Abstract
Lambdoid bacteriophages form a group of viruses that shares a common schema of genome organization and lifecycle. Some of them can play crucial roles in creating the pathogenic profiles of Escherichia coli strains. For example, Shiga toxin-producing E. coli (STEC) acquired stx genes, encoding Shiga toxins, via lambdoid prophages (Stx phages). The results obtained so far present the evidence for the relation between the exo-xis region of the phage genome and lambdoid phage development, however molecular mechanisms of activities of the exo-xis genes' products are still unknown. In view of this, we decided to determine the influence of the uncharacterized open reading frame orf63 of the exo-xis region on lambdoid phages development using recombinant prophages, λ and Stx phage Φ24B. We have demonstrated that orf63 codes for a folded protein, thus, it is a functional gene. NMR spectroscopy and analytical gel filtration were used to extend this observation further. From backbone chemical shifts, Orf63 is oligomeric in solution, likely a trimer and consistent with its small size (63 aa.), is comprised of two helices, likely intertwined to form the oligomer. We observed that the deletion of phage orf63 does not impair the intracellular lambdoid phage lytic development, however delays the time and decreases the efficiency of prophage induction and in consequence results in increased survival of E. coli during phage lytic development. Additionally, the deletion of phage orf63 negatively influences expression of the major phage genes and open reading frames from the exo-xis region during prophage induction with hydrogen peroxide. We conclude, that lambdoid phage orf63 may have specific functions in the regulation of lambdoid phages development, especially at the stage of the lysis vs. lysogenization decision. Besides, orf63 probably participates in the regulation of the level of expression of essential phage genes and open reading frames from the exo-xis region during prophage induction.
Keywords: Shiga toxin-producing Escherichia coli (STEC), lambdoid bacteriophages, lytic development, exo-xis region, open reading frames
Introduction
The significance of Shiga toxin-producing E. coli (STEC) as a public health problem was first recognized in 1982 during an investigation of an outbreak of hemorrhagic colitis associated with consumption of contaminated hamburgers (Riley et al., 1983). Since then, STEC strains have been implicated in many outbreaks of diarrhea world-wide. Quite recently (2011), the Shiga toxin-producing E. coli serotype O104:H4 was responsible for a serious epidemic outbreak in Germany (Bloch et al., 2012; Muniesa et al., 2012). STEC pathogens can cause serious food poisoning with bloody diarrhea in humans (Nataro and Kaper, 1998). Their main virulence factors are Shiga toxins, encoded by stx genes located in genomes of bacteriophages which occur in bacteria as prophages (Mizutani et al., 1999). These bacteriophages are called Shiga toxin-converting or Stx, for short, and belong to the lambdoid family of phages (Schmidt, 2001). All phages within this group indicate high similarities in the lifecycle and genomic organization to bacteriophage λ, the most reviewed member of this family (Wegrzyn et al., 2012). In the prophage state, most of phage genes, including stx genes, are not transcribed due to inhibition caused by the phage cI repressor. As a consequence, Shiga toxins are not produced under such conditions. Expression of stx as well as other phage genes occurs effectively only after prophage induction. In most cases, this process requires activation of the RecA-dependent bacterial S.O.S. response which is provoked by factors causing appearance of single-stranded DNA fragments. Activated RecA protein stimulates cleavage of the S.O.S. regulon repressor, the LexA protein, and the cI phage repressor. Prophage induction and subsequent phage lytic development lead to production of progeny phage particles and Shiga toxins, followed by their release from the lysed cell (Licznerska et al., 2016b). In the regulation of the lysis-vs. -lysogenization decision after infection of the host cell by a bacteriophage, both phage- and host-encoded proteins play important roles (for a review, see Wegrzyn et al., 2012). Among environmental factors influencing the decision, the crucial are temperature, nutrients availability and multiplicity of infection (m.o.i.). Lytic growth is supported by high temperature, rich medium and high m.o.i., while low temperature, starvation and low m.o.i. favor lysogenization. At the molecular level, the major players supporting lytic and lysogenic pathways are Cro and cI proteins, respectively. They are transcriptional regulators, and Cro represses expression of the cI gene, whereas cI downregulates transcription from two major “lytic” promoters (pL and pR, which provide mRNAs for cro and other “lytic” genes, encoding proteins involved in all processes during production of phage progeny) while stimulating its own expression by activation of the pM promoter. Thus, the result of the competition between Cro and cI is crucial for choosing one of the alternative developmental pathways. Since shortly after infection no cI protein is present, another transcription regulator, the cII protein (whose gene is transcribed from pR), is a key player in this game. This protein activates the second promoter for cI expression, pE. Therefore, cII activity decides on the Cro or cI predominance. In fact, cII is a subject of various regulatory mechanisms acting in response to different environmental conditions, including those playing major roles in the lysis-vs. -lysogenization decision (see Wegrzyn et al., 2012, for details).
An evolutionarily conserved region of lambdoid bacteriophage genome, located between exo and xis genes (so called “the exo-xis region”), contains several genes and open reading frames (Figure 1). Quite surprisingly, until recently, the role of this region in bacteriophage development was almost completely unknown. Recent studies indicated that overexpression of genes from the exo-xis region' impaired lysogenization of E. coli by bacteriophage λ (Loś et al., 2008b) and enhanced induction of prophages λ and Φ24B (one of Shiga toxin-converting phages) (Bloch et al., 2013). The Ea8.5 protein, encoded by a gene located in the exo-xis region, contains a fused homeodomain/zinc-finger fold (Kwan et al., 2013), which suggest a regulatory role for this protein. Interestingly, prophage induction with mitomycin C or hydrogen peroxide caused different expression patterns of genes from the exo-xis region; such differences were observed in both phages, λ and Φ24B (Bloch et al., 2014). Moreover, phages with deletions in the exo-xis region responded to the oxidative stress in a different manner relative to wild-type phages (Licznerska et al., 2016a). Therefore, it is important to determine structures and functions of particular proteins encoded in the exo-xis region.
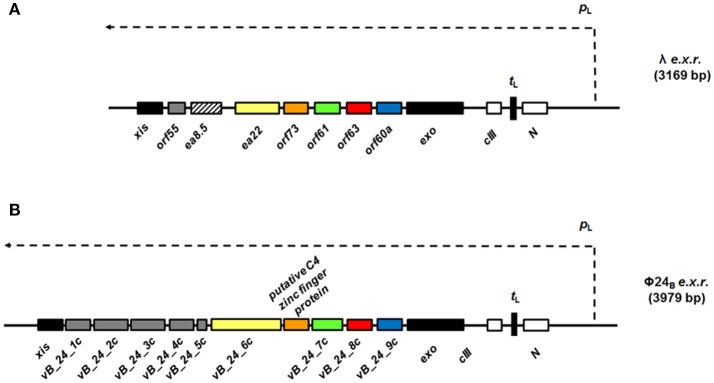
Figure 1.
Maps of genes and open reading frames (ORFs) from the region located between exo and xis genes (black rectangles), the exo-xis region or e.x.r., of lambdoid bacteriophages: λ (A) and Φ24B (B). In the case of phage λ (A), the exo-xis region consists of two recognized genes: ea22 and ea8.5, and five additional ORFs, named: orf60a, orf63, orf61, orf73, and orf55, which expression is under control of the pL promoter (thin dashed arrow). Comparatively, the exo-xis region of phage Φ24B (B) contains additional ORFs (gray rectangles), but there is no homolog of the ea8.5 gene of phage λ (A), rectangle with black stripes). Note, that some ORFs from the exo-xis region of phage Φ24B (B): vb_24B_9c (blue rectangles), vb_24B_8c (red rectangles), vb_24B_7c (green rectangles), putative C4 zinc finger protein (orange rectangles), and vb_24B_6c (yellow rectangles) are homologs of phage λ orf60a, orf63, orf61, orf73, ea22 (B), respectively. In spite of the differences in composition of both λ and Φ24B exo-xis regions, attention needs to paid to highly conserved sequences of the orf60a-orf73 regions among lambdoid bacteriophages (≥70% nucleotide and amino acid sequence identity) (Bloch et al., 2013). The regulatory genes: N and cIII are marked as white rectangles and tL terminator is indicated as black vertical rectangle.
