Abstract
Clostridium difficile infection (CDI) is associated with risk for severe disease and high mortality. Little is known about the extent of hospital-acquired CDI in Mainland China. In this study, we aimed to investigate the annual CDI incidence, bacterial genotypes, risk factors for severe CDI and survival over a 7-year period. A total of 307 hospital-acquired CDI patients were enrolled, and 70.7% of these cases were male. CDI incidence was 3.4 per 10,000 admissions. Thirty-three different sequence types (STs) were identified, among which ST-54 (18.2%), ST-35 (16.6%) and ST-37 (12.1%) were the most prevalent. During the follow-up period, 66 (21.5%) patients developed severe CDI and 32 (10.4%) patients died in 30 days. Multivariate analysis revealed that bloodstream infection, pulmonary infection and C-reactive protein were significantly associated with severe CDI. After adjustment for potential confounders, old age, bloodstream infection, fever, mechanical ventilation, connective tissue disease, macrolide use and hypoalbuminaemia were independently associated with 30-day mortality in patients with CDI. The CDI prevalence has been low and stable in our center, and STs of Clostridium difficile were different from dominant STs in Western countries. Our data emphasize the need of continued education and surveillance of CDI to reduce the CDI burden in China.
Introduction
Clostridium difficile infection (CDI) is increasingly common in health-care facilities, and represents 20–30% of antibiotic-associated diarrhea1. Clinical CDI manifestations vary from mild diarrhea to severe complications of pseudomembranous colitis, toxic megacolon and death. Outbreaks in North America and Europe coincided with the emergence of a hypervirulent strain, NAP1/BI/0272. This fluoroquinolone-resistant strain produces not only the two usual toxins (toxin A and B), but also a third toxin (binary toxin, CDT); and this was frequently associated with high risk for poor prognosis. However, additional strains were also associated with increased CDI incidence and unfavorable outcomes, and CDI is becoming a persistent challenge throughout the world. Overall, CDI incidence has exceeded that of methicillin-resistant Staphylococcus aureus (MRSA), and amounts to 12.1% of all health care-associated infections in America3, 4.
To meet the CDI challenge, several countries have sought to monitor CDI nationally; and many studies have analyzed CDI isolates to determine the molecular epidemiology of Clostridium difficile (C. difficile) in different regions of the world5–8. However, awareness and surveillance of CDI in Asia have remained poor9, 10. Limited studies performed in Asia over the recent decade indicate that CDI is a significant cause of nosocomial diarrhea in this region, but the true prevalence of CDI remains unknown. Few studies have also reported CDI in China, and the majority of these studies were focused on antimicrobial resistance or sporadic case reports11, 12. Due to the lack of awareness of increased CDI frequency, long-term clinical characteristics, prognosis and risk factors, clinicians may overlook or misdiagnose CDI in Mainland China. Considering the enormous differences in geographical location, genetics of a population, diet habits and even antibiotic use, we expected a different picture of CDI in Mainland China, compared with reports from Western countries.
To understand CDI in our center, we initiated this study and retrospectively analyzed the annual CDI incidence, bacterial genotypes and risk factors for severe CDI over a 7-year period.
Methods
Patients and Definitions
This retrospective study was carried out at The First Affiliated Hospital, Zhejiang University School of Medicine, which is a 2,500-bed tertiary teaching hospital in Hangzhou, Zhejiang, China. From September 1, 2009 to September 30, 2016, inpatients with confirmed hospital-acquired CDI were enrolled and followed up for at least 30 days. Patients aged eighteen years or older were included in the study. Stool samples were collected after written informed consent was obtained from all of the patients. This study was performed in accordance with the declaration of Helsinki and approved clinical practice guidelines and was approved by the Medical Ethics Committee of the First Affiliated Hospital, Zhejiang University School of Medicine.
Diarrhea was defined as three or more loose stools within 24 hours. Inpatients with diarrhea, whose stool samples were positive for both C. difficile culture and toxin gene tests, were diagnosed as CDI. Hospital-acquired CDI is defined as CDI diagnosis established 48 hours after admission, or within 28 days after discharge13, 14. A CDI case was classified as severe if at least one of the following specific manifestations is fulfilled: severe colitis or a complicated course of disease, with significant systemic toxin effects and shock (severe or bloody diarrhea, severe abdominal pain, vomiting, ileus, temperature >38.9 °C, white-cell count >20,000/mm3, albumin level <2.5 mg/dL, and acute kidney injury); resulting in admission to the Intensive Care Unit (ICU), colectomy, or death within 14 days15. Patients with CDI recurrences (defined as new episodes within eight weeks after the confirmation of the first positive sample) were also excluded16.
Microbiological data
Stool samples (semi-formed, unformed, or liquid) were submitted to the clinical microbiology laboratory and cultured anaerobically on cycloserine–cefoxitin–taurocholate agar (CCFA-TA; Oxoid) supplemented with 7% sheep serum at 35 °C for 48 hours. Strains were confirmed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry using the Microflex LT system (Bruker Daltonik). The tcdA, tcdB, cdtA and cdtB genes were detected by PCR in all strains, as previously described17, 18. Multilocus sequence typing (MLST) with seven housekeeping genes (adk, atpA, dxr, glyA, recA, sodA and tpi) was performed on all toxigenic isolates, as previously described19. DNA sequences were submitted to the MLST database (http://pubmlst.org/cdifficile/) to assign the sequence types (STs).
