Abstract
Children suffering from autism have been reported to have low bone mineral density and increased risk for fracture, yet the cellular origin of the bone phenotype remains unknown. Here we have utilized a mouse model of autism that duplicates 6.3 Mb region of chromosome 7 (Dp/+) corresponding to a region of chromosome 15q11-13, duplication of which is recurrent in humans to characterize the bone phenotype. Paternally inherited Dp/+ (patDp/+) mice showed expected increases in the gene expression in bone, normal postnatal growth and body weight acquisition compared to the littermate controls. Four weeks-old patDp/+ mice develop a low bone mass phenotype in the appendicular but not the axial skeleton compared to the littermate controls. This low bone mass in the mutant mice was secondary to a decrease in the number of osteoblasts and bone formation rate while the osteoclasts remained relatively unaffected. Further in vitro cell culture experiments and gene expression analysis revealed a major defect in the proliferation, differentiation and mineralization abilities of patDp/+ osteoblasts while osteoclast differentiation remained unchanged compared to controls. This study therefore characterizes the structural and cellular bone phenotype in a mouse model of autism that can be further utilized to investigate therapeutic avenues to treat bone fractures in children with autism.
Introduction
The skeleton is an organ system in the body that performs many vital functions1. On one hand, it protects many soft organs such as brain and heart, and on the other it serves as an endocrine organ to influence function of other organs such as pancreas, testis and brain1, 2. Major component of the mature skeleton are bones, which are continuously remodelled, through a process known as bone remodelling, throughout life to perform these vital functions. Bone remodelling is carried out by two cell types, osteoblasts and osteoclasts3, 4. Osteoblasts are the mono-nucleated cells of mesenchymal origin that deposit new bone matrix while the osteoclasts are multinucleated cells of hematopoietic origin that degrade the old bone matrix5–7. These two cells function as a bone remodelling unit to replace the old bone matrix with a new one to maintain the mechanical strength of the skeleton. Bone remodelling is tightly regulated via a complex process, which is only now beginning to be understood5–8. It is affected by variety of cues, which are environmental, autocrine/endocrine, and genetic in nature. An imbalance in these cues leads to bone diseases characterized by a low bone density and an increased risk of fractures1–8. Many diseases with complex aetiology have long been known to be associated with low bone density and increased risk of fractures yet the underlying mechanisms remain unclear.
Autism is a neurodevelopmental disorder characterized by impaired language skills and communication, as well as repetitive and stereotypic patterns of behavior9. Autism is one of the most heritable psychiatric disorder suggesting that genetic factors play a large role in its development or manifestation of symptoms10, 11. Psychiatric symptoms in autism appear during early life, as early as 6 months of age, and persist throughout life9. Besides the psychiatric problems children with autism are often found to have low bone density and increased risk of fractures12–15. The fact that like autism, genetics and heritability plays an important role in the establishment and maintenance of bone density suggests that common factors could be involved in regulating bone density and autism16. However, besides genetic factors environment also plays an important role in regulating different phenotypes in autism spectrum disorders viz., physical exercise and nutrition. In the past, several knockout mice have been created to investigate different phenotypes in autism, but the molecular mechanism responsible for the pathophysiology of autism and its associated metabolic diseases is far from complete17–25. The above facts underscore that we need to increase our knowledge of bone development in autism in an animal model so that we can optimize dysregulated bone homeostasis in children with autism.
A genetic mouse model had been created by us earlier to investigate the psychiatric symptoms in autism following duplication of a region on the chromosome 15 i.e., 15q11.2-13.126. Duplication of this chromosomal region is the recurrent cytogenetic aberration associated with autism, which occurs in up to 5% of patients9–11, 27–33. On the basis of conserved human/mouse linkage, a mouse model with a 6.3 Megabase (Mb) duplication of mouse chromosome 7, mirroring the human chromosome 15q11.2-13.1 duplication was created through sequential tragetting of the embryonic stem cells26. The paternally inherited Dp/+ mouse genetic model (referrred in this study as patDp/+ from now on) faithfully recapitulates several behavioural phenotypes observed in the autism patients and provides us an opportunity to characterize the bone phenotype in these mice. Here we report that patDp/+ mice display a low bone mass phentopye and charatcerise the architectural and cellular bone parameters in these mutant mice.
Results
Generation and analysis of mice with paternally duplicated autism loci on chromosome 7 (patDp/+ mice)
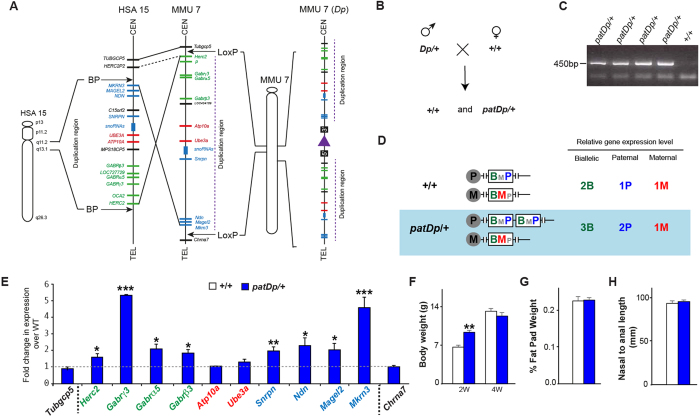
The duplication of the region of human chromosome 15q11-13 was modelled in mouse through interstitial duplication of mouse chromosome 7 corresponding to the region between common breakpoints in human chromosome 15q11-13 by chromosomal engineering (Fig. 1A) as described previously26. To generate the experimental cohort of mice for analysis male patDp/+ mice were breed with female +/+ C57Bl6/N mice and genotyped using PCR on the genomic DNA, these mice are labelled as patDp/+ mice from here on (Fig. 1B,C).
