Abstract
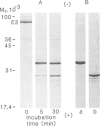
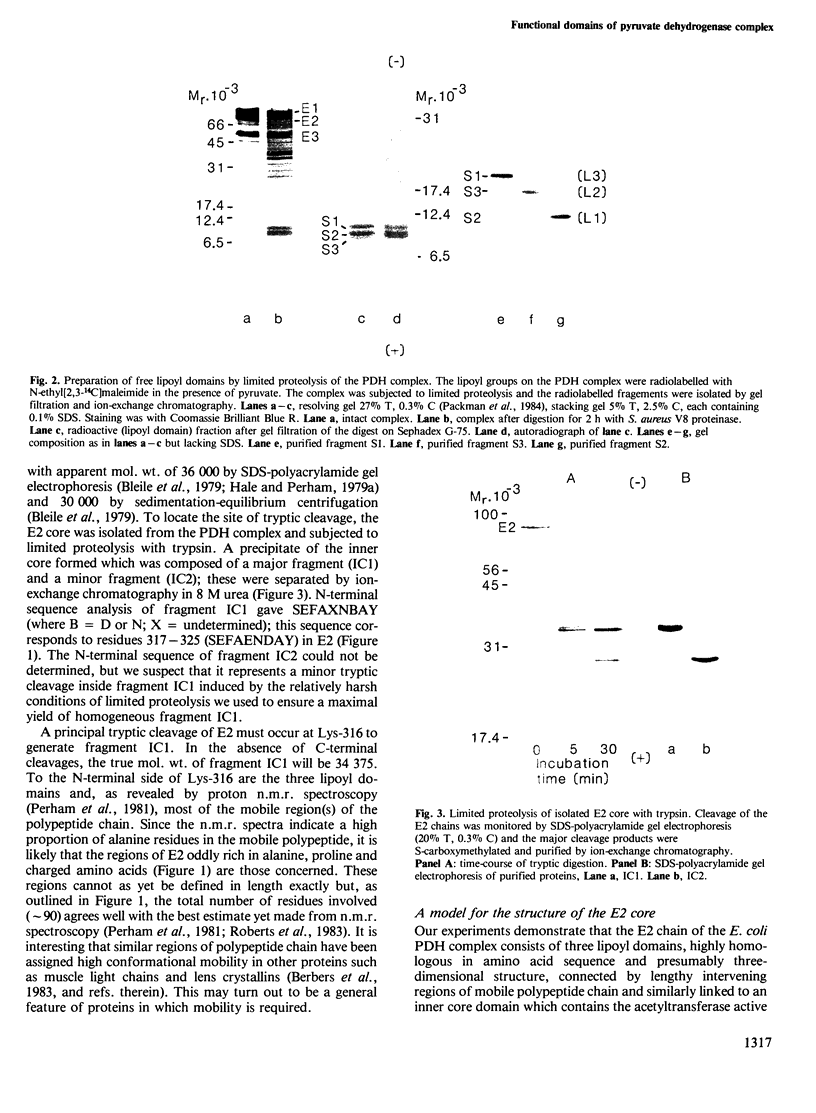
Each polypeptide chain in the lipoate acetyltransferase (E2) core of the pyruvate dehydrogenase complex from Escherichia coli contains three repeating sequences in the N-terminal half of the molecule. The repeats are highly homologous in primary structure and each includes a lysine residue that is a potential site for lipoylation. We have shown that all three sites are lipoylated, at least in part, and that the three lipoylated segments of the E2 chain can be isolated as distinct functional domains after limited proteolysis. Each domain becomes partly acetylated in the intact complex in the presence of substrate. In the primary structure, the domains are separated by regions of polypeptide chain oddly rich in alanine and proline residues. These regions are probably the conformationally mobile segments observed in the 1H-n.m.r. spectrum of the complex and which are removed by tryptic cleavage at Lys-316. The C-terminal half of the molecule contains the acetyltransferase active site and the binding sites for E1, E3 and other E2 subunits. The pyruvate dehydrogenase complex of E. coli, which has a heterogeneous quaternary structure, is thus far unique among the 2-oxo acid dehydrogenase complexes in possessing more than one lipoyl domain per E2 chain, but this may be a general feature of the enzyme from Gram-negative organisms.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bates D. L., Danson M. J., Hale G., Hooper E. A., Perham R. N. Self-assembly and catalytic activity of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Nature. 1977 Jul 28;268(5618):313–316. doi: 10.1038/268313a0. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Harrison R. A., Perham R. N. The stoichiometry of polypeptide chains in the pyruvate dehydrogenase multienzyme complex of E. coli determined by a simple novel method. FEBS Lett. 1975 Dec 15;60(2):427–430. doi: 10.1016/0014-5793(75)80764-2. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Thomas J. O. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 1981 Nov 25;9(22):5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbers G. A., Hoekman W. A., Bloemendal H., de Jong W. W., Kleinschmidt T., Braunitzer G. Proline- and alanine-rich N-terminal extension of the basic bovine beta-crystallin B1 chains. FEBS Lett. 1983 Sep 19;161(2):225–229. doi: 10.1016/0014-5793(83)81013-8. [DOI] [PubMed] [Google Scholar]
- Berman J. N., Chen G. X., Hale G., Perham R. N. Lipoic acid residues in a take-over mechanism for the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochem J. 1981 Dec 1;199(3):513–520. doi: 10.1042/bj1990513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleile D. M., Hackert M. L., Pettit F. H., Reed L. J. Subunit structure of dihydrolipoyl transacetylase component of pyruvate dehydrogenase complex from bovine heart. J Biol Chem. 1981 Jan 10;256(1):514–519. [PubMed] [Google Scholar]
- Bleile D. M., Munk P., Oliver R. M., Reed L. J. Subunit structure of dihydrolipoyl transacetylase component of pyruvate dehydrogenase complex from Escherichia coli. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4385–4389. doi: 10.1073/pnas.76.9.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma H. J., de Graaf-Hess A. C., de Kok A., Veeger C., Visser A. J., Voordouw G. Pyruvate dehydrogenase complex from Azotobacter vinelandii: structure, function, and inter-enzyme catalysis. Ann N Y Acad Sci. 1982;378:265–286. doi: 10.1111/j.1749-6632.1982.tb31202.x. [DOI] [PubMed] [Google Scholar]