Data Collection
The following data of all patients with hospital-acquired CDI were collected through the hospital database, including age, gender, date of onset of diarrhea, prior underlying diseases, prior medication, prior surgery, clinical data, laboratory parameters, in-hospital medications and prognosis. “Prior” refers to within two months before the CDI diagnosis. CDI characteristics at enrollment were also collected. Vital signs and laboratory findings within 12 hours before or 24 hours post-enrollment were tabularized. Charlson comorbidity index score was used to evaluate the basic conditions of underlying diseases20. Follow-up clinical data were initiated upon hospital admission. For discharged patients, prognostic information was obtained from medical records, telephone contact, or personal visits. The primary endpoint of the study was 30-day mortality.
Statistical Analysis
Annual CDI incidence was calculated as the number of events per 10,000 admissions. For the assessment of risk factors for severe CDI and 30-day mortality, continuous variables were expressed as median with interquartile range (IQR) or mean ± standard deviation (SD). Continuous data were compared in the univariate analyses by Student t-test or Mann-Whitney’s U-test. Nominal variables were expressed as number/percentage and compared using chi-square test. A Cox’s proportional hazard model was used to identify the predictors of time-dependent death of CDI patients. Candidate variables (P < 0.10) after a bivariate analysis were entered into a multivariate Cox’s model using a backward-forward approach. The survival curves of the different ST groups were plotted through the multivariate Cox’s model. In addition, a multivariate logistic regression analysis was performed to assess risk factors for severe CDI, in which the entry and removal probability for the stepwise-backward method was set as 0.05 and 0.10, respectively, and variables with P < 0.05 were retained in the final model. Hosmer-Lemeshow goodness of fit test was performed for logistic regression. Statistical analyses were performed using SPSS (version 23.0; SPSS Inc., Chicago, IL, USA).
Results
Incidence
During the 85-month study period, 307 of 0.91 million hospital admissions were diagnosed with hospital-acquired CDI (Fig. 1). The average incidence was 3.4 per 10,000 admissions (3.1 per 100,000 patient-days), which varied between 2.5 and 4.3 per 10,000 admissions annually. The average positive rate of toxigenic C. difficile in diarrhea patients was 7.3%, which varied between 5.2% and 8.9% annually (Supplementary Table 1).
Figure 1.
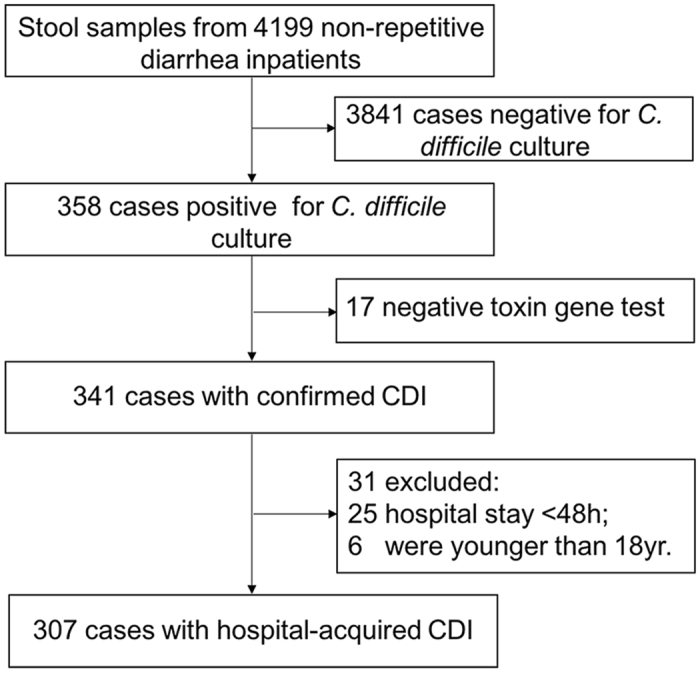
Flow diagram of the procedures used to identify hospital-acquired CDI patients.
Patient Characteristics
The age of patients in this cohort ranged between 18 and 97 years (median age: 56 years). Male accounted for 70.7%. Median hospital stay was 30 days (interquartile range [IQR], 18–61 days). Both median duration of hospitalization before and after CDI onset was 14 days (IQR, 6–29; 7–31 days). The most frequent comorbidities were liver disease (25.6%) and solid tumor (24.1%). Within eight weeks prior to enrollment, 219 patients (70.0%) were admitted to the hospital at least once, and 16.0% of them had a history of ICU admission prior to developing CDI. The 203 patients (66.1%) received at least 1acid suppression medication, while antibiotic use was noted in 257 patients (83.7%), which were most commonly cephalosporins (50.2%), fluoroquinolones (32.6%), or β-lactam/β-lactamase inhibitors (29.6%). Furthermore, only 110 patients (35.8%) were treated with metronidazole (intravenous or oral) or oral vancomycin that targeted CDI. As the lack of understanding of CDI among clinicians, nearly half of these patients (n = 142, 46.3%) only received antidiarrheal and probiotics treatment; and the remaining 55 patients (17.9%) were not treated at all (Table 1).