Figure 1.
Generation and analysis of a duplicated region of chromosome 7 (Dp/+) in mice. (A) Schematic representation of the genomic regions in the human and mouse genomes showing the details of conserved linkage in human 15q11-13 and mouse chromosome 7. The duplicated region is marked by the insertional site for the 2 LoxP (Adapted from Nakatani et al., Cell 2009). (B) Schematic represetation of the breeding strategy used to generate patDp/+ and corresponding control WT (+/+) mice. (C) Genotyping PCR analysis on the tail genomic DNA to identify the +/+ and patDp/+ mice. The gel is the full length gel and lanes have not been cropped or stitched. (D) Expected expression levels in the +/+ and patDp/+ mice for the genes present in the duplicated region. (E) qPCR analysis of genes in the duplicated region and 2 genes that lie outside of the duplicated region in the long bone from +/+ and patDp/+ mice. Paternally expressed genes are marked with blue, maternally expressed with red and nonimprinting genes are marked with green. Dotted lines represent boundaries of the chromosomal rearrangement. (F–H) Body weight (F) % Fat pad weight (G) and nasal to anal length (H) analysis in +/+ and patDp/+ mice. n = 8–10 mice per group.
The 6.3 Mb duplication region in the patDp/+ mice includes the region of genomic imprinting and has the following genes: Herc2, Gabrγ3, Gabrα5, Gabrβ3, Atp10a, Ube3a, Snrpn, Ndn, Magel2 and Mkrn3. Because the gene expression is expected to vary depending on the tissue type, imprinting status and mode of inheritance as shown in Fig. 1D, relative gene expression was assessed by quantitative RT-PCR in the bones from patDp/+ mice relative to WT mice (Fig. 1E). Moreover, the expression changes in these genes in the patDp/+ mice has been assessed earlier in the brain and therefore it was imperative for us to establish their expression levels in the bones of these mice to further investigate their possible involvement in the cellular phenotype in the bone. In the bone, Herc2, Gabrγ3, Gabrα5, Gabrβ3, Snrpn, Ndn, Magel2 and Mkrn3 genes in the duplicated region were increased in the expression whereas Atp10a and Ube3a did not show a significant change in the expression (Fig. 1E). As expected Tubgcp5 and Chrna7, which are located outside the duplicated region, did not show any significant difference in their expression levels (Fig. 1E). patDp/+ mice bred normally and were fertile and as reported previously26 showed normal body weight compared to wild-type (WT) mice (Fig. 1F–H). *P < 0.05, **p < 0.01, ***p < 0.001.
Analysis of bone volume and architecture in patDp/+ mice
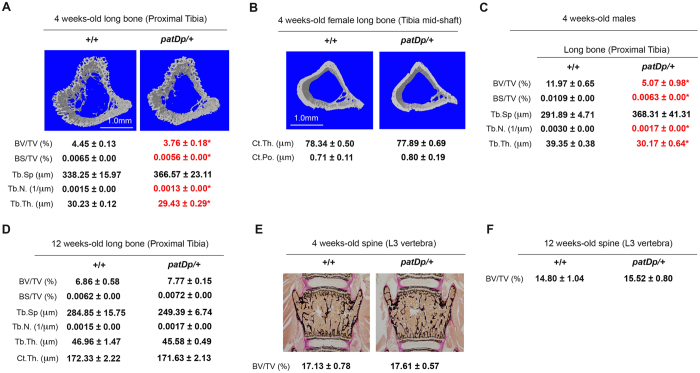
Having confirmed that the patDp/+ mice have the expected expression changes in bone for the genes present in the duplicated region we next analyzed whether significant differences in specific bone parameters exist between WT and patDp/+ mice. As the bone composition (cortical versus trabecular) in mammals differs depending on the skeletal site analyzed we decided to analyze two different skeletal sites, long bone and vertebra. We selected tibia as a representative long bone for μCT analysis and analyzed them at two different locations, proximal region for changes in the trabecular parameters and mid-shaft for changes in the cortical parameters. For trabecular bone analysis, we selected lumbar 4 vertebrae as a representative trabecular bone rich site. We first performed analysis on the tibia bones by μCT. As shown in Fig. 2A, the proximal tibias of 4 weeks-old female patDp/+ mice exhibited significant decreases in the bone volume over total volume % (BV/TV%), bone surface per total volume % (BS/TV%), trabecular number (Tb.N.) and thickness (Tb.Th.) and a tendency to have higher trabecular separation (Tb.Sp.) relative to WT littermate controls. The differences in these parameters ranged between 2.7% and 17%. We next assessed changes in cortical parameters, thickness and porosity, at tibia mid-shaft. Cortical thickness tended to be on the lower side (ns) and cortical porosity on the higher side (ns) in the patDp/+ relative to WT littermates (Fig. 2B). μCT analysis of 4 weeks-old male patDp/+ mice showed a compromised bone architecture like the female mice with significant decreases in the BV/TV%, BS/TV%, Tb.N. and Tb.Th. (Fig. 2C). The mutant animals also had a tendency towards higher Tb.Sp. (Fig. 2C). In contrast to the significant changes in the bone phenotype in the 4 weeks-old mice, 12 weeks-old patDp/+ mice did not display any changes in the bone architecture in tibia (Fig. 2D). We next analyzed vertebra through classical non-demineralized histology after methyl methacrylate-embedding, a standard technique used to assess vertebral histopathology. In contrast to the compromised bone structure in the tibia, analysis of vertebra collected from 4 weeks- and 12 weeks-old patDp/+ mice through histology and histomorphometry did not reveal any significant difference in the bone volume over total volume % between WT and mutant mice indicating that bone mass in the vertebral column is not affected in the patDp/+ mice (Fig. 2E,F).