In this work, we have focused on orf63. In our preliminary experiments with mutants in particular genes and ORFs from the exo-xis region, deletion of orf63 gave one of the strongest effects (Licznerska et al., 2016a). Moreover, the transcription factor YqhC has been recognized as a potential partner for interaction with Orf63 in yeast two-hybrid study of phage-host interactions (Blasche et al., 2013). Therefore, we decided to investigate structure and functions of orf63 and its product, the Ofr63 protein, in more detail.
Materials and methods
The orf63 gene expression and protein purification
A codon-optimized orf63 gene (NCBI ID: 2703507) for high level expression E. coli was synthesized by ATUM (Menlo Park, CA) and supplied in plasmid pD441-NH for direct transformation of a BL21 host strain (Novagen). Amino terminal 6xHis and Flag (DYKDDDDK) tags were included to facilitate affinity purification and detection. Milligram quantities of isotopically labeled 6xHis-Flag-Orf63 for NMR spectroscopy were obtained from a 1.0 L fermentation in a minimal medium containing 1 g 15NH4Cl, 3 g of 13C-glucose, and 1 g of 15N-13C Celtone algal extract (CIL; Cambridge, MA). The cell pellet was dissolved in T300 buffer (20 mM Tris-HCl, 300 mM NaCl, 0.05% NaN3) and lysed by French press and sonication. The Orf63 protein purified from the bacterial soluble fraction by Nickel-NTA affinity chromatography (Qiagen) that included a 10 mM imidazole wash step and a 20 mM EDTA elution step, all in T300 buffer. A subsequent gel filtration chromatography step (Sephacryl-100, HiLoad 16/60; GE Life Sciences) was employed to further purify the Orf63 protein and exchange it into NMR spectroscopy buffer (5 mM Tris-HCl, 0.15 M NaCl, 0.05% NaN3).
Gel filtration assay to estimate apparent molecular weight
Chromatograms of eleven proteins on the same gel filtration column used to purify Orf63 were compiled to produce a standard curve describing the relationship between retention volume and molecular weight. Specific proteins used for the standard curve included the HACS1 SH3 domain (10.6 kDa), the AIDA1 PTB domain (22.0 kDa), the monomeric and dimeric states CASKIN2 SAM domain tandem (20.2 / 40.5 kDa), the CASKIN2 SAM1 domain (10.3 kDa), the SHP2 adaptor SH2 domain (14.1 kDa), the AIDA1 SAM domain tandem (16.4 kDa), two deletion mutants of the La RRM domain (14.6 / 15.9 kDa), the Crk2 adaptor SH2 domain (15.5 kDa), dimeric glutathione S-transferase (52 kDa), and calmodulin (18.8 kDa).
NMR spectroscopy
A 0.5 mM sample of 13C,15N-labeled Orf63 was prepared for NMR spectroscopy in NMR buffer supplemented with 10% D2O. All experiments were performed at 310 K using a Bruker Avance 700 MHz NMR spectrometer equipped with a cryogenically cooled 5 mm probe at the York University Life Sciences Building Central Facility. Backbone (HN, N, CA, CB, C') assignments were achieved using a set of conventional triple resonance experiments (HNCA, HNCACB, CBCAcoNH, HNCO, HNcaCO) incorporating sparse sampling for the optimum sensitivity and resolution. Datasets were processed with NMRpipe (Delaglio et al., 1995) and istHMS (Hyberts et al., 2012) and interpreted with CCPN Analysis (Skinner et al., 2015).
Bacteria, bacteriophages and plasmids
The E. coli strains, bacteriophages and plasmids used in in vivo work are presented in Table 1. Work with these strains was approved by the Ministry of Environment (decision no. 189/2016). The E. coli lysogens were obtained using the following lambdoid phages: λ, λΔorf63, Φ24B or Φ24BΔorf63 (Bloch et al., 2013; Licznerska et al., 2016b). In the first step, phage lysates were prepared. Bacterial cultures were grown at 37°C to A600 = 0.1. Then, mitomycin C (Sigma—Aldrich) was added to all flasks to a final concentration of 1 μg/ml. The incubation with shaking was continued for about 12 h. To obtain lysates, bacterial debris were centrifuged (2,000 × g for 10 min at 4°C) and supernatants were filtered through the 0.22-μm-pore-size filters (Sigma—Aldrich). In the next step, the lysogenization procedure was carried out. Briefly, E. coli strain C600 was cultivated at 37°C to A600 = 0.2. Then, 4 ml of bacterial culture was centrifuged (2,000 × g for 5 min at RT), the pellet was washed with TCM buffer (10 mM Tris-HCl, 10 mM MgSO4, 10 mM CaCl2, pH 7.2; Sigma—Aldrich) and suspended in LB medium (Sigma—Aldrich) supplemented with MgSO4 (phages λ and λΔorf63) or with MgSO4 and CaCl2 (phages Φ24B and Φ24BΔorf63) to a final concentration of 10 mM. Bacteriophages were added to the suspensions to m.o.i. of 10. Following incubation at 37°C, the mixtures were spread on LB agar plates. After overnight incubation at 37°C, bacterial colonies were tested for the presence of prophages by using UV irradiation (this procedure is described in detail in the next section). For construction of the plasmid pSB_orf63_λ, nucleotide sequence of orf63 from phage λ was amplified by PCR with primers: Fλorf63_EcoRI (5′GGA GAA TTC GGC TGT ATG CAC AAA GC) and Rλorf63_BamHI (5′ GAG GAT CCT GCA TTC CGT GGT TGT C), and phage DNA as a template, which was isolated by using MasterPure™ Complete DNA and RNA Purification Kit (Epicenter). Then, the λorf63 was ligated with fragment of plasmid pUC18 (insert and vector were digested with EcoRI and BamHI restrictions endonucleases; Thermo Scientific), bearing an ampicillin resistance gene and sequence of plac promoter. The plasmid pSB_orf63_Φ24B was constructed according to similar procedure. To amplify a DNA fragment containing vb_24B_8c sequence (the homolog of λorf63) by PCR method, two primers: FΦ24Borf63_EcoRI (5′GGA GAA TTC GGC TGT ATG CAC AAA GC) and RΦ24Borf63_BamHI (5′GTA GGA TCC TTG TCA TGC CGG GTC) were used. Next, plasmid pUC18 and insert were cut with EcoRI and BamHI enzymes and ligate by the T4 DNA ligase (Thermo Scientific). The construction of pUC18 derivatives was confirmed by DNA sequencing (Genomed).
Table 1.
