Abstract
Epigenetic gene inactivation by microRNAs (miRNAs) is crucial in malignant transformation, prevention of apoptosis, development of drug resistance, and metastasis. miR-204 dysregulation has been reported in prostate cancer (PC). It is considered to exert tumor suppressor functions and is associated with the development of chemoresistance. However, the detailed mechanisms underlying the role of miR-204 in PC, particularly in chemoresistance, remain to be fully elucidated. In this study, analysis using miRNA microarray showed that miR-204 is downregulated in chemoresistant PC tissues with respect to its expression in chemosensitive PC tissues and benign prostatic hyperplasia tissues. Microarray results were validated via qPCR. The changes in miR-204 expression levels were also observed in vitro. Forced overexpression of miR-204 evidently attenuated docetaxel chemoresistance and promoted apoptosis in PC-3-R cells, whereas miR-204 knockdown effectively reduced docetaxel-induced cell death and inhibited cell apoptosis. Mechanistically, miR-204 directly targets the 3’-untranslated region of zinc-finger E-box-binding homeobox 1 (ZEB1) and inhibits its protein expression via translational repression. Furthermore, suppression of ZEB1 could effectively improve miR-204 deficiency-triggered chemoresistance in PC cells. Our results collectively indicate that miR-204 expression is downregulated in chemoresistant PC tissues and cells and that miR-204/ZEB1 could potentially be used as adjunct therapy for patients with advanced/chemoresistant PC.
Keywords: Prostate cancer, chemoresistant, docetaxel, miR-204, zinc-finger E-box-binding homeobox 1
Introduction
Prostate cancer (PC) is one of the most common cancers that affects males and represents the second leading cause of cancer-related mortality [1]. The incidence of PC has increased dramatically in China [2]. The majority of PC-associated mortality arises from metastatic castration-resistant PC (CRPC). Although treatment with taxanes, such as docetaxel, cabazitaxel, and paclitaxel has been reported to improve survival in patients with metastatic CRPC, the prognosis of the disease remains dismal [3]. Docetaxel is currently the first line of treatment for patients with CRPC and provides symptomatic and survival benefits over other anti-cancer agents [4]. However, chemotherapy resistance can eventually develop through a variety of mechanisms that remain largely unclear [5,6]. In this study, we attempted to characterize the molecular mechanisms underlying docetaxel resistance in patients with PC.
Increasing evidence supports the role of microRNAs (miRNAs) as tumor suppressors or oncogenes. miRNAs are small, non-coding, single-stranded RNAs involved in post-translational regulation of gene expression and have been implicated in a wide range of essential biological activities [7]. Aberrant miRNA expression is strongly correlated with the development of chemoresistance in various cancers, including PC [8]. The unique role of miRNAs likely facilitates the predictive and prognostic markers of treatments, as well as molecular targets for drug resistance in PC. miR-204 downregulation has been reported in PC cells with respect to expression in normal prostate epithelial cells [9]. Previous studies have also reported that miR-204 is downregulated in various other carcinomas, thereby suggesting a common role for miR-204 in human tumorigenesis [10-12]. Importantly, a recent study suggested the therapeutic potential of miR-204 against chemoresistance of colorectal cancer [13]. However, the exact role of miR-204 in PC and its target genes remains to be elucidated.
In this study, miR-204 was found to be significantly downregulated in chemoresistant PC tissues and cells. MiR-204 could sensitize PC cells to docetaxel and promote cell apoptosis. In addition, we identified zinc-finger E-box-binding homeobox 1 (ZEB1) as a novel direct target of miR-204. The ZEB1 gene encodes a zinc-finger transcription factor that is essential to normal embryonic development [14]. ZEB1 expression has been reported to be upregulated in epithelial cancers and shown to correlate with poor PC prognosis [15]. Interestingly, studies have determined that ectopic ZEB1 plays an important role in chemoresistant prostate cancer [16,17]. Our subsequent functional analyses showed that the miR-204/ZEB1 axis significantly influences the sensitivity of PC cells to docetaxel. These data provide evidence that miR-204 regulates docetaxel resistance by targeting ZEB1 signaling and could thus serve as a novel therapeutic target against PC chemoresistance.
Materials and methods
Study population
Patients admitted to the Urological Department of Peking Union Medical College Hospital between January 2012 and October 2015 were evaluated. The study was approved by the Research Ethics Committee of the Peking Union Medical College Hospital, and all participants provided written informed consent. Diagnosis of prostate cancer (PC) was performed according to the criteria set by the World Health Organization. Specimens from 30 patients with benign prostatic hyperplasia and biopsies from patients with chemosensitive PC (n=80) or chemoresistant PC (n=44) were included. Patients with chemoresistant PC were defined as patients with PC who had tumor progression after four cycles of first-line docetaxel-based chemotherapy [18]. The following exclusion criteria were used: >75 years of age, urinary infection, bladder stones, catheterization, and no informed consent. Clinicopathological information for all patients were recorded.