Figure 2.
Low bone mass in the long bone but not spine of 4 week-old patDp/+ mice. (A) μCT analysis of proximal tibia in 4 week-old +/+ and patDp/+ mice. (B) μCT analysis of tibial mid-shaft in 4 weeks-old +/+ and patDp/+ mice. (C) μCT analysis of proximal tibia in 4 weeks-old +/+ and patDp/+ male mice. (D) μCT analysis of proximal tibia in 12 weeks-old +/+ and patDp/+ mice. (E,F) Von Kossa staining of histological sections of lumber 3 vertebra in 4 weeks (E)- and 12 weeks-old (F) +/+ and patDp/+ mice. *P < 0.05. Values are mean ± SEM. n = 8–10 mice were utilized per group. All mice were females except in the panel C where male mice have been used. Abbreviations used are Bone volume over total volume % (BV/TV%); Bone surface over total volume % (BS/TV%); Trabecular separation (Tb.Sp); Trabecular number (Tb.N.); Trabceular thickness (Tb.Th.); Cortical thickness (Ct.Th.) and Cortical porosity (Ct.Po.).
Histomorphometric analysis of 4 weeks-old WT and patDp7/+ female mice
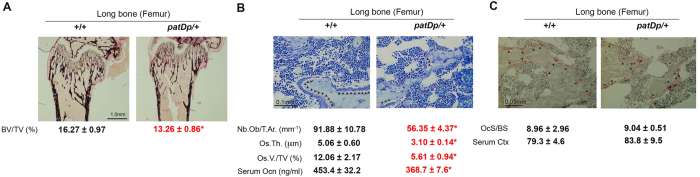
Because we observed a compromised bone volume, surface and trabecular parameters in the patDp/+ mice in the long bones, it was of interest to further examine this bone phenotype at the cellular level. Histological and histomorphometric analysis was performed on femurs at the femoro-tibial joint of WT and patDp/+ mice at 4 weeks of age. This analysis, as shown in Fig. 3A, clearly demonstrates that there is a significant decrease in the trabecular bone of patDp/+ mice relative to WT littermates. In light of the striking defects observed in the histological analysis of femurs from female patDp/+ mice with regard to bone mass, we next determined if this defect was at least in part due to differences in the number of osteoblasts or osteoclasts lining the bone surface of these animals (Fig. 3B,C).
Figure 3.
Isolated osteoblast defect contributes to the low bone mass phenotype in the long bone of patDp7/+ mice. (A) Von Kossa staining of histological section and bone volume over total volume % (BV/TV%) analysis in +/+ and patDp/+ mice. (B) Analysis of osteoblast numbers, osteoid thickness and osteoid volume per total volume % in +/+ and patDp/+ mice. (C) Analysis of osteoclast surface per bone surface and Ctx levels in 4 week-old +/+ and patDp/+ mice. *P < 0.05. Arrow heads indicate location of respective cells in the sections. Values are mean ± SEM. n = 8–10 mice per group. All mice were females.
We first analyzed changes in the osteoblast numbers and function. As shown in Fig. 3B, there was a significant decline in the number of osteoblasts per trabecular area. This accounted for a significant decrease in the osteoid parameters as well. Osteoid is the protein matrix secreted by osteoblasts. Both osteoid thickness and osteoid volume/total volume % was significantly decreased in the patDp/+ mice relative to WT littermates (Fig. 3B). This decrease in the osteoblast numbers and function was also reflected in the serum marker of osteoblast function, osteocalcin, which was decreased by 19% in the patDp/+ mice (Fig. 3B). In contrast to the observed defect in the osteoblast parameters there was no significant difference in the osteoclast surface/bone surface in the patDp/+ mice relative to WT littermates (Fig. 3C). Accordingly, serum marker of bone resorption, collagen type 1 cross-linked C-telopeptide (Ctx) was not altered in the patDp/+ mice relative to WT littermates (Fig. 3C).