Bacterial strains, bacteriophages and plasmids used for in vivo experiments.
| E. coli strains, bacteriophages or plasmids | Relevant genotype or description | References |
|---|---|---|
| E. coli LABORATORY STRAINS | ||
| MG1655 | F– λ– ilvG rfb-50 rph-1 | Jensen, 1993 |
| MG1655 (λ) | MG1655 bearing λ prophage | Bloch et al., 2013 |
| MG1655 (λΔorf63) | MG1655 bearing λ prophage with deletion of orf63 | Licznerska et al., 2016a |
| MG1655 (Φ24B) | MG1655 bearing Φ24B prophage | Bloch et al., 2013 |
| MG1655 (Φ24BΔorf63) | MG1655 bearing Φ24B prophage with deletion of vb_24B_8c, the homolog of λorf63 | Licznerska et al., 2016a |
| C600 | F– tonA21 thi-1 thr-1 leuB6 lacY1 glnV44 rfbC1 fhuA1 λ− | Appleyard, 1954 |
| C600 (λ) | C600 bearing λ prophage | This study, by lysogenization |
| C600 (λΔorf63) | C600 bearing λ prophage with deletion of orf63 | This study, by lysogenization |
| C600 (Φ24B) | C600 bearing Φ24B prophage | This study, by lysogenization |
| C600 (Φ24BΔorf63) | C600 bearing Φ24B prophage with deletion of vb_24B_8c, the homolog of λorf63 | This study, by lysogenization |
| BACTERIOPHAGES | ||
| λ | carries a frameshift mutation relative to Ur-lambda | Hendrix and Duda, 1992 |
| λΔorf63 | Φ24B phage with deletion of orf63 | Licznerska et al., 2016 |
| Φ24B | λstx2::cat | Allison, 2003 |
| Φ24BΔorf63 | Φ24B phage with deletion of vb_24B_8c, the homolog of λorf63 | Licznerska et al., 2016 |
| PLASMIDS | ||
| pUC18 | ori pMB1 (pBR322 derivative), bla, AmpR | Thermo fisher scientific |
| pSB_orf63_λ | as pUC18 but bearing the orf63 from phage λ | This study |
| pSB_orf63_Φ24B | as pUC18 but bearing the orf63 from phage Φ24B | This study |
Media and growth conditions
All in vivo experiments were performed in LB liquid medium (Sigma—Aldrich) supplemented with 10 mM MgSO4 (phage λ or phage λΔorf63) or with 10 mM MgSO4 and 10 mM CaCl2 (phage Φ24B or phage Φ24BΔorf63), and with 50 μg/ml ampicillin (if necessary) (Sigma—Aldrich). To stimulate Orf63 protein production from the recombinant pUC18 derivatives, overnight bacterial cultures were diluted 1:100 in fresh LB medium and treated with IPTG (A&A Biotechnology) to a final concentration of 1 mM. Then, host bacteria were grown in aeration condition, achieved by shaking, at 30°C to A600 = 0.1 or 0.2 (the optical density of bacterial cells was dependent on the experimental conditions described in the following chapters).
Double overlay plaque assay
Bacteriophage titration was performed on the standard Petri dishes (Alchem) filled with 25 ml of LB agar (1.5% agar; Sigma—Aldrich), according to a procedure described by Sambrook and Russell (2001), with some modification. The top layer was prepared by mixing 2 ml of LB agar (0.7% agar; Sigma—Aldrich) with 1 ml of the overnight bacterial cell culture. To obtain visible plaques formed by Stx phages, the bottom agar was supplemented with sublethal concentration of chloramphenicol (Sigma—Aldrich). This antibiotic was effective in increasing of size of plaques of phage Φ24B and its derivative, which possessed in genomes chloramphenicol resistance gene (Table 1). As described previously (Loś et al., 2008a), the cm gene expression, especially after phage infection of E. coli bacteria, may have the positive influence on cellular productivity by decreasing the inhibitory effects of the antibiotic on protein synthesis. To determine the number of phages per ml of suspension (PFU/ml), serial 10-fold dilutions were prepared in TM buffer (10 mM Tris–HCl, 10 mM MgSO4; pH 7.2). Then, appropriate volume of each dilution of phage lysate was spotted onto double agar layer. The plates were incubated at 37°C overnight, plaques were counted, and the phage titer was calculated.
One-step growth experiments in phage-infected bacteria
To investigate the intracellular lytic development of lambdoid phages the one-step-growth experiment was prepared using the method described by Wegrzyn et al. (1995), with a minor modification (Bloch et al., 2014; Nejman-Falenczyk et al., 2015). Host bacteria were grown in LB medium at 30°C to A600 = 0.2. In the next step, 10 ml of a bacterial culture was centrifuged (2,000 × g for 10 min at 4°C). The pellet was suspended in 1 ml of LB medium supplemented with 3 mM sodium azide (Sigma—Aldrich). Bacteriophages were added to E. coli cells to m.o.i. of 0.05. After 10 min incubation at 30°C, unadsorbed phages were removed by three times washing in LB medium with 3 mM sodium azide (2,000 × g for 10 min at 4°C). Then, 25 μl of the suspension was added to 25 ml of LB medium prewarmed to 30°C (time 0) and cultivated in an incubator shaker. The number of infection centers were determined at times: 5, 10, 15 min after infection by mixing 0.1 ml of the sample with 0.9 ml of an overnight culture of appropriate indicator bacteria and 2 ml of top agar. Next, the mixture was poured onto LB agar plate (phages λ and λΔorf63) or LB agar plate with 2.5 μg/ml chloramphenicol (phage Φ24B and Φ24BΔorf63). Samples taken at later times were treated with chloroform (POCH), shaken vigorously and cleared by centrifugation (2,000 × g for 5 min at RT). The phage lysate was diluted in TM buffer and titrated under permissive condition. Plates were incubated at 37°C overnight. The number of viruses released from each infected cell (burst size) was calculated as a ratio of phage titer to the titer of infection centers.
Prophage induction with hydrogen peroxide
Bacteria lysogenic for lambdoid phages were grown in LB medium at 30°C to A600 = 0.1. Next, the culture was divided into two aliquots. One of them was treated with 1 mM hydrogen peroxide (Sigma—Aldrich) to provoke the prophage induction. The second one was a control without an induction agent. The cultivation was continued at 30°C. At indicated times samples were harvested, mixed with chloroform and vortexed for 1 min. The suspension was centrifuged for 5 min in a microfuge at RT. The supernatant was diluted in TM buffer and 2.5 μl of each serial dilution was dropped onto a freshly prepared double-layer LB agar in plastic Petri dishes. Plates were incubated at 37°C overnight. The relative phage titer was estimated by subtracting the phage titer determined in non-induced cultures from the phage titer estimated in induced cultures.
Survival of host bacteria after bacteriophage infection
To estimate the percentage of surviving cells after bacteriophage infection the procedure created by Sambrook and Russell (2001) was used, with a minor modification. A bacterial culture was grown at 30°C to A600 = 0.2. Then, 4 ml of the sample was centrifuged (2,000 × g for 10 min at 4°C). The supernatant was discarded and the pellet was washed with 0.85% sodium chloride (POCH) (2,000 × g for 10 min at 4°C). Finally, the bacterial pellet was suspended in 1 ml of LB medium supplemented with MgSO4 (phage λ and phage λΔorf63) or with MgSO4 and CaCl2 (phage Φ24B and Φ24BΔorf63) to a final concentration of 10 mM. The suspension was incubated for 30 min at 30°C and then phage particles were added to m.o.i. of 1, 5, 10. The mixture was kept for 15 min (phage λ and phage λΔorf63) or 30 min (phage Φ24B and Φ24BΔorf63) at 30°C. In the next step, serial dilutions in 0.85% sodium chloride were prepared and 40 μl of each dilution was spread on LB agar plates. After overnight incubation at 37°C, percentage of surviving E. coli bacteria was calculated relative to bacterial culture in which TM buffer was added instead of phage particles.