Table 1.
Demographic and clinical characteristics of patients with hospital-acquired Clostridium difficile infection.
| Patient characteristics | Value or No. (%, N = 307) |
|---|---|
| Age | 56.77 ± 20.21 |
| Gender | |
| Male | 217 (70.7) |
| Female | 90 (29.3) |
| Area | |
| Rural | 152 (49.5) |
| Urban | 155 (50.5) |
| Smoke | 86 (28.0) |
| Alcohol intake | 69 (22.5) |
| Medication exposures prior to developing CDI a | |
| Proton pump inhibitor | 203 (66.1) |
| Chemotherapy | 58 (18.9) |
| Use of any antibiotic not directed at CDI | 257 (83.7) |
| Fluoroquinolones | 100 (32.6) |
| β-lactam/β-lactamase inhibitor combinations | 91 (29.6) |
| Cephalosporin | 154 (50.2) |
| Carbapenem | 79 (25.7) |
| Aminoglycoside | 10 (3.3) |
| Glycopeptides | 41 (13.4) |
| Macrolide | 5 (1.6) |
| Co-trimoxazole | 24 (7.8) |
| Other antibiotics | 23 (7.5) |
| Antifungal agent | 61(19.5) |
| Charlson comorbidity index score, median (IQR) | 2 (1–3) |
| Myocardial infarction | 1 (0.3) |
| Congestive heart failure | 6 (2.0) |
| Chronic obstructive pulmonary disease | 18 (5.8) |
| Cerebrovascular disease | 23 (7.5) |
| Peptic ulcer disease | 5 (1.6) |
| Connective tissue disease | 9 (2.9) |
| Diabetes | 51 (16.3) |
| Chronic kidney disease | 40 (13.0) |
| Dementia | 5 (1.6) |
| Leukemia | 52 (16.9) |
| Malignant lymphoma | 10 (3.3) |
| Solid tumor | 74 (24.1) |
| Liver disease | 79 (25.7) |
| AIDS | 6 (1.0) |
| Prior hospitalization | 215 (70.0) |
| Length of hospital stay, days, median (IQR) | 30 (18–61) |
| Length of hospital stay before CDI, days, median (IQR) | 14 (6–29) |
| Length of hospital stay after CDI, days, median (IQR) | 14 (7–31) |
| Surgery prior to developing CDI | 63 (20.5) |
| Abdominal surgery prior to developing CDI | 41 (13.4) |
| Mechanical ventilation prior to developing CDI | 43 (14.0) |
| Intensive care unit admission prior to developing CDI | 49 (16.0) |
| Ward of admission at onset of CDI | |
| Medical unit | 43 (14.0) |
| Surgical unit | 144 (46.9) |
| Intensive care unit | 58 (18.9) |
| Haemato-oncological unit | 22 (2.2) |
| Geriatric unit | 40 (16.0) |
| Any infection concomitant to CDI | 116 (37.8) |
| Bloodstream infection concomitant to CDI | 21 (6.8) |
| Pulmonary infection concomitant to CDI | 86 (27.7) |
| Other infection concomitant to CDIb | 23 (7.5) |
| Fever >38·5 °C | 90 (29.3) |
| Leukocyte (cells × 109/L), median (IQR) | 7.1 (4.0–10.7) |
| Haemoglobin (g/dL), median (IQR) | 99 (79–116) |
| Platelet (109/L), median (IQR) | 158 (76–243) |
| Albumin (g/dL), median (IQR) | 33.1 (29.0–37.9) |
| Creatinine (μmol/L), median (IQR) | 58 (44–79) |
| Alanine transferase (u/L), median (IQR) | 22 (13–45) |
| C-reactive protein (mg/L), median (IQR) | 28.7 (10.7–68.1) |
| Therapeutic management | |
| No therapy | 56 (17.9) |
| Symptomatic treatment | 142 (46.3) |
| Vancomycin (oral) | 42 (13.4) |
| Metronidazole (intravenous or oral) | 54 (17.6) |
| Vancomycin (oral) and metronidazole (intravenous or oral) | 15 (4.9) |
| Outcome of CDI | |
| Intensive care unit admission | 12 (3.9) |
| Recurrence within 54 days | 13 (4.2) |
| Development of severe CDI | 66 (21.5) |
| 30-day all-cause mortality | 32 (10.4) |
Interquartile range (IQR); Clostridium difficile infection(CDI); aPrior was defined as within two months before the diagnosis of CDI; bAbdominal infection = 12; urinary tract = 4; upper respiratory tract = 2; surgical site = 3; intracranial infection = 2.