Overall, these findings suggest that female patDp/+ mice are osteopenic mainly due to a significant decrease in osteoblast numbers and function and not likely due to a large difference in the osteoclast parameters.
Cell autonomous decrease in osteoblast parameters in patDp/+ mice
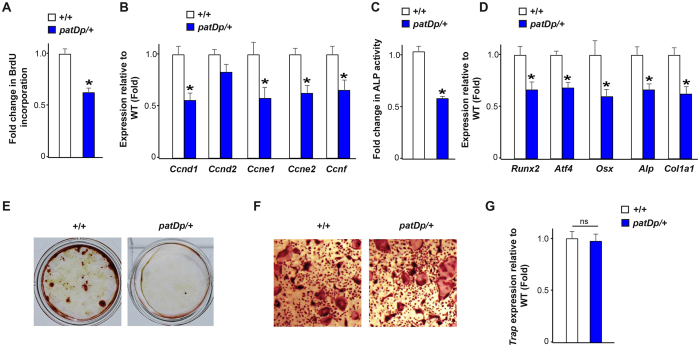
The decrease in osteoblast numbers and functions observed in the patDp/+ mice in vivo through skeletal histology and histomorphometry raises the question whether osteoblasts in these mice have a cell autonomous defect. This osteoblast defect could be either in the ability of these mutant osteoblasts to proliferate, differentiate or deposit the mineral matrix or all of the aforementioned parameters. To test this contention, we next utilized primary osteoblasts isolated from calvarias of the WT and patDp/+ mice and tested them for proliferation, differentiation and mineralization using in vitro assays (Fig. 4).
Figure 4.
Osteoblast proliferation, differentiation and function is compromised in patDp/+ mice. (A) BrdU incorporation assay in +/+ and patDp/+ calvarial osteoblasts. (B) Real-time PCR analysis of Cyclins in WT and patDp/+ calvarial osteoblasts. (C) Alkaline phosphatase activity assay in +/+ and patDp /+ calvarial osteoblasts. (D) Real-time PCR analysis of osteoblast marker genes in +/+ and patDp/+ osteoblasts. (E) Alizarin red staining in +/+ and patDp/+ calvarial osteoblasts cultured for 21 days. (F,G) Trap staining analysis (F) and Trap gene expression (G) in differentiated osteoclasts from +/+ and patDp/+ mice. *P < 0.05. Values are mean ± SEM. n = 5–8 per group.
To determine if osteoblast proliferation was affected by patDp/+ mutation, we measured the number of cells actively synthesizing DNA by BrdU incorporation in vitro (Fig. 4A). Analysis of proliferation of osteoblasts isolated from WT and patDp/+ mice revealed a 40% decrease in proliferation of patDp/+ osteoblasts compared to their WT counterparts (Fig. 4A). Molecularly this decrease in proliferation of patDp/+ osteoblasts was associated with a decrease in the expression of Cyclins in these cells (Fig. 4B).
We next determined whether differentiation potential of osteoblasts was affected by patDp/+ mutation using two different assays, alkaline phosphatase (ALP) activity assay and marker gene expression analysis. ALP activity was decreased by ~50% in the patDp/+ mice osteoblasts compared to the WT counterparts (Fig. 4C). Analysis of gene expression markers of osteoblast differentiation6 revealed a 35–50% decrease in the expression of Runx2, Atf4, Osx, Alp and Col1a1 (Fig. 4D).
Lastly, we looked at the effect of patDp/+ mutation on the osteoblasts function by analyzing changes in mineralization ability of osteoblasts isolated from the WT and patDp/+ mice. As shown in Fig. 4E qualitative analysis of mineralization using Alizarin red assay revealed a major decline in the mineralization potential of patDp/+ osteoblasts compared to the WT counterparts (Fig. 4E).
We next assayed for the ability of patDp/+ mutation to affect differentiation of bone marrow precursor cells into tartrate-resistant acid phosphatase (TRAP)-positive osteoclasts in vitro. As can be seen in Fig. 4F-G there was no significant difference in the osteoclast differentiation at the level of the multinucleated osteoclast formation (Fig. 4F) or the expression Trap in these cells (Fig. 4G).
Taken together these in vitro results demonstrate that patDp/+ mutation directly, and very potently, affects various aspects of osteoblast biology, but does not affect differentiation of osteoclasts.
Discussion
In humans, it is well known that autism and autism associated disorders are often associated with a low bone density, but the bone phenotype has not been characterized in an animal model to pave a way towards future therapeutic developments to treat bone disorders in this disease12–15. The principal aim of our study was to characterize the bone phenotype in a mouse genetic model of autism at the structural, cellular and molecular levels.
The Dp/+ mice have been created with a 6.3 Mb duplication in chromosome 7 in a region that is the recurrent genetic abnormality in autism. Using a battery of tests, it has been documented that patDp/+ mice display many of the characteristic features of autism viz., social and anxiety abnormalities26. We therefore used patDp/+ mice for the detailed characterization of skeletal phenotype. We report that 4 weeks-old patDp/+ mice display structural defects in the long bones in the trabecular compartment and show a tendency towards reduced cortical integrity in the cortical compartment. This low bone mass phenotype was found to be associated with a defect in the bone formation parameters while bone resorbing cell numbers remained relatively unaltered. In contrast to changes in the long bone, bone mass was not affected in the vertebral column with the patDp/+ mutation. In vitro analysis of osteoblast and osteoclast cultures showed the patDp/+ mutant osteoblasts have intrinsic defects in multiple parameters compared to their WT counterparts. The above facts point towards the genes present in the patDp/+ region affecting the osteoblast functions through their expression in the osteoblasts.