Efficiency of prophage formation after bacterial virus infection
Efficiency of lysogenization was estimated according to Arber et al. (1983) and Wegrzyn et al. (1992), with some modification. Host bacteria were cultured at 30°C to A600 = 0.2. Next, 1 ml of the sample was centrifuged (2,000 × g for 10 min at 4°C). Bacterial culture was washed with TCM buffer twice, and then pellet was suspended in the same buffer. Bacteriophages were added to bacterial cells to m.o.i. of 1, 5, 10. The mixture was incubated at 30°C. Then, serial dilutions were prepared and 20 μl of each suspension was spread on LB agar plates prior to overnight incubation at 37°C. The next day, 96 colonies were passaged in each well of a 96-well plate with 200 μl of LB medium and shaken at 37°C to A600 = 0.1. To estimate a percent of lysogens among survivors, bacterial cultures were treated with UV light at 50 J/m2 (the dose used routinely for lambdoid prophage induction) and incubated at 37°C for 2 h. Following induction, putative lysogens were mixed with chloroform, centrifuged (2,000 × g for 10 min at 4°C) and the water phase was spotted onto a double-layer LB agar (phage λ and phage λΔorf63) or a double-layer LB agar supplemented with chloramphenicol to a final concentration of 2.5 μg/ml (phage Φ24B and Φ24BΔorf63). Efficiency of lysogenization was calculated as a percent of lysogens relative to all tested bacterial cells. Lysogens were also infected with the same phage to check their resistance to superinfection, as described previously (Wegrzyn et al., 1992).
Prophage induction and extraction of RNA
Induction of tested prophages was provoked in lysogenic bacteria by addition of hydrogen peroxide to a final concentration of 1 mM. At the appropriate time, 109 bacterial cells were harvested, treated with 10 mM sodium azide and deep frozen in liquid nitrogen (this procedure was necessary to inhibit the growth of host bacteria). Total RNA from all samples were isolated with the High Pure RNA Isolation Kit (Roche Applied Science). To remove DNA from RNA preparations the TURBO DNA-free™ Kit (Life Technologies) was used. The quality and quantity of total isolated RNA were analyzed by a NanoDrop spectrophotometer and agarose gel electrophoresis. The contamination of DNA from RNA samples was also tested by routine PCR and qRT-PCR.
cDNA synthesis from an RNA template
To synthesize cDNA from an RNA template, the Transcriptor Reverse Transcriptase and random hexamer primers (Roche Applied Science) were used, according to the protocol supplied from the provider. 1.25 μg of the total RNA was taken for each reaction. Finally, mixture was diluted 10-fold and tested in qRT-PCR.
qRT-PCR assay and data analysis
The pattern of genes expression after prophage induction was performed by using the LightCycler® 480 Real-Time PCR System (Roche Applied Science), LightCycler® 480 SYBR Green I Master (Roche Applied Science) and cDNA samples. Transcription rates of genes of lambdoid bacteriophages were compared in parallel to the 16S rRNA housekeeping gene (according to a procedure described by Strauch et al. (2008), which expression was stable during prophage induction provoked by hydrogen peroxide. All primers were created by Primer3web version 4.0.0 and are listed in Table 2. Each reaction mixture consisted of: 2x SYBR Green I Master Mix, 6.25 ng/μl cDNA and 200 nM specific primers. qRT-PCR amplifications were performed for 55 cycles. To confirm the specificity of primers, melting curve for each product was analyzed. The relative changes in gene expressions were determined by E-Method and calculated by the following formula: Normalized relative ratio = Et CT (target) calibrator−CT (target) sample / Er CT (reference) calibrator−CT (reference) sample, where Et is the PCR efficiency of target and Er means the PCR efficiency of reference. The sample before the addition of the inductor (the time point “zero”) was a calibrator. The raw run data for tested lambdoid phages were transferred using the “LC480 Conversion: conversion of raw LC480 data” software and then, PCR efficiency for each gene was calculated by LinRegPCR program, which was successfully used previously (Bloch et al., 2014, 2015; Nejman-Falenczyk et al., 2015; Licznerska et al., 2016a).
Table 2.
Primers used for RT-Qpcr.
| Primer name | Sequence (5′ → 3′) |
|---|---|
| pF_λ_ea8.5 | GGGCAAGTATCGTTTCCACC |
| pR_ λ_ea8.5 | GCAATGTGCGAGAAATGACTG |
| pF_λ_ea22 | GCAGTTCCAGCACAATCGAT |
| pR_ λ_ea22 | AATGCATGACGACTGGGGAT |
| pF_λ_orf73 | CACTTCGAACCTCTCTGTTTACTG |
| pR_ λ_orf73 | CAGGGTTGTCGGACTTGTG |
| pF_λ_orf61 | TTAGCCTGACGGGCAATG |
| pR_ λ_orf61 | CCGACATGGGACTTGTTCA |
| pF_λ_orf60a | GCATACAGCCCCTCGTTTAT |
| pR_ λ_orf60a | CCGAAATCCACTGAAAGCAC |
| pF_λ_cIII | ATTCTTTGGGACTCCTGGCTG |
| pR_ λ_cIII | GTAAATTACGTGACGGATGGAAAC |
| pF_λ_N | CTCGTGATTTCGGTTTGCGA |
| pR_ λ_N | AAGCAGCAAATCCCCTGTTG |
| pF_λ_cro | ATGCGGAAGAGGTAAAGCCC |
| pR_ λ_cro | TGGAATGTGTAAGAGCGGGG |
| pF_λ_cII | TCGCAATGCTTGGAACTGAGA |
| pR_ λ_cII | CCCTCTTCCACCTGCTGATC |
| pF_λ_Q | TTCTGCGGTAAGCACGAAC |
| pR_ λ_Q | TGCATCAGATAGTTGATAGCCTTT |
| pF_λ_R | ATCGACCGTTGCAGCAATA |
| pR_ λ_R | GCTCGAACTGACCATAACCAG |
| pF_Φ24B_ea22 | TCAGCAACATGGCATTCACT |
| pR_ Φ24B_ea22 | GGTTGGGAAGCTGAGAGTTG |
| pF_Φ24B_orf73 | CGAACCTCTCTGTTTACTGATAAGC |
| pR_ Φ24B_orf73 | TTCAGGGTTGTCGGACTTGT |
| pF_Φ24B_orf61 | TTAGCCTGACGGGCAATG |
| pR_ Φ24B_orf61 | CCGACATGGGACTTGTTCA |
| pF_Φ24B_orf60a | CATACAGCCCCTCGTTTAT |
| pR_ Φ24B_orf60a | CCGAAATCCACTGAAAGCAC |
| pF_Φ24B_cIII | ATTCTTTGGGACTCCTGGCTG |
| pR_Φ24B_cIII | GTAAATTACGTGACGGATGGAAAC |
| pF_Φ24B_N | AGGCGTTTCGTGAGTACCTT |
| pR_ Φ24B_N | TTACACCGCCCTACTCTAAGC |
| pF_Φ24B_cro | CGAAGGCTTGTGGAGTTAGC |
| pR_ Φ24B_cro | GTCTTAGGGAGGAAGCCGTT |
| pF_Φ24B_cII | TGATCGCGCAGAAACTGATTTAC |
| pR_Φ24B_cII | GACAGCCAATCATCTTTGCCA |
| pF_Φ24B_O | AAGCGAGTTTGCCACGAT |
| pR_Φ24B_O | GAACCCGAACTGCTTACCG |
| pF_Φ24B_Q | GGGAGTGAGGCTTGAGATGG |
| pR_ Φ24B_Q | TACAGAGGTTCTCCCTCCCG |
| pF_Φ24B_R | GGGTGGATGGTAAGCCTGT |
| pR_ Φ24B_R | TAACCCGGTCGCATTTTTC |
| pF_E.coli_16SrRNA | CCTTACGACCAGGGCTACAC |
| pR_ E.coli_16SrRNA | TTATGAGGTCCGCTTGCTCTC |
Statistical analysis
Each experiment was repeated three times and variation among replicates was presented as the error bars indicating the standard deviation (SD). All data comparisons were made by using Student's t-test. Significant differences were marked by asterisks when P < 0.05 (*) or P < 0.01 (**).