RNA isolation and miRNA microarray assay
All specimens from patients were stored at -80°C. Total RNA was isolated from tissues using TRIzol reagent according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). Samples were randomly selected from different treatment groups. miRNA expression analysis was performed using miRCURY LNA (Locked Nucleic Acid) microRNA Arrays (Exiqon, A/S, Vedbaek, Denmark) version 10.0. RNA quality and quantity were determined via standard electrophoresis and spectrophotometric methods. RNA samples were labeled using the miRCURY Hy3/Hy5 Power labeling kit (Exiqon Inc., Woburn, MA, USA) and hybridized on the miRCURY LNA Array station. Images were then scanned in an Agilent G2565BA Microarray Scanner System (Agilent Technologies, Santa Clara, CA, USA). Results were analyzed using ImaGene™ software (BioDiscovery, CA, USA). Data were analyzed by first subtracting the background and normalizing the signals using a LOWESS filter (locally weighted regression) [19]. Data were visualized as heat maps using Heml [20].
Real-time quantitative PCR
Quantitative reverse transcription PCR (qPCR) was performed on an iCycler iQ™ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). Reverse transcription was performed using One Step PrimeScript miRNA cDNA Synthesis Kit (Takara, Dalian, China) following the manufacturer’s instructions. Primers for miR-204 (forward, 5’-GTCCCTGTGTCATCCT-3’ and reverse, 5’-CAGTGCAGGGTCCGAGGTAT-3’) were purchased from Exiqon (microRNA LNA™ PCR primer set) and and ZEB1 primers were purchased from GeneCopoeia Co., (Rockville, MD, USA) (forward, 5’-CTCGCTTCGGCAGCACA-3’ and reverse, 5’-AACGCTTCACGAATTTGCGT-3’). qPCR reactions were run based on the following profile: 95°C for 2 min; in 40 cycles at 95°C for 15 s; and 60°C for 1 min. Small nuclear RNA U6 (forward, 5’-CTCTCTGCGGCAGCACA-3’ and reverse, 5’-AACGCTGTACGAATGTGAGT-3’) and β-actin (forward, 5’-CTGGATCGGTGAGAGTGACA-3’ and reverse, 5’-AAGGGACTTCATGTAACAGTGCA-3’) mRNA were used as internal controls for calculating the relative expression levels of miR-204 and ZEB1 via the 2-∆∆Ct method.
Cell culture and treatment
Normal human prostate epithelial cells (PECs) and the human PC cell line PC-3 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). These cells were cultured in RPMI media with 10% fetal bovine serum (Invitrogen). Docetaxel was purchased from Cayman Chemical (MI, USA). Resistant PC-3 sub-lines (PC-3-R) were generated as previous described [21]. Cells were seeded at 1×104 per well in 96-well plates and treated with varying concentrations of docetaxel for different incubation times as indicated. Cells were transfected with 20 nM miRNA mimics (miR-204 or negative control mimic; Shanghai GenePharma, Shanghai, China), 50 nM miRNA inhibitors (miR-204 or negative control inhibitor), siRNA against ZEB1 (HSS110548, HSS11050, and HSS110554), or control siRNA (Invitrogen, Carlsbad, CA, USA) using RNAiMAX transfection reagent according to the manufacturer’s instructions (Life Technologies, Gaithersburg, MD, USA).
Plasmid construction and luciferase assays
To construct the ZEB1 expression vector, the ZEB1 gene (forward, 5’-AGTAGATATACAGAATATTAACTATGTG-3’ and reverse, 5’-TCTAGGCTTATGTTTCTCTGTCTC-3’) was amplified via PCR and cloned into pcDNA3.1+HA using Life Technologies (GENECHEM, Shanghai, China). Sequencing was performed to verify the clones; the positive clone was designated pZEB1. A fragment of the ZEB1 3’-UTR and a mutated ZEB1 3’-UTR of ZEB1 were cloned into the downstream region of the luciferase gene in the pGL3-REPORT luciferase vector (Invitrogen). For the luciferase assay, PC-3-R cells were co-transfected with pGL3-3’-UTR and negative control plasmid, miR-204, or NC mimics and miR-204 inhibitors or NC inhibitors. Reporter activity was measured using a dual-luciferase reporter assay system (Promega, Madison, Wisconsin, USA). Firefly luciferase activity was normalized against Renilla luciferase activity.