We observed a decrease in the bone mass and architecture in the long bone and not in the vertebra of the patDp/+ mice whereas in humans with this disease the bone phenotype is present in both the sites12–16. There are important differences in the forces that determine bone deposition at different skeletal sites in mice and humans that may explain this underlying phenotypic differences observed34, 35. In humans both the long bones and the vertebral column are load bearing bones and show changes in bone structure and mass with changes in load34. In contrast in rodents the load bearing bones are only the long bones and not the vertebral column34. Changes in the endocrine environment and intrinsic osteoblast defects often lead to changes in both the long bones and vertebral bone mass; in contrast if the mutation in osteoblasts is affecting their ability to respond to mechanical stimuli coupled with hormonal/local changes then there is often an isolated change in bone mass in the long bone34–36. Our observation of a decreased bone mass in the long bone and not the vertebral column in the patDp/+ mice therefore points toward the intrinsic defect in patDp/+ osteoblasts that hampers their ability to proliferate, differentiate and mineralize and likely compromises their ability to respond to mechanical stimuli.
There are 10 genes present in the region that is duplicated in the patDp/+ mice and although it is difficult at the present time to point towards any particular gene ultimately responsible for the low bone mass phenotype in this model, prior studies point towards few of these genes being involved in the bone phenotype. We will now discuss the genes that may possibly be associated with a low bone mass in patDp/+ mice. Loss of function of Mkrn3 has been shown to be associated with advanced bone age in humans and therefore an increase in its expression, as observed in our model, may negatively affect bone mass37. Mouse and human genetic studies have shown that NECDIN (NDN), MAGEL2, and SNRPN contribute to the complex neurogenetic imprinting disorder Prader–Willi-Syndrome (PWS). The symptoms of PWS, which include hyperphagia, severe obesity, hypogonadism, and behavioral abnormalities have been shown to be caused by loss of expression of these genes38. Based on the two PWS associated phenotypes, i.e., obesity and hypogonadism that are known to affect bone mass, few or all of these genes could be implicated in bone growth disorders. Indeed, PWS is associated with a decrease in bone density and an increased risk of fractures37. A decreased expression of these genes in the PWS leads to osteopenia and given that in our autism model there is ~2-fold increase in the expression of these genes, these mice would have been expected to have an increase in bone mass. In contrast to the above mentioned expected phenotype we observed osteopenia in patDp/+ mouse model a fact consistent with the bone phenotype observed in the autism patients. Therefore, it seems likely that effect of genes involved in the PWS is dominated by the negative effect of another gene(s) in this locus, the identity of which can only be speculated at the present time.
Three genes, Gabrα5, Gabrγ3, Gabrβ3, that are part of the Gamma-Aminobutyric Acid (GABA) receptor complex are most upregulated in the patDp/+ mice. GABA receptors mediate signals of the neurotransmitter GABA39. Osteoblasts constitutively express metabotropic GABAB receptor subunits and not the majority of the GABAA receptor subunits to which the Gabrα5, Gabrγ3, Gabrβ3 in the patDp/+ locus belong39, 40. However, at least one of these genes, Gabrγ3 has been shown to be induced in expression when osteoblasts are exposed to external stimuli41. Furthermore, it has been demonstrated that GABA can inhibit alkaline phosphatase activity and calcium accumulation in osteoblasts42. These evidences suggest that an increased local GABA signaling due to the observed increase in the expression of Gabrα5, Gabrγ3, Gabrβ3 may negatively regulate the osteoblast functions in the patDp/+ mice. Although ours and earlier studies point toward a cell autonomous defect in the osteoblasts we cannot rule out the central signaling, perhaps through GABA, contributing towards the bone phenotype observed in the patDp/+ mice. Further studies are needed to identify which of the genes in the patDp/+ locus are expressed/induced in the osteoblasts and account for the cell autonomous decrease in osteoblast proliferation or function. This will require duplication or overexpression of genes present in this locus, either alone or in combination, to mimic the in vivo expression levels observed in patDp/+ mice.
In summary, we have shown the existence of a low bone mass phenotype in a mouse model of autism. Our results reveal an isolated decrease in osteoblast function and bone formation that accounts for this low bone mass phenotype and that osteoblasts from the patDp/+ mice have a cell autonomous decrease in their proliferation and differentiation. Given that autism patients often show low bone density and increased risk of fractures these results lay a foundation for development of therapies that can be used to treat bone disorders using this mouse model.