- Chang J. Y. Manual micro-sequence analysis of polypeptides using dimethylaminoazobenzene isothiocyanate. Methods Enzymol. 1983;91:455–466. doi: 10.1016/s0076-6879(83)91043-1. [DOI] [PubMed] [Google Scholar]
- Coggins J. R., Hooper E. A., Perham R. N. Use of dimethyl suberimidate and novel periodate-cleavable bis(imido esters) to study the quaternary structure of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochemistry. 1976 Jun 15;15(12):2527–2533. doi: 10.1021/bi00657a006. [DOI] [PubMed] [Google Scholar]
- Collins J. H., Reed L. J. Acyl group and electron pair relay system: a network of interacting lipoyl moieties in the pyruvate and alpha-ketoglutarate dehydrogenase complexes from Escherichia coli. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4223–4227. doi: 10.1073/pnas.74.10.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danson M. J., Fersht A. R., Perham R. N. Rapid intramolecular coupling of active sites in the pyruvate dehydrogenase complex of Escherichia coli: mechanism for rate enhancement in a multimeric structure. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5386–5390. doi: 10.1073/pnas.75.11.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danson M. J., Hale G., Johnson P., Perham R. N., Smith J., Spragg P. Molecular weight and symmetry of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J Mol Biol. 1979 Apr 25;129(4):603–617. doi: 10.1016/0022-2836(79)90471-6. [DOI] [PubMed] [Google Scholar]
- Danson M. J., Perham R. N. Evidence for two lipoic acid residues per lipoate acetyltransferase chain in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochem J. 1976 Dec 1;159(3):677–682. doi: 10.1042/bj1590677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert G. A., Gilbert L. M. Detection in the ultracentrifuge of protein heterogeneity by computer modelling, illustrated by pyruvate dehydrogenase multienzyme complex. J Mol Biol. 1980 Dec 15;144(3):405–408. doi: 10.1016/0022-2836(80)90099-6. [DOI] [PubMed] [Google Scholar]
- Gray J. C., Hooper E. A., Perham R. N. Subunit stoichiometry of tobacco ribulose 1.5-bisphosphate carboxylase. FEBS Lett. 1980 Jun 2;114(2):237–239. doi: 10.1016/0014-5793(80)81123-9. [DOI] [PubMed] [Google Scholar]
- Hackert M. L., Oliver R. M., Reed L. J. A computer model analysis of the active-site coupling mechanism in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Proc Natl Acad Sci U S A. 1983 May;80(10):2907–2911. doi: 10.1073/pnas.80.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale G., Hooper E. A., Perham R. N. Amidination of pyruvate dehydrogenase complex of Escherichia coli under denaturing conditions. Biochem J. 1979 Jan 1;177(1):136–137. doi: 10.1042/bj1770136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale G., Hooper E. A., Perham R. N. Amidination of pyruvate dehydrogenase complex of Escherichia coli under denaturing conditions. Biochem J. 1979 Jan 1;177(1):136–137. doi: 10.1042/bj1770136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale G., Perham R. N. Amino acid sequence around lipoic acid residues in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochem J. 1980 Jun 1;187(3):905–908. doi: 10.1042/bj1870905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale G., Perham R. N. Primary structure of the swinging arms of the pyruvate dehydrogenase complex of Escherichia coli. FEBS Lett. 1979 Sep 15;105(2):263–266. doi: 10.1016/0014-5793(79)80625-0. [DOI] [PubMed] [Google Scholar]