Microbiological Characteristics
A total of 307 toxigenic isolates, isolated from 4,199 stool samples, were available for detailed microbiological characterization. Seventy-two isolates (23.5%) were A− B+ CDT− strains, 13 (4.2%) isolates were A+ B+ CDT+ strains and the rest (222, 72.3%) were A+ B+ CDT− strains. Toxigenic C. difficile strains were analyzed by MLST and divided into 33 different STs. The most common STs are listed in Table 2. ST-37 (PCR-ribotypes, RT 017) and ST-81 accounted for 63.9% (46/72) and 18.1% (13/72) of the A− B+ strains, respectively. For CDT+ strains, nine of these isolates belonged to ST-5 (RT 023), two belonged to ST-201, and one belonged to ST-1 (RT 027) and ST-11 (RT 078). ST-54 (56/222, 25.2%, RT 012) and ST-35 (51/222, 23.0%, RT 046) represented the most prevalent A+ B+ isolate in this cohort. We also found a total of 17 nontoxigenic isolates, eight of these isolates belonged to ST-37, five belonged to ST-39 (RT 085), two belonged to ST-2 (RT 014), and one belonged to ST-35 and ST-26 (RT 140).
Table 2.
30-day mortality and disease severity stratified by age and sequence types.
| Stratification | Total | Mortality | Severity |
|---|---|---|---|
| % (N = 307) | % (N = 32) | % (N = 66) | |
| Age group, y | |||
| 18–29 | 38 (12.4) | 2 (6.3) | 10 (16.7) |
| 30–39 | 32 (10.4) | 1 (3.1) | 1 (1.5) |
| 40–49 | 41 (13.4) | 3 (9.4) | 5 (7.6) |
| 50–59 | 50 (16.3) | 4 (12.5) | 8 (12.1) |
| 60–69 | 47 (15.3) | 4 (12.5) | 13 (19.7) |
| 70–79 | 46 (15.0) | 5 (15.6) | 12 (18.2) |
| 80–89 | 47 (15.3) | 11 (34.4) | 15 (22.7) |
| ≥90 | 6 (2.0) | 2 (6.3) | 1 (1.5) |
| Microbiological characteristics | |||
| 54 | 56 (18.2) | 8 (25.0) | 10 (15.2) |
| 35 | 51 (16.6) | 6 (18.8) | 9 (13.6) |
| 37 | 46 (15.0) | 4 (12.5) | 10 (15.2) |
| 3 | 37 (12.1) | 1 (3.1) | 6 (9.1) |
| 2 | 19 (6.2) | 2 (6.3) | 5 (7.6) |
| 81 | 13 (4.2) | 2 (6.3) | 5 (7.6) |
| 8 | 10 (3.3) | 1 (3.0) | 3 (4.5) |
| 5 | 9 (2.9) | 2 (6.3) | 3 (4.5) |
| 39 | 9 (2.9) | 2 (6.3) | 1 (1.5) |
| 102 | 9 (2.9) | 0 (0) | 2 (3.0) |
| Presence of either or both binary toxin genes in toxigenic isolates | 13 (4.2) | 2 (6.3) | 4 (6.1) |
| Toxin A negative and toxin B positive strains in toxigenic isolates | 72 (23.5) | 8 (25.0) | 17 (25.8) |
Mortality and CDI severity were analyzed by age groups, respectively. Additionally, stratification displayed the 10 most frequently found sequence types (STs) among toxigenic isolates.
Risk Factors for Severe CDI
Sixty-six of 307 CDI episodes were classified as severe CDI (21.5%). The multivariate analyses of risk factors for severe CDI are shown in Table 3 (Candidate variables were selected by univariate logistic regression, Supplementary Table 2). The P value of Hosmer-Lemeshow test is 0.792. In the multivariate logistic regression, concomitant bloodstream infection (OR: 8.48, 95% CI: 1.65–43.49; P = 0.010), pulmonary infection (OR: 6.03, 95% CI: 1.16–31.36; P = 0.033) and CRP ≥100 mg/L (OR: 2.93, 95% CI: 1.34–6.42; P = 0.007) remained significantly associated with severe CDI.
Table 3.
Risk factors associated with severe hospital-acquired Clostridium difficile infection analyzed by multivariate logistic regression.