Materials and Methods
Animals
Studies were performed in accordance with appropriate guidelines of the Wellcome Trust Sanger Institute and the United Kingdom Home Office. All the experiments were performed under the home office project license number PPL 80/2479 and were approved by the Wellcome Trust Sanger Institute Animal Ethics Committee. Generation of Dp/+ mice has been described previously26. Heterozygous Dp/+ female or male mice were used for all experiments. These were generated using a Dp/+ male mouse and a wild type C57Bl/6 N female mouse and littermate controls were used for all analysis. The wild type C57Bl/6 N mice for breeding were obtained from in-house wild type colonies maintained at the Wellcome Trust Sanger Institute research support facility.
Micro-computed tomography (µCT) analysis of long bones
Trabecular bone and cortical architecture of the proximal tibia (secondary spongiosa) was assessed using a µCT system (Skyscan 1172). Tibia bone specimen was stabilized with gauze in a 2 ml centrifuge tube filled with 70% ethanol and fastened in the specimen holder of the µCT scanner. One hundred µCT slices, corresponding to a 1.05 mm region distal from the growth plate, were acquired at an isotropic spatial resolution of 10.5 µm. A global thresholding technique was applied to binarize gray-scale µCT images where the minimum between the bone and bone marrow peaks in the voxel gray value histogram was chosen as the threshold value. The trabecular bone compartment was segmented by a semi-automatic contouring method and subjected to a model-independent morphological analysis43 by the standard software provided by the manufacturer of the µCT scanner. 3D morphological parameters, including model independent measures by distance transformation (DT) of bone volume fraction (BV/TV), Tb.Th. (trabecular thickness), Tb.N. (trabecular number), Tb.Sp. (trabecular separation) and connectivity density (Conn.D) were evaluated. The Conn.D is a quantitative description of the trabecular connection44, 45.
Histology and histomorphometry of vertebra and long bone
Histological analyses were performed on femur and vertebral column specimens collected from 4 weeks-old mice using undecalcified sections stained for bone mass, osteoblasts and osteoclasts analysis as described previously46. Static and dynamic histomorphometric analyses were performed according to standard protocols using the Osteomeasure Analysis System (Osteometrics, Atlanta).
Cell Cultures
Bromodeoxy Uridine (BrdU) cell proliferation assay
Primary calvarial osteoblasts from WT and patDp/+ mice at 70% to 80% confluence were trypsinized, and 5000 cells/well were seeded in 96-well plate in alpha-MEM supplemented with 10% fetal bovine serum (FBS). After 24 hours cells were left for another 24 hours in alpha-MEM supplemented with 0.5% FBS. For the last 4 hours of the 24-hour stimulation period, the cells were pulsed with BrdU. BrdU incorporation was measured using ELISA kit (Roche, IN, USA).
Alkaline phosphatase (ALP) activity assay
Primary calvarial osteoblasts from WT and patDp/+ mice at approximately 80% confluence were trypsinized, and 5000 cells/well were seeded onto 96-well plates. Cells from WT and patDp/+ mice were cultured for 48 hours in osteoblast differentiation medium containing alpha-MEM supplemented with 10% FBS, 10 mM beta-glycerophosphate, 50 mg/mL of ascorbic acid, and 1% penicillin/streptomycin. At the end of incubation period, total ALP activity was measured using p-nitrophenylphosphate (PNPP) as substrate, and absorbance was read at 405 nm (SensoLyte® pNPP Alkaline Phosphatase Assay kit, AnaSpec, Inc.).
Mineralization assay
Primary osteoblast cells were seeded onto 12-well plates (25,000 cells/well) in osteoblast differentiation medium. Osteoblasts from WT and patDp/+ mice were cultured for 21 days with a medium change every 48 hours. At the end of the experiment, cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 15 minutes. Alizarin red-S was used for staining mineralized nodules.
Bioassays
Serum was prepared using BD Vacutainer® SSTTM, snap frozen in liquid nitrogen and stored at −80 °C until analyzed. Serum osteocalcin and Ctx levels were measured using ELISA assays as described previously47.
Molecular studies
Total RNA was isolated from either long bone flushed of bone marrow or primary calvarial osteoblasts. Real-time PCR was performed on DNase I-treated total RNA converted to cDNA using appropriate primers and standard protocols. β-Actin amplification was used as an internal control. Genotypes of all the mice were determined by PCR. All primer sequences are listed on Supplementary Table 1.
Statistical analysis
Results are given as mean ± standard error. Statistical Analysis was performed by Student’s t-test between two groups and one-way ANOVA followed by Newman-keuls posthoc test for more than two groups. All panels in Figs 1–4 *p < 0.05 and **p < 0.01 versus WT.
Data availability
All the mouse strains, experimental tools and raw data present in the manuscript are available upon request.
Electronic supplementary material
Acknowledgements
We would like to thank Professor Allan Bradley for facilitating access to the patDp/+ mice. This work was supported by The Wellcome Trust (Grant Number: 098051) and Ramalingaswamy Fellowship from the Department of Biotechnology, Government of India and National Institute of Immunology Core funding.