Cell viability and chemosensitivity assay
PC-3 cells and PC-3-R cells were added into six-well plates and transfected with oligonucleotides for 48 h. Subsequently, cells were reseeded in 96-well plates at a density of 5×103 cells/well. After 24 h, cells were treated with 20 nM docetaxel for varying incubation periods. Cell viability was determined using the MTT assay. Absorbance was measured at 450 nm using a Laboratory Weighing UV-VIS spectrophotometer (KINO Industry Co., USA). Six wells were analyzed for cell viability in each treatment group.
Cell apoptosis assay
Cells were harvested at approximately 48 h post-transfection. Annexin V-FITC Apoptosis Detection Kit (Abcam, Cambridge, UK) was used to detect early stages of apoptosis following the manufacturer’s instructions. Cells were washed with phosphate-buffered saline (PBS) and stained with Annexin V-fluorescein isothiocyanate and propidium iodide. Stained cells were measured by flow cytometry (FACSCalibur, BD Bioscience, Heidelberg, Germany) using Cell Quest Pro software (BD Bioscience).
Western blotting
Total protein was extracted with RIPA lysis buffer (Beyotime, Jiangsu, China), and protein concentrations were measured following the BCA method (Beyotime). Cell lysates (30 μg/lane) were separated via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After being transferred to polyvinylidene fluoride membranes (Millipore, MA, USA), the proteins were incubated with primary antibodies against ZEB1 or GAPDH (Sigma, Santa Clara, CA, USA) at 4°C overnight, and further incubated in HRP-linked secondary antibodies (Santa Cruz Biotechnology, USA) for 1 h at room temperature. Alpha Innotech (San Leandro, CA) imaging software was used to quantify western blotting data.
Statistical analysis
Data were analyzed using SPSS version 20.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism5 (GraphPad Software, San Diego, CA, USA). Data for continuous variables were presented as median ± standard deviation of data obtained from at least three independent experiments. Differences between groups were analyzed using student’s t-test or one-way ANOVA. Correlation between miR-204 and ZEB1 expression levels was determined using Pearson correlation test. Results were considered statistically significant at P<0.05.
Results
miR-204 expression is downregulated in chemoresistant PC tissues and cells
To explore the potential role of miRNAs in PC chemoresistance, miRNA microarray was performed on the obtained human PC specimens to identify differentially expressed miRNAs. Surprisingly, miR-204 was found to be downregulated in chemosensitive PC tissues than in benign prostatic hyperplasia tissues, and miR-204 levels were further decreased in chemoresistant PC tissues (Figure 1). Microarray data were then validated via qPCR. The clinicopathological significance of miR-204 expression in patients with PC is shown in Table 1. There was no statistical significance between clinicopathological parameters and the expression level of miR-204 except for response to docetaxel treatment. Results in Figure 2A indicate that chemosensitive PC samples (n=80) have lower miR-204 expression levels than do benign prostatic hyperplasia specimens (n=30), with the minimum expression values detected in the chemoresistant counterparts (n=44). PC-3-R cells showed stronger cell viability than PC-3 cells upon treatment with increasing docetaxel concentrations (Figure 2B) and at long incubation periods (Figure 2C). Subsequent qPCR analysis demonstrated that miR-204 expression was upregulated in human prostate epithelial cells (PECs), downregulated in PC-3 cells, and strongly downregulated in PC-3-R cells (Figure 2D). The above findings suggest that miR-204 expression is decreased in chemoresistant PC tissues and cells.
Figure 1.