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | |
| Age ≥65 | 2.035(1.173–3.529) | 0.011 | ||
| Ward of admission at onset of CDI | 0.047 | |||
| Medical | Reference | |||
| Surgical | 0.904 (0.311–2.632) | |||
| Intensive care | 1.000 (0.287–3.488) | |||
| Haemato-oncological | 2.571 (0.818–8.080) | |||
| Geriatric | 0.783 (0.233–2.632) | |||
| Fever >38·5 °C | 2.313 (1.313–4.074) | 0.004 | ||
| ICU admission prior to CDI | 2.540 (1.313–4.915) | 0.006 | ||
| Mechanical ventilation prior to CDI | 2.869 (1.445–5.694) | 0.003 | ||
| Infection concomitant to CDI | 2.230 (1.284–3.875) | 0.004 | ||
| Bloodstream infection concomitant to CDI | 3.734 (1.511–9.228) | 0.004 | 8.481 (1.654–43.491) | 0.010 |
| Pulmonary infection concomitant to CDI | 2.379 (1.343–4.212) | 0.003 | 6.026 (1.158–31.363) | 0.033 |
| Chronic kidney disease | 2.226 (1.087–4.559) | 0.029 | ||
| Antibiotic | 2.215 (0.901–5.445) | 0.083 | ||
| Cephalosporin | 1.796 (1.035–3.118) | 0.037 | ||
| Leukocyte ≥15 × 109/L | 3.125 (1.397–6.991) | 0.006 | ||
| Serum creatinine increase >50% | 2.218 (1.059–4.642) | 0.035 | ||
| CRP ≥100 mg/L | 3.352 (1.633–6.882) | 0.001 | 2.931 (1.339–6.417) | 0.007 |
OR, Odds ratio; CI, confidence interval; variables entering the multivariate logistic analysis were selected by univariate logistic regression (Supplementary Table 2).
Risk factors for 30-Day Mortality (all causes)
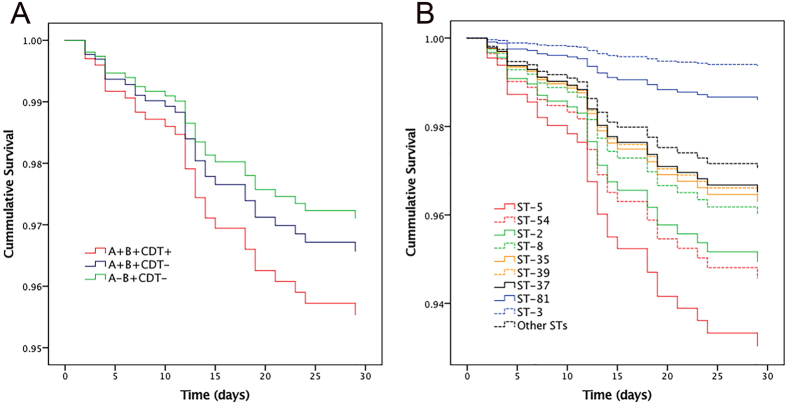
A total of 32 patients died within 30 days, accounting for 10.4% of the CDI population. The 30-day mortality increased with age, and the highest case fatality was observed among patients aged between 80 and 89 years (11/47, 23.4%) and >90 years (2/6, 33.3%). ST-5 (2/9, 22.2%) and ST-39 (2/9, 22.2%) were associated with higher 30-day mortalities (Table 2). A multivariate Cox’s model was performed to identify risk factors for 30-day mortality (Candidate variables were selected by bivariate analysis, Supplementary Table 3). Age ≥65 years (hazard ratio [HR]: 2.98, 95% CI: 1.27–7.02; P = 0.012), concomitant bloodstream infection (HR: 3.53, 95% CI: 1.50–8.32; P = 0.004), fever ≥38.5 °C (HR: 3.03, 95% CI: 1.45–6.31; P = 0.003), mechanical ventilation prior to CDI (HR: 3.20, 95% CI: 1.46–7.00; P = 0.004), connective tissue disease (HR: 5.53, 95% CI: 1.39–22.00; P = 0.015), macrolide use (HR: 10.17, 95% CI: 2.25–45.93; P = 0.003) and albumin ≤2.5 mg/dL (HR: 3.94, 95% CI: 1.38–11.25; P = 0.011) were independently associated with poor prognosis in CDI patients (Table 4). After adjustment of baseline differences by the multivariate Cox’s model, the 30-day mortality was found to be associated with changes in toxin profiles (Fig. 2A) and STs (Fig. 2B).
Table 4.
Risk Factors associated with 30-day mortality analyzed by multivariable Cox’s proportional hazard mode in hospital-acquired Clostridium difficile infection patients.
| Variable | B | Standard error | Wald chi-square | Hazard ratio | 95% CI | P value |
|---|---|---|---|---|---|---|
| Age ≥65 | 1.092 | 0.437 | 6.251 | 2.981 | 1.266–7.017 | 0.012 |
| Bloodstream infection concomitant to CDI | 1.262 | 0.437 | 8.332 | 3.532 | 1.499–8.321 | 0.004 |
| Fever >38·5 °C | 1.108 | 0.375 | 8.725 | 3.027 | 1.452–6.312 | 0.003 |
| Mechanical ventilation prior to developing CDI | 1.164 | 0.399 | 8.491 | 3.201 | 1.464–7.002 | 0.004 |
| Connective tissue disease | 1.710 | 0.704 | 5.895 | 5.531 | 1.391–22.000 | 0.015 |
| Macrolide | 2.320 | 0.769 | 9.099 | 10.174 | 2.254–45.929 | 0.003 |
| Albumin ≤2.5 g/dL | 1.370 | 0.536 | 6.534 | 3.935 | 1.376–11.250 | 0.011 |
Statistical analysis was performed using a multivariable Cox’s proportional hazard model. Variables entering the multivariate analysis were selected by binary analysis (Supplementary Table 3).
Figure 2.