Results
The oligomeric state of Orf63
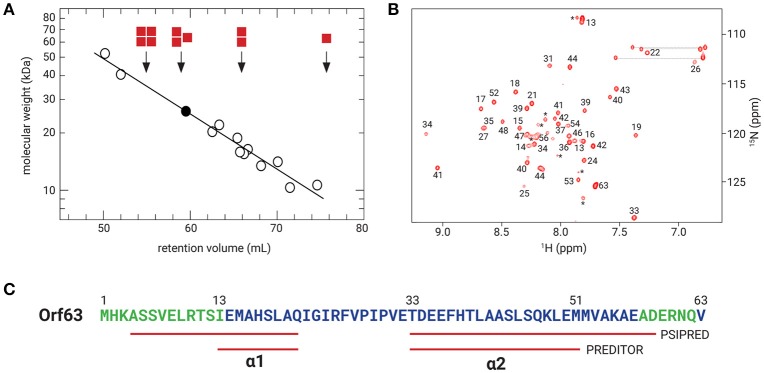
Samples from four independent preparations of 6xHis-Flag tagged Orf63 (15 aa. tag + 63 aa. protein) eluted as one peak on a preparative gel filtration column with an average retention volume of 59.5 mL corresponding to an apparent molecular weight of ~26 kDa (Figure 2A). Since affinity-tagged Orf63 is only 9 kDa, the gel filtration results suggest that Orf63 is oligomeric with a trimer as the most plausible configuration. This estimate is most accurate if Orf63 has the characteristics of a globular protein to match the standards used.
Figure 2.
Orf63 structural insights. (A) The retention volumes for a set of proteins ranging from 10–67 kDa were plotted against molecular weight to produce the log-linear relationship shown (open circles). The Kav value observed for Orf63 corresponds to an apparent molecular weight of ~28 kDa (closed circle). For reference, red squares denote the expected retention volumes of monomeric, dimeric, trimeric, and tetrameric species of the 9 kDa affinity tagged Orf63 protein. (B) A representative 1H-15N HSQC spectrum of Orf63 at 37°C demonstrating the characteristic resonance dispersion of a folded protein. Unassigned residues are labeled with an asterisk. Lines denote the amino resonances of three asparagine and glutamine side chains. (C) A conventional strategy was used to assign the backbone (HN, N, CA, CB, C') chemical shifts of Orf63. The extent of these assignments is colored blue on the sequence. The chemical shift data were then used as input to PREDITOR (Berjanskii et al., 2006) which identified two helices labeled α1 and α2. Using sequence data alone as input to PSIPRED (McGuffin et al., 2000), longer helices are predicted.
The orf63 gene encodes a folded protein
Consistent with the observation that Orf63 is oligomeric in solution, NMR spectra of Orf63 at 298 K (25°C) suffered from considerable resonance line broadening that was characteristic of proteins >20 kDa in overall molecular weight. Consequently, a higher temperature of 310 K (37°C) was chosen for all NMR studies to increase the tumbling time of the protein that, in turn, improves the sensitivity of triple resonance experiments. In Figure 2B, a 1H-15N HSQC spectrum is presented. The amide resonances in this two-dimensional spectrum are disperse indicating that the protein is folded. The combined analysis of several triple resonance (1H, 13C, 15N) spectra lead to the determination of backbone (HN, N, CA, CB, C') chemical shift assignments for residues 14–52 of Orf63. Resonances for the amino terminal affinity tags and from residue 53 onwards to the carboxy-terminus were either not observed or unassignable. Thus, the NMR data suggest that the folded region of Orf63 includes from residues 14–52.
Structural characteristics of Orf63
Several statistical methods are available to predict secondary structure from backbone chemical shift data with a high degree of accuracy. As shown in Figure 2C, two helices are predicted (α1: 13–21; α2: 33–50). The secondary structure determined from chemical shift data is consistent with the secondary structure of Orf63 predicted from sequence information alone, although the helical boundaries are different.
The sequence of putative orf63 products is conserved among lambdoid bacteriophages
Since experiments shown in Figure 2 indicated that orf63 encodes a protein, we have tested similarity of the putative proteins encoded by orf63 of different lambdoid bacteriophages. Thus, scores of pairwise alignments of the predicted amino acid sequences of orf63 from six such phages have been calculated. As demonstrated in Table 3, all these putative proteins are similar to each other. This indicate that the high similarity is kept at the protein level of Orf63 of lambdoid bacteriophages.
Table 3.
Scores of pairwise alignments of the predicted amino acid sequences of orf63 from six analyzed lambdoid phages: λ phage (NC_001416), Φ24B phage (HM208303), 933W phage (NC_000924), VT2 Sakai phage (AP000422), Stx1 converting phage (NC_004913), and Stx2 converting phage II (NC_004914).
| λ | Φ24B | 933W | VT2 Sakai | Stx1 | Stx2_II | |
|---|---|---|---|---|---|---|
| λ | 86 | 86 | 87 | 87 | 87 | |
| Φ24B | 100 | 98 | 98 | 98 | ||
| 933W | 98 | 98 | 98 | |||
| VT2 Sakai | 100 | 100 | ||||
| Stx1 | 100 | |||||
| Stx2_II |
Pairwise scores are simply the number of identities between the two sequences, divided by the length of the alignment, and represented as a percentage. The multiple sequence alignment was performed using the ClustalW algorithm.
Efficiency of lysogenization and prophage induction in the absence of orf63
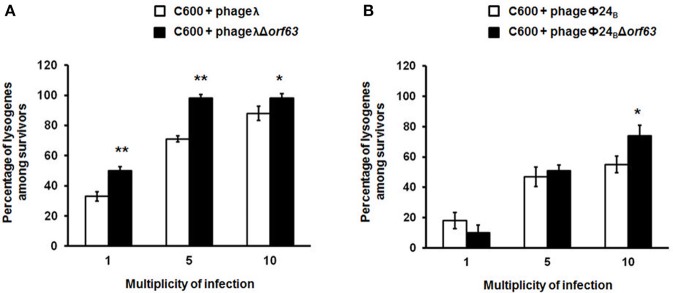
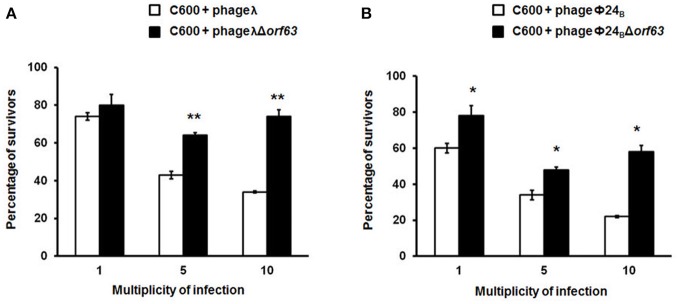
Since previous studies suggested that genes from the exo-xis region might be involved in the regulation of bacteriophage development (Bloch et al., 2013, 2014; Licznerska et al., 2016a), we have tested two crucial controlled steps in the lambdoid phage life cycle, the lysis-vs.-lysogenization decision, and prophage induction. We found that lysogenization efficiency was significantly increased in bacteriophages λ and Φ24B devoid of orf63 (Figures 3A,B, respectively) though this phenomenon was more pronounced in λ (the effects were seen at all tested m.o.i.) (Figure 3A) than in Φ24B (significant effects observed only at m.o.i. = 10) (Figure 3B). Also, survival rates of bacterial cells (i.e., cells lysogenized and not infected) in populations infected with Δorf63 or Φ24B Δorf63 were higher than those in experiments with wild-type λ or Φ24B (Figures 4A,B, respectively), supporting the conclusion that lysogenization is more effective for the mutant, indeed.
