan (25.03079). S.E. is supported by a Japan Society for the Promotion of Science (JSPS) Fellowship for Foreign Researchers (P13079). We would like to thank Dr Horie for supplying the pGLIP vector and Mr Haraguchi, Ms Matsuyama, Mr Kanda, Ms Nakagawa, Mr Okuya, Mr Ueno, Mr Handa, Dr Ozawa, and Dr Chimene for their assistance with animal care.
Author Contributions
M.E.H.K., S.E., H.C., K.R., and B.K. performed experiments; A.M. supervised GLIPS assays; N.M. performed pathological analyses; T.S., Y.H., and T.H. performed silver staining and updated H&E analysis; and M.E.H.K., S.E., M.K., and K.T.-K. designed and performed experiments and wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Mohammad Enamul Hoque Kayesh and Sayeh Ezzikouri contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-10329-7
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Michinori Kohara, Email: kohara-mc@igakuken.or.jp.
Kyoko Tsukiyama-Kohara, Email: kkohara@vet.kagoshima-u.ac.jp.
References
- 1.Lavanchy D. The global burden of hepatitis C. Liver international: official journal of the International Association for the Study of the Liver. 2009;29(Suppl 1):74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 2.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 3.Horner SM, Gale M., Jr. Regulation of hepatic innate immunity by hepatitis C virus. Nature medicine. 2013;19:879–888. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thimme R, Binder M, Bartenschlager R. Failure of innate and adaptive immune responses in controlling hepatitis C virus infection. FEMS microbiology reviews. 2012;36:663–683. doi: 10.1111/j.1574-6976.2011.00319.x. [DOI] [PubMed] [Google Scholar]
- 5.Cashman SB, Marsden BD, Dustin LB. The Humoral Immune Response to HCV: Understanding is Key to Vaccine Development. Frontiers in immunology. 2014;5 doi: 10.3389/fimmu.2014.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 7.Claro da Silva T, Polli JE, Swaan PW. The solute carrier family 10 (SLC10): beyond bile acid transport. Molecular aspects of medicine. 2013;34:252–269. doi: 10.1016/j.mam.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verrier ER, et al. Solute Carrier NTCP Regulates Innate Antiviral Immune Responses Targeting Hepatitis C Virus Infection of Hepatocytes. Cell reports. 2016;17:1357–1368. doi: 10.1016/j.celrep.2016.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korenaga M, et al. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. The Journal of biological chemistry. 2005;280:37481–37488. doi: 10.1074/jbc.M506412200. [DOI] [PubMed] [Google Scholar]
- 10.Tsukiyama-Kohara K. Role of oxidative stress in hepatocarcinogenesis induced by hepatitis C virus. International journal of molecular sciences. 2012;13:15271–15278. doi: 10.3390/ijms131115271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valavanidis A, Vlachogianni T, Fiotakis C. 8-hydroxy-2′-deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. Journal of environmental science and health. Part C, Environmental carcinogenesis & ecotoxicology reviews. 2009;27:120–139. doi: 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- 12.Waterham HR, et al. Mutations in the 3beta-hydroxysterol Delta24-reductase gene cause desmosterolosis, an autosomal recessive disorder of cholesterol biosynthesis. American journal of human genetics. 2001;69:685–694. doi: 10.1086/323473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takano T, et al. Augmentation of DHCR24 expression by hepatitis C virus infection facilitates viral replication in hepatocytes. Journal of hepatology. 2011;55:512–521. doi: 10.1016/j.jhep.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Ezzikouri S, et al. Serum DHCR24 Auto-antibody as a new Biomarker for Progression of Hepatitis C. EBioMedicine. 2015;2:604–612. doi: 10.1016/j.ebiom.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barth H. Hepatitis C virus: Is it time to say goodbye yet? Perspectives and challenges for the next decade. World journal of hepatology. 2015;7:725–737. doi: 10.4254/wjh.v7.i5.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdelwahab SF. Cellular immune response to hepatitis-C-virus in subjects without viremia or seroconversion: is it important? Infectious agents and cancer. 2016;11 doi: 10.1186/s13027-016-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chinchilla-Lopez P, Qi X, Yoshida EM, Mendez-Sanchez N. The Direct-Acting Antivirals for Hepatitis C Virus and the Risk for Hepatocellular Carcinoma. Annals of hepatology. 2017;16:328–330. doi: 10.5604/16652681.1235473. [DOI] [PubMed] [Google Scholar]
- 18.Mercer DF, et al. Hepatitis C virus replication in mice with chimeric human livers. Nature medicine. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 19.Dorner M, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsukiyama-Kohara K, Kohara M. Tupaia belangeri as an experimental animal model for viral infection. Experimental animals/Japanese Association for Laboratory Animal Science. 2014;63:367–374. doi: 10.1538/expanim.14-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends in genetics: TIG. 2007;23:158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Fan Y, et al. Genome of the Chinese tree shrew. Nature communications. 2013;4 doi: 10.1038/ncomms2416. [DOI] [PubMed] [Google Scholar]