Mortality rates of all patients with Clostridium difficile infection up to 30 days after onset of diarrhea stratified by different toxins (A) or sequence types (B).
Discussion
In this retrospective study, a standard diagnosis procedure was used to determine hospital-acquired CDI frequencies between 2009 and 2016 in our center. Our analysis revealed an annually stable, low CDI prevalence in our center, and the average CDI rate was 3.4 per 10,000 admissions or 7.3% among diarrhea patients. We also analyzed the STs of all isolates, and the dominant STs were different from that reported in North America and Europe.
A recent meta-analysis of CDI incidence in Mainland China suggested that the pooled incidence of toxigenic C. difficile among patients with diarrhea was 14%, but it varied significantly from 23% to 3% in different regions11. A 8.4% (19/227) −15.6% (125/801) high CDI among diarrhea patients was reported in Western countries21–27. An average of 1.33 nosocomial CDI per 1,000 admissions (range: 0.74–2.30) was reported in 13 European hospitals7. The 7.3% CDI incidence among diarrhea patients or 3.4 per 10,000 admissions in this study was low.
We have reason to believe that a CDI diagnosis may have been missed in a portion of patients with diarrhea because of the poor awareness of CDI among physicians, representing the main reason for the relatively low CDI incidence. In addition, a portion of patients who developed CDI after discharge within 28 days were not included due to incomplete clinical data. Another possible reason was the different dominant CD strains compared in other countries.
Age appears to be important for developing CDI. The median age among patients in this study was 56 years, which was slightly younger than that reported in Western countries6–8. However, this study clearly revealed that the age composition of CDI patients was biased toward an older population: more than half of the cases were 50 years or older, and one in four cases was 70 years or older. The association of old age with CDI may suggest that weakened immunity is a condition for CDI.
Another finding in this study was that 70.7% of CDI patients were male, and 9.2% (20/217) of these male patients died within 30 days, which was in contrast with the findings reported in Western countries, where females were dominant among CDI patients6, 7. The population of CDI patients was also not predominant female in several studies from East Asian countries, we suggest that the difference in CDI patient gender may be related to geographical location, genetic disposition and diet habits28–30.
Furthermore, as high as 83.7% of CDI patients had a history of previous use of antibiotics in this study, which was in line with the rates reported by others7, 27, suggesting that the use of antibiotics may have caused imbalance of bacterial flora in the gut. It was interesting to note that the highest CDI occurred in patients admitted to the Internal Medicine Department, since these patients were most likely exposed to antibiotics for long periods and had a long hospital stay.
Due to the lack of awareness of CDI among physicians, the treatment of diarrhea did not follow the CDI treatment guidelines. Interestingly, when 30-day mortality was correlated with the treatment, mortality was 9.9% (14/142) in patients who accepted symptomatic treatment and 9.1% (5/55) in patients with no therapy; and both were lower than the overall mortality. The lower mortality in patients without treatment may suggest mild CDI, and only severe diarrhea brought the attention of physicians to possible CDI and initiated the CDI treatment. Furthermore, among patients who accepted symptomatic treatment, 100 of 142 (70.4%) patients received probiotics. Most of these probiotics contained clostridium butyricum, which has been shown to be effective in restoring CDI affected colonies both in vitro and in vivo 31–33.
In this study, ST-54 (RT 012) and ST-35 (RT 046) were the most prevalent, followed by ST-37 (RT 017) and ST-3 (RT 001), which differed from that identified by Bauer and colleagues7. Among the isolates obtained from the 73 hospitals in 26 countries presented by Bauer et al., PCR-ribotypes (RT) 014 and 001 were the most prevalent, followed by 078 and 018. Lower rates of A− B+ strains were noted in Europe7, while 72/307 (23.4%) of isolates were A− B+ strains in our study. The A− B+ strains appeared to be more prevalent in East Asia. For instance, several reports from Mainland China also revealed that A− B+ strains accounted for 30% of all isolates17, 34.
Marjolein and colleagues6 reported that CDT+ strains accounted for 23% (90/389) of the total isolates, and 19/90 (21.1%) of these isolates were RT 027. However, CDT+ strains in our study were rare (4.2%, 13/307), and most of the strains were ST-5 (9/13, 69.2%). Similarly, Rupnik and colleagues35 only found five isolates (1.6%) positive for binary toxin among the 310 isolates obtained from nine hospitals in Japan and Korea. In the present study, only a single isolate for ST-1 (RT 027) and ST-11 (RT 078) was identified, respectively, in 2012 and 2016. Due to the association of ST-1 with the CDI outbreak, the CDI incidence in the Estrie region of Quebec increased from 22.2 per 100,000 population in 1991 to 92.2 per 100,000 population in 200336. This first detection of ST-1 in our hospital alerts us that this type is frequently associated with CDI outbreaks and dismal prognosis in the Western world2.