Figure 3.
Efficiency of lysogenization of E. coli C600 strain with lambdoid bacteriophages: λ and Φ24B (□ in (A,B), respectively) or their deletion mutants λΔorf63 and Φ24BΔorf63 (■ in A,B, respectively). Results are presented as mean values ±SD from three independent experiments. Statistical analysis (t test) was performed for results from each m.o.i. (multiplicity of infection) between wild type phage and its deletion mutant. Significant differences are marked by asterisks P < 0.05 (*) or P < 0.01 (**).
Figure 4.
Survival (%) of the wild-type strain E. coli C600, after infection with lambdoid bacteriophages: λ and Φ24B (□ in (A,B), respectively) or their deletion mutants λΔorf63 and Φ24BΔorf63 (■ in A,B, respectively). Mean values from three independent experiments ±SD are shown. Statistical analysis were performed for each m.o.i. by t test. The significance of differences between fractions of bacterial cells surviving the infection with λ and λΔorf63 as well as Φ24B and Φ24BΔorf63 are observed and marked by asterisks P < 0.05 (*) or P < 0.01 (**).
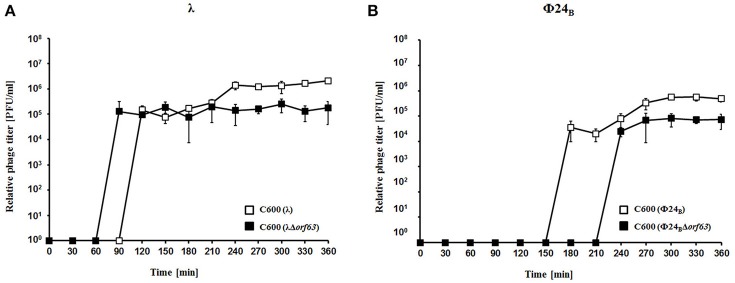
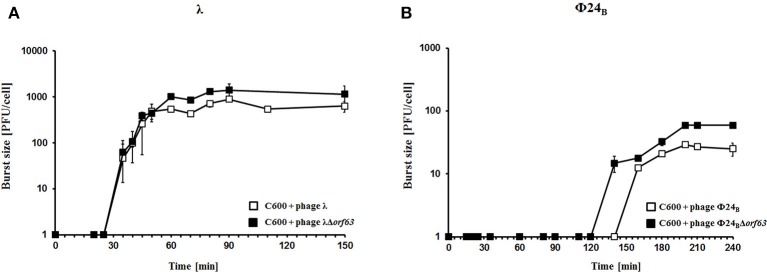
To test efficiency of prophage induction, we have estimated the number of phages appearing after prophage induction with hydrogen peroxide (one of natural prophage inducers occurring in human intestine, the common habitat of E. coli). Deletion of orf63 caused a lower phage titer after prophage induction for both λ and Φ24B (Figures 5A,B, respectively). However, when measured kinetics of phage development following infection of E. coli cells at low m.o.i. (0.05), we found that phages λ and Φ24B devoid of orf63 gave even more progeny per infected cell than their wild-type counterparts (Figures 6A,B, respectively). Therefore, combining results of experiments presented in Figures 5, 6, one can conclude that deletion of orf63 influences efficiency of prophage induction in both λ and Φ24B. Since formation of progeny phages is definitely not impaired in the absence of orf63 when lytic development starts after infection, we suggest that lower phage titer after prophage induction indicates lower efficiency of this process in phages devoid of this gene.
Figure 5.
Development of λ and λΔorf63 (A) or Φ24B and Φ24BΔorf63 (B) bacteriophages after prophage induction with hydrogen peroxide at 30 °C. E. coli C600 bacteria were either lysogenic with wild-type phages λ and Φ24B (□ in A,B, respectively) or their deletion mutants λΔorf63 and Φ24BΔorf63 (■ in (A,B, respectively). Phage lytic development was initiated by addition of H2O2 to final concentration of 1 mM at time 0. The presented results are mean values from three independent experiments with error bars indicating SD (note that in the most cases, the bars are smaller than sizes of symbols). Results are shown as PFU (plaque forming units) per one ml.
Figure 6.
Development of λ and λΔorf63 (A) or Φ24B and Φ24BΔorf63 (B) bacteriophages following phage infection of E. coli bacteria. Host E. coli strains were infected with wild-type phages λ and Φ24B (□ in A,B, respectively) or their deletion mutants λΔorf63 and Φ24BΔorf63 (■ in A,B, respectively) at time 0. The presented results are mean values ±SD from three independent experiments. Results are shown as PFU (plaque forming units) per cell.
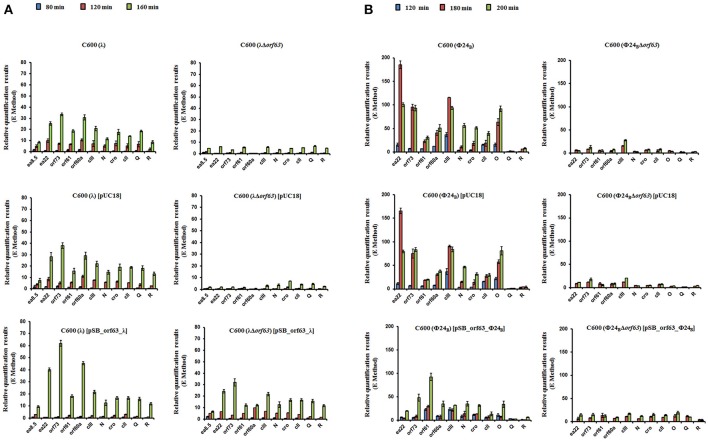
Deletion of Orf63 influences expression of genes of λ and Φ24B phages
Since experiments described above indicated that orf63 function is involved in the regulation of lysogenization and prophage induction in both λ and Φ24B, we aimed to measure expression of selected bacteriophage genes in E. coli cells after hydrogen peroxide-provoked prophage induction. Reverse transcription quantitative real time PCR (RT-qPCR) was used to assess abundance of particular transcripts. We have measured expression levels of genes from the exo-xis region (ea8.5, ea22, orf73, orf61, orf60a) and some key regulatory genes of λ and Φ24B, i.e., N, cro, cII, Q, R. We found that expression of all tested genes was significantly impaired in Δorf63 mutants of both λ and Φ24B relative to wild-type phages at all tested times after prophage induction (Figures 7A,B, respectively). These results confirm that prophage induction is significantly impaired in the absence of orf63, and suggest a regulatory role for the orf63 gene product in the control of expression of phage genes. In the case of phage λ, complementation of the Δorf63 mutation by overexpression of wild-type orf63 from a plasmid was successful, at least at certain times after prophage induction (Figure 7A). However, we failed to obtain such a complementation in phage Φ24B (Figure 7B). This might suggest that specific ratio(s) of Orf63 is/are required for accurate regulation of phage development.
Figure 7.
Levels of transcripts of indicated genes of wild type phages λ and Φ24B and their deletion mutants in the absence or presence of control plasmid [pUC18] or plasmid bearing the deleted orf63 [pSB_orf63], assessed by quantitative reverse RT-PCR analysis, aftser prophage induction with 1 mM H2O2 in E. coli C600 host at 30°C. Levels of transcripts corresponding to particular genes were determined at following times after induction: 80 (blue), 120 (red) or 160 (green) minutes in case of phages λ and λΔorf63 (A), and 120 (blue), 180 (red) or 200 (green) minutes in case of phages Φ24B and Φ24BΔorf63 (B). Results are presented as mean values from three independent experiments with error bars indicating SD.