Author Contributions
V.K.Y. designed and coordinated the studies. K.S. and K.E.L. performed primary cultures of osteoblasts. K.E.L. and K.S. generated mutant mice. V.K.Y. performed bone sectioning. K.S. and K.E.L. stained and analyzed all the histomorphometry of the bone sections in a double-blind manner. T.T. provided reagents. K.S. and K.E.L. prepared the figures and V.K.Y. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Kirsty E. Lewis and Kunal Sharan contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-09921-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Karsenty G, Ferron M. The contribution of bone to whole-organism physiology. Nature. 2012;481:314–20. doi: 10.1038/nature10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clemens TL, Karsenty G. The osteoblast: an insulin target cell controlling glucose homeostasis. J Bone Miner Res. 2011;26:677–80. doi: 10.1002/jbmr.321. [DOI] [PubMed] [Google Scholar]
- 3.Rodan GA, Martin TJ. Therapeutic approaches to bone diseases. Science. 2000;289:1508–14. doi: 10.1126/science.289.5484.1508. [DOI] [PubMed] [Google Scholar]
- 4.McClung M, Baron R, Bouxsein M. An update on osteoporosis pathogenesis, diagnosis, and treatment. Bone. 2017;98 doi: 10.1016/j.bone.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4:638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- 6.Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–48. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- 7.Manolagas SC. Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr Rev. 2000;21:115–137. doi: 10.1210/edrv.21.2.0395. [DOI] [PubMed] [Google Scholar]
- 8.Zaidi M. Skeletal remodeling in health and disease. Nat Med. 2007;13:791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- 9.Lord C, Cook EH, Leventhal BL, Amaral DG. Autism spectrum disorders. Neuron. 2000;28:355–63. doi: 10.1016/S0896-6273(00)00115-X. [DOI] [PubMed] [Google Scholar]
- 10.Folstein SE, Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet. 2001;2:943–55. doi: 10.1038/35103559. [DOI] [PubMed] [Google Scholar]
- 11.Cook EH, Jr., Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–23. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 12.Hediger ML, et al. Reduced bone cortical thickness in boys with autism or autism spectrum disorder. J Autism Dev Disord. 2008;38:848–56. doi: 10.1007/s10803-007-0453-6. [DOI] [PubMed] [Google Scholar]
- 13.Calarge, C. A. & Schlechte, J. A. Bone Mass in Boys with Autism Spectrum Disorder. J Autism Dev Disord. J Autism Dev Disord. doi: 10.1007/s10803-017-3097-1 (2017). [DOI] [PMC free article] [PubMed]
- 14.Neumeyer AM, et al. Bone microarchitecture in adolescent boys with autism spectrum disorder. Bone. 2017;97:139–146. doi: 10.1016/j.bone.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekhlaspour L, et al. Bone Density in Adolescents and Young Adults with Autism Spectrum Disorders. J Autism Dev Disord. J Autism Dev Disord. 2016;46:3387–3391. doi: 10.1007/s10803-016-2871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karasik D, Rivadeneira F, Johnson ML. The genetics of bone mass and susceptibility to bone diseases. Nat Rev Rheumatol. 2016;12:323–34. doi: 10.1038/nrrheum.2016.48. [DOI] [PubMed] [Google Scholar]
- 17.DiCicco-Bloom E, et al. The develop- mental neurobiology of autism spectrum disorder. J. Neurosci. 2006;26:6897–6906. doi: 10.1523/JNEUROSCI.1712-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamain S, et al. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc. Natl. Acad. Sci. USA. 2008;105:1710–1715. doi: 10.1073/pnas.0711555105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon CH, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lijam N, et al. Social interaction and sensorimotor gating abnormalities in mice lacking Dvl1. Cell. 1997;90:895–905. doi: 10.1016/S0092-8674(00)80354-2. [DOI] [PubMed] [Google Scholar]
- 21.Moretti P, Bouwknecht JA, Teague R, Paylor R, Zoghbi HY. Abnormalities of social interactions and home-cage behavior in a mouse model of Rett syndrome. Hum. Mol. Genet. 2005;14:205–220. doi: 10.1093/hmg/ddi016. [DOI] [PubMed] [Google Scholar]
- 22.Nomura, J., Kannan, G. & Takumi T. Rodent models of Genetic and Chromosomal Variations in Psychiatric disorders. Psychiatry Clin Neurosci., doi:10.1111/pcn.12524 (2017). [DOI] [PubMed]
- 23.Tabuchi K, et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winslow JT, Insel TR. The social deficits of the oxytocin knockout mouse. Neuropeptides. 2002;36:221–229. doi: 10.1054/npep.2002.0909. [DOI] [PubMed] [Google Scholar]