Furthermore, as many as 21.1% of cases were diagnosed as severe CDI, and this was in line with the reported percentages in literatures8, 37. Several studies had identified pulmonary infection as independent risk factors for severe CDI, and other risk factors were mentioned occasionally27. CDI patients that were accompanied by concomitant bloodstream and pulmonary infection may have weakened immunity, aggravating the CDI disease. High CRP likely reflects the severity of colonic inflammation, which represents a candidate marker for prognosis. Strain types have been suggested as an additional factor for CDI severity. In the present study, the disease in patients with ST-81 (5/13, 38.5%) and ST-5 (3/9, 33.3%) infection was likely more severe, compared with other STs.
Most risk factors identified in the present study for 30-day mortality were consistent with previous studies37. For instance, a pervious study suggested that CDI-related mortality occurred mainly within 30 days after onset, and was low after 90 days, as demonstrated by long-term follow-ups6. The 30-day CDI-related mortality in this cohort was 10.4%, which was comparable to the reported mortalities in North America and Europe6, 7, 38, 39. It was noted in our study that CDT+ strains or ST-5 infected patients were predisposed to higher mortality than other STs. Thus, different extents of pathologic changes caused by different strains may have different impacts on mortality. Interestingly, A+ B+ strains were associated with more fatal outcomes than A− B+ strains, except for ST-3. This difference in mortality between A− B+ and A+ B+ strains requires further investigation. In addition, differences in the severity of the primary disease in each CDI patient in hospitals may primarily contribute to death. Unexpectedly, we found that connective tissue disease was associated with increased mortality. Patients with connective tissue disease may have immune dysfunction, which may have made it difficult to contain CDI.
Few limitations in this study warrants discussion. First, only 30-day follow-ups were carried out, since many of the patients in this cohort resided outside of Hangzhou metropolitan boundaries. Second, our results derived a single-center study, and the main findings require verification through multicenter long-term studies in the future.
In conclusion, hospital-acquired CDI frequency in our region appears to be relatively moderate, and no clear spike was observed during the 7-year period. CDI outcomes appeared to be less severe compared to other regions. The strain types of C. difficile infection in our center also markedly differed from the strains reported from Western countries. We identified bloodstream infection and connective tissue disease as independent risks for severe CDI and 30-day mortality.
Electronic supplementary material
Acknowledgements
This work was supported by the National Basic Research Program of China (973 program) (No. 2013CB531401).
Author Contributions
L.L., Q.X. conceived and designed the experiments. Y.C., S.G., and T.L., and performed the experiments. J.Q., Y.F. collected samples and performed DNA extractions. B.Z., P.S., performed library construction, sequencing and designed the analysis. Y.C., and S.G., analysed the data. Q.X. drew the all figures and wrote the paper. L.L. revised the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Qiaomais Xu, Yunbo Chen and Silan Gu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-09961-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bartlett JG. Clinical practice. Antibiotic-associated diarrhea. N Engl J Med. 2002;346:334–339. doi: 10.1056/NEJMcp011603. [DOI] [PubMed] [Google Scholar]
- 2.Loo VG, et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353:2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 3.Magill SS, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey N, Dennison E, Cooper C. Osteoporosis: impact on health and economics. Nat Rev Rheumatol. 2010;6:99–105. doi: 10.1038/nrrheum.2009.260. [DOI] [PubMed] [Google Scholar]
- 5.Redelings MD, Sorvillo F, Mascola L. Increase in Clostridium difficile-related mortality rates, United States, 1999–2004. Emerg Infect Dis. 2007;13:1417–1419. doi: 10.3201/eid1309.061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hensgens MP, Goorhuis A, Dekkers OM, van Benthem BH, Kuijper EJ. All-cause and disease-specific mortality in hospitalized patients with Clostridium difficile infection: a multicenter cohort study. Clin Infect Dis. 2013;56:1108–1116. doi: 10.1093/cid/cis1209. [DOI] [PubMed] [Google Scholar]
- 7.Bauer MP, et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 8.Abou Chakra CN, et al. Factors Associated With Complications of Clostridium difficile Infection in a Multicenter Prospective Cohort. Clin Infect Dis. 2015;61:1781–1788. doi: 10.1093/cid/civ749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins DA, Hawkey PM, Riley TV. Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control. 2013;2 doi: 10.1186/2047-2994-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawkey PM, et al. Molecular epidemiology of Clostridium difficile infection in a major chinese hospital: an underrecognized problem in Asia? J Clin Microbiol. 2013;51:3308–3313. doi: 10.1128/JCM.00587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang C, et al. The incidence and drug resistance of Clostridium difficile infection in Mainland China: a systematic review and meta-analysis. Sci Rep. 2016;6 doi: 10.1038/srep37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin K, Wang S, Huang Z, Lu S. Clostridium difficile infections in China. J Biomed Res. 2010;24:411–416. doi: 10.1016/S1674-8301(10)60055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debast SB, Bauer MP, Kuijper EJ. European Society of Clinical, M. & Infectious, D. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2014;20(Suppl 2):1–26. doi: 10.1111/1469-0691.12418. [DOI] [PubMed] [Google Scholar]