Discussion
Considering that the complete 48,502 bp genome of λ was achieved in 1983, it is astounding that this well-investigated model virus still contains uncharacterized open reading frames, many of which lie between the exo, and xis genes. We began this investigation by demonstrating that orf63 found within the exo-xis region, encodes a bona fide protein both in structural, and functional terms. Consistent with its name, this small 63 aa. protein is comprised of only two helices that cover most of the available sequence. Analytical gel filtration of purified Orf63 suggests that the oligomeric state is a trimer. If Orf63 deviates significantly from a globular shape, it is possible that the molecular weight may be overestimated by the gel filtration assay, the oligomeric state could be a dimer. However, given the significant line broadening observed during a series of initial NMR based surveys performed at 25°C that could only be alleviated by performing all of the studies subsequently at 37°C, the NMR data tend to corroborate the gel filtration findings.
Since Orf63 appears to be a functional protein, we tested effects of deletion of orf63 on development of bacteriophages λ and Φ24B. In the absence of functional Orf63, we observed a significant increase in the efficiency of lysogenization, and considerable lower efficiency of hydrogen peroxide-mediated prophage induction. These results may suggest that Orf63 is involved in the regulation of expression of specific phage genes. Studies with the use of RT-qPCR revealed that expression of vast majority of crucial regulatory genes, as well as genes from the exo-xis region, is significantly influenced by the absence of orf63. Moreover, perhaps specific ratio of Orf63 to other regulators is required, as it was impossible to obtain complementation with the wild-type orf63 expressed from a plasmid in Φ24B, though it was successful in λ. The hypothesis about the requirement of specific Orf63 ratio(s) for accurate regulation of phage development is supported by impaired expression of Φ24B genes during overexpression of orf63.
A previous yeast two-hybrid study identified the transcription factor YqhC as a possible protein partner of Orf63 (Blasche et al., 2013). YqhC is interesting because it is a transcription factor that promotes the synthesis of YqhD, the major enzyme responsible for detoxifying compounds produced from glucose under conditions of oxidative stress (Lee et al., 2010). Among the compounds that YqhD acts upon are 2-oxoaldehydes, toxic and highly reactive products of oxidative stress on the bacterium formed from glucose. It has been proposed that oxoaldehydes are one class of compound that is capable of inducing a wider stress response through SoxRS (Benov and Fridovich, 2002). Since neutrophils mount a vigorous oxidative attack during a STEC infection, Orf63 may be beneficial to the bacteriophage by manipulating the microenvironment of the bacterial host. Orf63 by binding the transcriptional activator YqhC, may prevent it from attenuating the stress response created by neutrophil mediated attack on EHEC strains in the gut, and promoting a transition to the lytic phase commensurate with the activation of associated phage genes in that response. A putative mechanism for Orf63-mediated modulation of prophage induction and phage genes' expression is presented in Figure 8. Notwithstanding its role in protein-protein interactions, it is still possible that Orf63 itself could function as a transcriptional regulator, although its small size, lack of a known DNA binding domain, and relatively few basic amino acids argue against this possibility. A high-resolution structure of Orf63 alone, or in complex with a possible interactor like YqhC, will resolve these outstanding questions.
Figure 8.
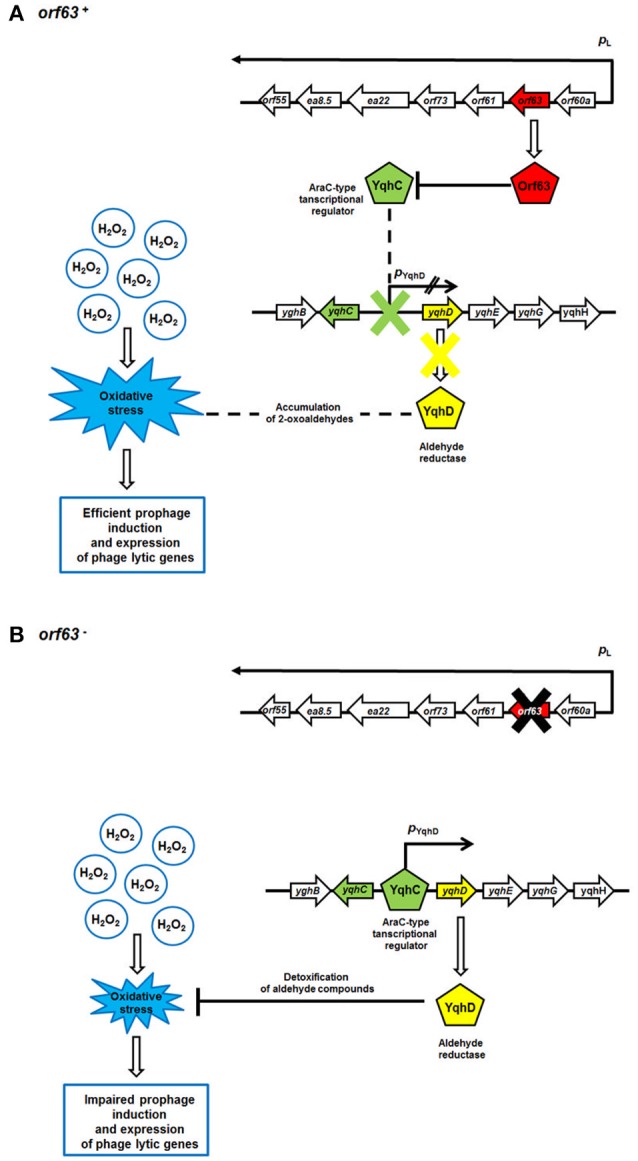
A model for Orf63-mediated modulation of bacteriophage development. Mechanisms operating in the presence (A) or absence (B) of the orf63 function are shown. Transcripts are shown as black arrows, with arrowheads indicating directionality of transcription. The lack of the activity of the pYghD promoter is marked as two black inclined lines. Phage and E. coli (host) proteins are presented as pentagons. Stimulations of processes or protein production/activity are shown by open arrows. Negative regulations of processes or protein functions are represented by blunt-ended lines.
In conclusion, Orf63 is a folded protein and has structural properties suggesting its regulatory role. Analyses of mutants of bacteriophages λ and Φ24B devoid of orf63 indicated its function in the control of bacteriophage development at the stages of lysis vs. lysogenization decision and prophage induction. Further studies will include determination of biochemical properties of Orf63 and its possible interactions with phage- and/or host-encoded proteins, as well as with phage DNA, to understand molecular mechanisms of its function as a modulator of phage development.
Author contributions
AD and SB contributed equally to this work. AD, SB, BN, GT, TG, and AN took part in the physiological studies on bacteria and phages and data processing. AD, SB, and BN participated also in writing the manuscript and planning of the study. AR, SP, and LD performed the protein analyses. GW, LD, and AW were engaged in writing the manuscript and took part in planning of the study and discussions.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer RO and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
This work was supported by the National Science Center (Poland), grants no. UMO-2013/09/B/NZ2/02366 and UMO-2015/17/B/NZ9/01724 to AW, and by the Faculty of Biology, University of Gdansk, grant no. 538-L140-B539-17 to AD. The Natural Sciences and Engineering Research Council (NSERC) is acknowledged for support to LD (Discovery Operating Grant 238924), and AR/SP (USRA training awards).