- Henderson C. E., Perham R. N. Purificaton of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus and resolution of its four component polypeptides. Biochem J. 1980 Jul 1;189(1):161–172. doi: 10.1042/bj1890161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packman L. C., Perham R. N., Roberts G. C. Domain structure and 1H-n.m.r. spectroscopy of the pyruvate dehydrogenase complex of Bacillus stearothermophilus. Biochem J. 1984 Jan 1;217(1):219–227. doi: 10.1042/bj2170219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packman L. C., Stanley C. J., Perham R. N. Temperature-dependence of intramolecular coupling of active sites in pyruvate dehydrogenase multienzyme complexes. Biochem J. 1983 Aug 1;213(2):331–338. doi: 10.1042/bj2130331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perham R. N., Duckworth H. W., Roberts G. C. Mobility of polypeptide chain in the pyruvate dehydrogenase complex revealed by proton NMR. Nature. 1981 Jul 30;292(5822):474–477. doi: 10.1038/292474a0. [DOI] [PubMed] [Google Scholar]
- Perham R. N. Self-assembly of biological macromolecules. Philos Trans R Soc Lond B Biol Sci. 1975 Nov 6;272(915):123–136. doi: 10.1098/rstb.1975.0075. [DOI] [PubMed] [Google Scholar]
- Roberts G. C., Duckworth H. W., Packman L. C., Perham R. N. Mobility and active-site coupling in 2-oxo acid dehydrogenase complexes. Ciba Found Symp. 1983;93:47–71. doi: 10.1002/9780470720752.ch4. [DOI] [PubMed] [Google Scholar]
- Runswick M. J., Walker J. E. The amino acid sequence of the beta-subunit of ATP synthase from bovine heart mitochondria. J Biol Chem. 1983 Mar 10;258(5):3081–3089. [PubMed] [Google Scholar]
- Schmitt B., Cohen R. The structure of the Escherichia coli pyruvate dehydrogenase complex is probably not unique. Biochem Biophys Res Commun. 1980 Apr 14;93(3):709–712. doi: 10.1016/0006-291x(80)91135-3. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the dihydrolipoamide acetyltransferase component. Eur J Biochem. 1983 Jul 1;133(3):481–489. doi: 10.1111/j.1432-1033.1983.tb07490.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the pyruvate dehydrogenase component. Eur J Biochem. 1983 Jun 1;133(1):155–162. doi: 10.1111/j.1432-1033.1983.tb07441.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Lewis H. M., Darlison M. G., Guest J. R. Nucleotide sequence of the lipoamide dehydrogenase gene of Escherichia coli K12. Eur J Biochem. 1983 Oct 3;135(3):519–527. doi: 10.1111/j.1432-1033.1983.tb07683.x. [DOI] [PubMed] [Google Scholar]
- Stepp L. R., Bleile D. M., McRorie D. K., Pettit F. H., Reed L. J. Use of trypsin and lipoamidase to study the role of lipoic acid moieties in the pyruvate and alpha-ketoglutarate dehydrogenase complexes of Escherichia coli. Biochemistry. 1981 Aug 4;20(16):4555–4560. doi: 10.1021/bi00519a007. [DOI] [PubMed] [Google Scholar]
- Vogel O., Hoehn B., Henning U. Molecular structure of the pyruvate dehydrogenase complex from Escherichia coli K-12. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1615–1619. doi: 10.1073/pnas.69.6.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenknecht T., Francis N., DeRosier D. J. alpha-Ketoglutarate dehydrogenase complex may be heterogeneous in quaternary structure. J Mol Biol. 1983 Apr 15;165(3):523–539. doi: 10.1016/s0022-2836(83)80217-4. [DOI] [PubMed] [Google Scholar]
- White R. H., Bleile D. M., Reed L. J. Lipoic acid content of dihydrolipoyl transacylases determined by isotope dilution analysis. Biochem Biophys Res Commun. 1980 May 14;94(1):78–84. doi: 10.1016/s0006-291x(80)80190-2. [DOI] [PubMed] [Google Scholar]