- 14.Steele SR, et al. Practice parameters for the management of Clostridium difficile infection. Dis Colon Rectum. 2015;58:10–24. doi: 10.1097/DCR.0000000000000289. [DOI] [PubMed] [Google Scholar]
- 15.Leffler DA, Lamont JT. Clostridium difficile infection. N Engl J Med. 2015;372:1539–1548. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- 16.Cohen SH, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 17.Kato H, et al. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol. 1998;36:2178–2182. doi: 10.1128/jcm.36.8.2178-2182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stubbs S, et al. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett. 2000;186:307–312. doi: 10.1111/j.1574-6968.2000.tb09122.x. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths D, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48:770–778. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 21.Vonberg RP, Schwab F, Gastmeier P. Clostridium difficile in discharged inpatients, Germany. Emerg Infect Dis. 2007;13:179–180. doi: 10.3201/eid1301.060611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyytikainen O, et al. Hospitalizations and deaths associated with Clostridium difficile infection, Finland, 1996–2004. Emerg Infect Dis. 2009;15:761–765. doi: 10.3201/eid1505.081154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soes, L. et al. The emergence of Clostridium difficile PCR ribotype 027 in Denmark–a possible link with the increased consumption of fluoroquinolones and cephalosporins? Euro Surveill14 (2009). [PubMed]
- 24.Soler P, Nogareda F, Cano R. Rates of Clostridium difficile infection in patients discharged from Spanish hospitals, 1997–2005. Infect Control Hosp Epidemiol. 2008;29:887–889. doi: 10.1086/590392. [DOI] [PubMed] [Google Scholar]
- 25.Hubert B, et al. A portrait of the geographic dissemination of the Clostridium difficile North American pulsed-field type 1 strain and the epidemiology of C. difficile-associated disease in Quebec. Clin Infect Dis. 2007;44:238–244. doi: 10.1086/510391. [DOI] [PubMed] [Google Scholar]
- 26.Gravel D, et al. Health care-associated Clostridium difficile infection in adults admitted to acute care hospitals in Canada: a Canadian Nosocomial Infection Surveillance Program Study. Clin Infect Dis. 2009;48:568–576. doi: 10.1086/596703. [DOI] [PubMed] [Google Scholar]
- 27.Alicino C, et al. Increasing incidence of Clostridium difficile infections: results from a 5-year retrospective study in a large teaching hospital in the Italian region with the oldest population. Epidemiol Infect. 2016;144:2517–2526. doi: 10.1017/S0950268816000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi M, Mori N, Bito S. Multi-institution case-control and cohort study of risk factors for the development and mortality of Clostridium difficile infections in Japan. BMJ Open. 2014;4 doi: 10.1136/bmjopen-2014-005665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong D, et al. Clinical and microbiological characterization of Clostridium difficile infection in a tertiary care hospital in Shanghai, China. Chin Med J (Engl) 2014;127:1601–1607. [PubMed] [Google Scholar]
- 30.Choi HY, et al. The epidemiology and economic burden of Clostridium difficile infection in Korea. Biomed Res Int. 2015;2015 doi: 10.1155/2015/510386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woo TD, et al. Inhibition of the cytotoxic effect of Clostridium difficile in vitro by Clostridium butyricum MIYAIRI 588 strain. J Med Microbiol. 2011;60:1617–1625. doi: 10.1099/jmm.0.033423-0. [DOI] [PubMed] [Google Scholar]
- 32.Imase K, et al. Efficacy of Clostridium butyricum preparation concomitantly with Helicobacter pylori eradication therapy in relation to changes in the intestinal microbiota. Microbiol Immunol. 2008;52:156–161. doi: 10.1111/j.1348-0421.2008.00026.x. [DOI] [PubMed] [Google Scholar]
- 33.Seki H, et al. Prevention of antibiotic-associated diarrhea in children by Clostridium butyricum MIYAIRI. Pediatr Int. 2003;45:86–90. doi: 10.1046/j.1442-200X.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- 34.Huang H, et al. Clostridium difficile infections in a Shanghai hospital: antimicrobial resistance, toxin profiles and ribotypes. Int J Antimicrob Agents. 2009;33:339–342. doi: 10.1016/j.ijantimicag.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 35.Rupnik M, Kato N, Grabnar M, Kato H. New types of toxin A-negative, toxin B-positive strains among Clostridium difficile isolates from Asia. J Clin Microbiol. 2003;41:1118–1125. doi: 10.1128/JCM.41.3.1118-1125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pepin J, et al. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ. 2004;171:466–472. doi: 10.1503/cmaj.1041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abou Chakra CN, Pepin J, Sirard S, Valiquette L. Risk factors for recurrence, complications and mortality in Clostridium difficile infection: a systematic review. PLoS One. 2014;9 doi: 10.1371/journal.pone.0098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hota SS, et al. Determining mortality rates attributable to Clostridium difficile infection. Emerg Infect Dis. 2012;18:305–307. doi: 10.3201/eid1802.101611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mlangeni DA, et al. Death certificates provide a poor estimation of attributable mortality due to Clostridium difficile when compared to a death review panel using defined criteria. J Hosp Infect. 2011;77:370–371. doi: 10.1016/j.jhin.2010.12.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.