References
- Allison H. E. (2003). Immunity profiles of wild-type and recombinant Shiga-like toxin-encoding bacteriophages and characterization of novel double lysogens. Infect. Immun. 71, 3409–3418. 10.1128/IAI.71.6.3409-3418.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleyard R. K. (1954). Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics 39, 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber W., Enquist L., Hohn B., Murray N. E., Murray K. (1983). Lambda II, in Experimental Methods for Use with λ, eds Roberts J. W., Stahl F. W., Weisberg R. A. (New York, NY: Cold Spring Harbor Laboratory Press; ), 433–466. [Google Scholar]
- Benov L., Fridovich I. (2002). Induction of the soxRS regulon of Escherichia coli by glycolaldehyde. Arch. Biochem. Biophys. 407, 45–48. 10.1016/S0003-9861(02)00498-8 [DOI] [PubMed] [Google Scholar]
- Berjanskii M. V., Neal S., Wishart D. S. (2006). PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 34, W63–W69. 10.1093/nar/gkl341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasche S., Wuchty S., Rajagopala S. V., Uetz P. (2013). The protein interaction network of bacteriophage lambda with its host, Escherichia coli. J. Virol. 87, 12745–12755. 10.1128/JVI.02495-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch S. K., Felczykowska A., Nejman-Falenczyk B. (2012). Escherichia coli O104:H4 out break- have we learnt a lesson from it? Acta Biochim. Pol. 59, 483–488. [PubMed] [Google Scholar]
- Bloch S., Nejman-Falenczyk J., Loś J. M., Baranska S., Lepek K., Felczykowska A., et al. (2013). Genes from the exo-xis region of λ and Shiga toxin-converting bacteriophages influence lysogenization and prophage induction. Arch. Microbiol. 195, 693–703. 10.1007/s00203-013-0920-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch S., Nejman-Falenczyk B., Dydecka A., Loś J. M., Felczykowska A., Wegrzyn A., et al. (2014). Different expression patterns of genes from the exo-xis region of bacteriophage λ and Shiga toxin-converting bacteriophage Φ24B following infection or prophage induction in Escherichia coli. PLoS ONE 9:e108233 10.1371/journal.pone.0108233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch S., Nejman-Falenczyk B., Topka G., Dydecka A., Licznerska K., Narajczyk M., et al. (2015). UV-Sensitivity of Shiga Toxin-Converting Bacteriophage Virions Φ24B, 933W, P22, P27 and P32. Toxins 7, 3727–3739. 10.3390/toxins7093727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. 10.1007/BF00197809 [DOI] [PubMed] [Google Scholar]
- Hendrix R. W., Duda R. L. (1992). Bacteriophage lambda PaPa: not a mother of all lambda phages. Science 258, 1145–1148. 10.1126/science.1439823 [DOI] [PubMed] [Google Scholar]
- Hyberts S. G., Milbradt A. G., Wagner A. B., Arthanari H., Wagner G. (2012). Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR 52, 315–327. 10.1007/s10858-012-9611-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. F. (1993). The Escherichia coli K-12 wild types W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bact. 175, 3401–3407. 10.1128/jb.175.11.3401-3407.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan J. J., Smirnova E., Khazai S., Evanics F., Maxwell K. L., Donaldson L. W. (2013). The solution structures of two prophage homologues of the bacteriophage λ Ea8.5 protein reveal a newly discovered hybrid homeodomain/zinc-finger fold. Biochemistry 52, 3612–3614. 10.1021/bi400543w [DOI] [PubMed] [Google Scholar]
- Lee C., Kim I., Lee J., Lee K. L., Min B., Park C. (2010). Transcriptional activation of the aldehyde reductase YqhD by YqhC and its implication in glyoxal metabolism of Escherichia coli K-12. J. Bacteriol. 192, 4205–4214. 10.1128/JB.01127-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licznerska K., Dydecka A., Bloch S., Topka G., Nejman-Falenczyk B., Wegrzyn A., et al. (2016a). The Role of the exo-xis region in oxidative stress-mediated induction of Shiga toxin-converting prophages. Oxid. Med. Cell. Longev. 2016:8453135. 10.1155/2016/8453135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licznerska K., Nejman-Falenczyk B., Bloch S., Dydecka A., Topka G., Gasior T., et al. (2016b). Oxidative stress in Shiga toxin production by enterohemorrhagic Escherichia coli. Oxid. Med. Cell. Longev. 2016:3578368. 10.1155/2016/3578368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loś J. M., Golec P., Wegrzyn G., Wegrzyn A., Loś M. (2008a). Simple method for plating Escherichia coli bacteriophages forming very small plaques or no plaques under standard conditions. Appl. Environ. Microbiol. 74, 5113–5120. 10.1128/AEM.00306-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loś J. M., Loś M., Wegrzyn A., Wegrzyn G. (2008b). Role of the bacteriophage λ exo-xis region in the virus development. Folia Microbiol. 53, 443–450. 10.1007/s12223-008-0068-0 [DOI] [PubMed] [Google Scholar]
- McGuffin L. J., Bryson K., Jones D. T. (2000). The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405. 10.1093/bioinformatics/16.4.404 [DOI] [PubMed] [Google Scholar]
- Mizutani S., Nakazono N., Sugino Y. (1999). The so-called chromosomal verotoxin genes are actually carried by defective prophages. DNA Res. 6, 141–143. 10.1093/dnares/6.2.141 [DOI] [PubMed] [Google Scholar]
- Muniesa M., Hammerl J. A., Hertwig S., Appel B., Brüssow H. (2012). Shiga toxin-producing Escherichia coli O104:H4: a new challenge for microbiology. Appl. Environ. Microbiol. 78, 4065–4073. 10.1128/AEM.00217-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejman-Falenczyk B., Bloch S., Licznerska K., Dydecka A., Felczykowska A., Topka G., et al. (2015). A small, microRNA-size, ribonucleic acid regulating gene expression and development of Shiga toxin-converting bacteriophage Φ24B. Sci. Rep. 5:10080 10.1038/srep1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro J. P., Kaper J. B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley L. W., Remis R. S., Helgerson S. D., McGee H. B., Wells J. G., Davis B. R. (1983). Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308, 681–685. 10.1056/NEJM198303243081203 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schmidt H. (2001). Shiga-toxin-converting bacteriophages. Res. Microbiol. 152, 687–695. 10.1016/S0923-2508(01)01249-9 [DOI] [PubMed] [Google Scholar]
- Skinner S. P., Goult B. T., Fogh R. H., Boucher W., Stevens T. J., Laue E. D., et al. (2015). Structure calculation, refinement and validation using CcpNmr Analysis. Acta Crystallogr. D Biol. Crystallogr. 71, 154–161. 10.1107/S1399004714026662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch E., Hammerl J. A., Konietzny A., Schneiker-Bekel S., Arnold W., et al. (2008). Bacteriophage 2851 is a prototype phage for dissemination of the Shiga toxin variant gene 2c in Escherichia coli O157:H7. Infect. Immun. 76, 5466–5477. 10.1128/IAI.00875-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn G., Glass R. E., Thomas M. S. (1992). Involvement of the Escherichia coli RNA polymerase α subunit in transcriptional activation by the bacteriophage λ CI and CII proteins. Gene 122, 1–7. 10.1016/0378-1119(92)90025-K [DOI] [PubMed] [Google Scholar]
- Wegrzyn G., Wegrzyn A., Konieczny I., Bielawski K., Konopa G., Obuchowski M., et al. (1995). Involvement of the host initiator function dnaA in the replication of coliphage lambda. Genetics 139, 1469–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn G., Licznerska K., Wegrzyn A. (2012). Phage λ–new insights into regulatory circuits. Adv. Virus. Res. 82, 155–178. 10.1016/B978-0-12-394621-8.00016-9 [DOI] [PubMed] [Google Scholar]