- 23.Yang C, et al. Chronic hepatitis B virus infection and occurrence of hepatocellular carcinoma in tree shrews (Tupaia belangeri chinensis) Virology journal. 2015;12 doi: 10.1186/s12985-015-0256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanada T, et al. Property of hepatitis B virus replication in Tupaia belangeri hepatocytes. Biochemical and biophysical research communications. 2016;469:229–235. doi: 10.1016/j.bbrc.2015.11.121. [DOI] [PubMed] [Google Scholar]
- 25.Xu X, Chen H, Cao X, Ben K. Efficient infection of tree shrew (Tupaia belangeri) with hepatitis C virus grown in cell culture or from patient plasma. The Journal of general virology. 2007;88:2504–2512. doi: 10.1099/vir.0.82878-0. [DOI] [PubMed] [Google Scholar]
- 26.Amako Y, et al. Pathogenesis of hepatitis C virus infection in Tupaia belangeri. Journal of virology. 2010;84:303–311. doi: 10.1128/JVI.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu W, et al. Characterization of hepatitis E virus infection in tree shrew (Tupaia belangeri chinensis) BMC infectious diseases. 2016;16 doi: 10.1186/s12879-016-1418-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Maria N, et al. Association between reactive oxygen species and disease activity in chronic hepatitis C. Free radical biology & medicine. 1996;21:291–295. doi: 10.1016/0891-5849(96)00044-5. [DOI] [PubMed] [Google Scholar]
- 29.Valgimigli L, Valgimigli M, Gaiani S, Pedulli GF, Bolondi L. Measurement of oxidative stress in human liver by EPR spin-probe technique. Free radical research. 2000;33:167–178. doi: 10.1080/10715760000300721. [DOI] [PubMed] [Google Scholar]
- 30.Valgimigli M, et al. Oxidative stress EPR measurement in human liver by radical-probe technique. Correlation with etiology, histology and cell proliferation. Free radical research. 2002;36:939–948. doi: 10.1080/107156021000006653. [DOI] [PubMed] [Google Scholar]
- 31.Mahmood S, et al. Immunohistochemical evaluation of oxidative stress markers in chronic hepatitis C. Antioxidants & redox signaling. 2004;6:19–24. doi: 10.1089/152308604771978318. [DOI] [PubMed] [Google Scholar]
- 32.Fujita N, Kaito M, Tanaka H, Horiike S, Adachi Y. Reduction of serum HCV RNA titer by bezafibrate therapy in patients with chronic hepatitis C. The American journal of gastroenterology. 2004;99 doi: 10.1111/j.1572-0241.2004.40695_3.x. [DOI] [PubMed] [Google Scholar]
- 33.Saito M, Kohara M, Tsukiyama-Kohara K. Hepatitis C virus promotes expression of the 3beta-hydroxysterol delta24-reductase through Sp1. Journal of medical virology. 2012;84:733–746. doi: 10.1002/jmv.23250. [DOI] [PubMed] [Google Scholar]
- 34.Ishii K, et al. [Characterization of antibodies against core, NS3, NS4, NS5 region of hepatitis C virus in patients with hepatitis C] Rinsho byori. The Japanese journal of clinical pathology. 1997;45:1156–1162. [PubMed] [Google Scholar]
- 35.Hoare M, et al. Histological changes in HCV antibody-positive, HCV RNA-negative subjects suggest persistent virus infection. Hepatology. 2008;48:1737–1745. doi: 10.1002/hep.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature immunology. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 37.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Yang DR, Zhu HZ. Hepatitis C virus and antiviral innate immunity: who wins at tug-of-war? World journal of gastroenterology: WJG. 2015;21:3786–3800. doi: 10.3748/wjg.v21.i13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metz P, Reuter A, Bender S, Bartenschlager R. Interferon-stimulated genes and their role in controlling hepatitis C virus. Journal of hepatology. 2013;59:1331–1341. doi: 10.1016/j.jhep.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 40.Lee J, et al. TNF-alpha Induced by Hepatitis C Virus via TLR7 and TLR8 in Hepatocytes Supports Interferon Signaling via an Autocrine Mechanism. PLoS pathogens. 2015;11 doi: 10.1371/journal.ppat.1004937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foy E, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sklan EH, Charuworn P, Pang PS, Glenn JS. Mechanisms of HCV survival in the host. Nature reviews. Gastroenterology & hepatology. 2009;6:217–227. doi: 10.1038/nrgastro.2009.32. [DOI] [PubMed] [Google Scholar]
- 43.Li K, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bode JG, et al. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2003;17:488–490. doi: 10.1096/fj.02-0664fje. [DOI] [PubMed] [Google Scholar]
- 45.Polyak SJ, et al. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. Journal of virology. 2001;75:6095–6106. doi: 10.1128/JVI.75.13.6095-6106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. The Journal of clinical investigation. 2009;119:1745–1754. doi: 10.1172/JCI39133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekiguchi S, et al. Immunization with a recombinant vaccinia virus that encodes nonstructural proteins of the hepatitis C virus suppresses viral protein levels in mouse liver. PloS one. 2012;7 doi: 10.1371/journal.pone.0051656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeuchi T, et al. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology. 1999;116:636–642. doi: 10.1016/S0016-5085(99)70185-X. [DOI] [PubMed] [Google Scholar]
- 49.Matsuu A, et al. Genetic and serological surveillance for non-primate hepacivirus in horses in Japan. Veterinary microbiology. 2015;179:219–227. doi: 10.1016/j.vetmic.2015.05.028. [DOI] [PubMed] [Google Scholar]
- 50.Kayesh MEH, et al. Susceptibility and initial immune response of Tupaia belangeri cells to dengue virus infection. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2017;51:203–210. doi: 10.1016/j.meegid.2017.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.