- 25.Young LJ. Regulating the social brain: a new role for CD38. Neuron. 2007;54:353–356. doi: 10.1016/j.neuron.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 26.Nakatani J, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137:1235–46. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belmonte MK, et al. Autism as a disorder of neural information processing: directions for research and targets for therapy. Mol. Psychiatry. 2004;9:646–663. doi: 10.1038/sj.mp.4001499. [DOI] [PubMed] [Google Scholar]
- 28.Bolton PF, et al. Chromosome 15q11-13 abnormalities and other medical conditions in individuals with autism spectrum disorders. Psychiatr. Genet. 2004;14:131–137. doi: 10.1097/00041444-200409000-00002. [DOI] [PubMed] [Google Scholar]
- 29.Dykens EM, Sutcliffe JS, Levitt P. Autism and 15q11-q13 disorders: behavioral, genetic, and pathophysiological issues. Ment. Retard. Dev. Disabil. Res. Rev. 2004;10:284–291. doi: 10.1002/mrdd.20042. [DOI] [PubMed] [Google Scholar]
- 30.Maestrini E, Paul A, Monaco AP, Bailey A. Identifying autism susceptibility genes. Neuron. 2000;28:19–24. doi: 10.1016/S0896-6273(00)00081-7. [DOI] [PubMed] [Google Scholar]
- 31.Veenstra-VanderWeele J, Christian SL, Cook EH., Jr. Autism as a paradigmatic complex genetic disorder. Annu. Rev. Genomics Hum. Genet. 2004;5:379–405. doi: 10.1146/annurev.genom.5.061903.180050. [DOI] [PubMed] [Google Scholar]
- 32.Veenstra-VanderWeele J, Cook EH., Jr. Molecular genetics of autism spectrum disorder. Mol. Psychiatry. 2004;9:819–832. doi: 10.1038/sj.mp.4001505. [DOI] [PubMed] [Google Scholar]
- 33.Vorstman JA, et al. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol. Psychiatry. 2006;11:18–28. doi: 10.1038/sj.mp.4001757. [DOI] [PubMed] [Google Scholar]
- 34.Forwood MR, Turner CH. Skeletal adaptations to mechanical usage: results from tibial loading studies in rats. Bone. 1995;17:197S–205S. doi: 10.1016/8756-3282(95)00292-l. [DOI] [PubMed] [Google Scholar]
- 35.Forwood MR, et al. Modification of the in vivo four-point loading model for studying mechanically induced bone adaptation. Bone. 1995;23:307–10. doi: 10.1016/S8756-3282(98)00090-8. [DOI] [PubMed] [Google Scholar]
- 36.Raisz LG. Pathogenesis of osteoporosis: concepts, conflicts, and prospects. J Clin Invest. 2005;115:3318–25. doi: 10.1172/JCI27071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dimitrova-Mladenova MS, et al. Males with Paternally Inherited MKRN3 Mutations May Be Asymptomatic. J Pediatr. 2005;179:263–265. doi: 10.1016/j.jpeds.2016.08.065. [DOI] [PubMed] [Google Scholar]
- 38.Buiting K. Prader-Willi syndrome and Angelman syndrome. Am J Med Genet C Semin Med Genet. 2010;154:365–76. doi: 10.1002/ajmg.c.30273. [DOI] [PubMed] [Google Scholar]
- 39.Fujimori S, Hinoi E, Yoneda Y. Functional GABA (B) receptors expressed in culturedcalvarial osteoblasts. Biochem Biophys Res Commun. 2002;293:1445–1452. doi: 10.1016/S0006-291X(02)00405-9. [DOI] [PubMed] [Google Scholar]
- 40.Fujimori S, Osawa M, Iemata M, Hinoi E, Yoneda Y. Increased GABA transport activity in rat calvarial osteoblasts cultured under hyperglycemic conditions. Biol Pharm Bull. 2006;29:297–301. doi: 10.1248/bpb.29.297. [DOI] [PubMed] [Google Scholar]
- 41.Kirschner G, et al. Effects of imatinib and nilotinib on the whole transcriptome of cultured murine osteoblasts. Mol Med Rep. 2016;14:2025–37. doi: 10.3892/mmr.2016.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mentink A, et al. Predicting the therapeutic efficacy of MSC in bone tissue engineering using the molecular marker CADM1. Biomaterials. 2013;34:4592–4601. doi: 10.1016/j.biomaterials.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Hildebrand T, Laib A, Muller R, Dequeker J, Ruegsegger P. Direct three-dimensional morphometric analysis of human cancellous bone: microstructural data from spine, femur, iliac crest, and calcaneus. J Bone Miner Res. 1999;14:1167–1174. doi: 10.1359/jbmr.1999.14.7.1167. [DOI] [PubMed] [Google Scholar]
- 44.Feldkamp LA, Goldstein SA, Parfitt AM, Jesion G, Kleerekoper M. The direct examination of three-dimensional bone architecture in vitro by computed tomography. J Bone Miner Res. 1989;4:3–11. doi: 10.1002/jbmr.5650040103. [DOI] [PubMed] [Google Scholar]
- 45.Gundersen HJ, Boyce RW, Nyengaard JR, Odgaard A. The Conneulor: unbiased estimation of connectivity using physical disectors under projection. Bone. 1993;14:217–222. doi: 10.1016/8756-3282(93)90144-Y. [DOI] [PubMed] [Google Scholar]
- 46.Roman-Garcia P, et al. Vitamin B12-dependent taurine synthesis regulates growth and bone mass. J Clin Invest. 2014;124:2988–3002. doi: 10.1172/JCI72606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharan, K., Lewis, K., Furukawa, T., and Yadav, V.K. Regulation of bone mass through pineal-derived melatonin-MT2 receptor pathway. J Pineal Res. 2017, doi:10.1111/jpi.12423 (2017). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the mouse strains, experimental tools and raw data present in the manuscript are available upon request.