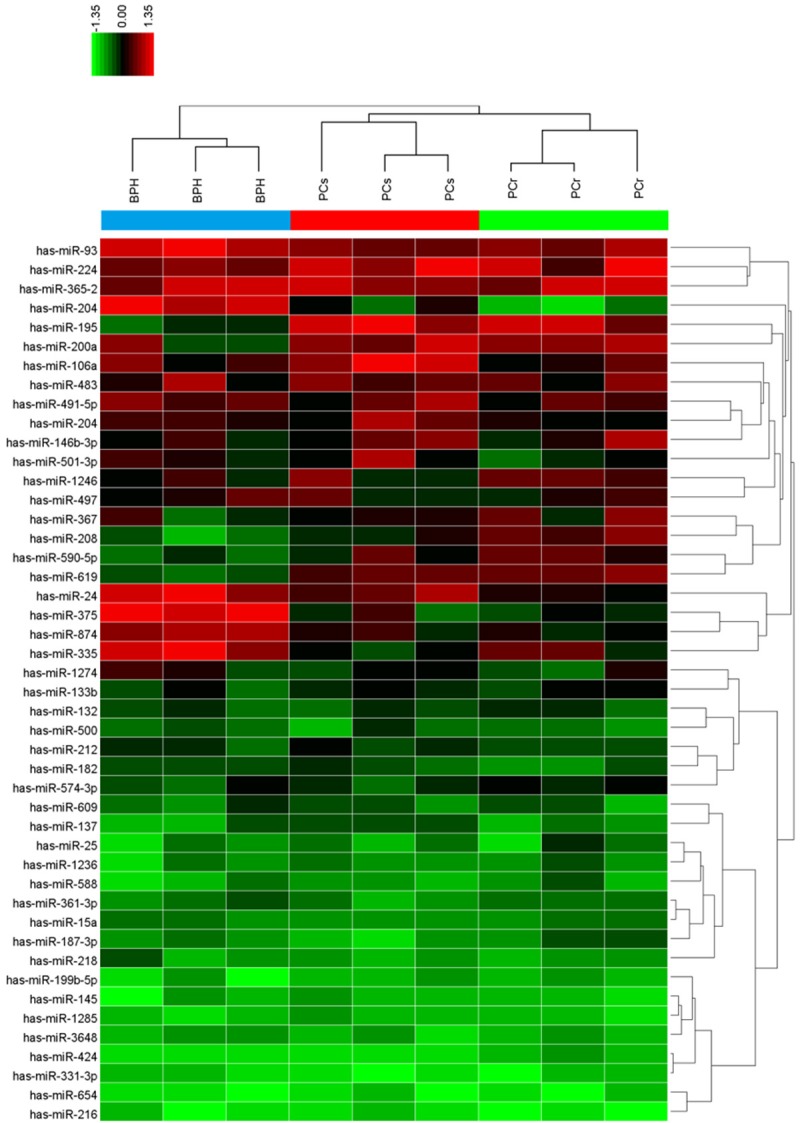
Heat map showing miRNA expression patterns in the three treatment groups. Three tissue samples were randomly chosen from 30 patients with benign prostatic hyperplasia (BPH), 80 with chemosensitive PC (PCs), and 44 with chemoresistant PC (PCr). Red areas represent mean expression levels of upregulated miRNAs, and green bars represent mean expression levels of downregulated miRNAs.
Table 1.
The analyzed relationship between miR-204 expression levels and clinicopathological significance in subjects
| Characteristic | Case | miR-204 expression | P valuea |
|---|---|---|---|
| Type | 0.001 | ||
| Chemoresistant prostate cancer | 80 | 1.129±0.294 | |
| Chemosensitive prostate cancer | 44 | 2.461±0.718 | |
| Age (years) | 0.365 | ||
| <60 | 73 | 1.986±0.436 | |
| ≥60 | 51 | 2.016±0.553 | |
| Lymph node metastasis | 0.534 | ||
| Yes | 81 | 2.101±0.635 | |
| No | 43 | 1.971±0.736 | |
| T classification | 0.298 | ||
| T1-2 | 75 | 1.957±0.569 | |
| T3-4 | 49 | 2.042±0.607 | |
| Angiolymphatic invasion | 0.108 | ||
| Positive | 69 | 1.648±0.436 | |
| Negative | 55 | 2.046±0.733 |
One-way ANOVA was used to analyze the correlation between the expression of miR-204 and clinicopathological features of the patients.
Figure 2.
miR-204 expression is downregulated in chemoresistant prostate cancer (PC) tissues and cells. Different expression levels of miR-204 in human benign prostatic hyperplasia specimens (n=30), chemoresistant PC samples (n=44), and chemosensitive PC samples (n=80) (A). PC-3 cells and docetaxel-resistant PC-3-R cells were incubated with 20 nM docetaxel for varying incubation periods (B) or treated with different docetaxel concentrations (2.5 nM to 100 nM) (C) Cell viabilities were then recorded. miR-204 expression levels in human prostate epithelial cell (PEC), PC-3 cells and PC-3-R (D). *P<0.05 vs. normal group; **P<0.05 vs. chemosensitive group; #P<0.05 vs. PC-3 group; ##P<0.05 vs. PEC group.
MiR-204 sensitizes PC cells to docetaxel and promotes cell apoptosis
To determine the role of miR-204 in docetaxel-induced cytotoxicity in PC-3-R cells, we transfected PC-3-R cells with miR-204 mimics or corresponding negative controls (NC) and miR-204 inhibitors or NC inhibitors, respectively. Based on the qPCR results shown in Figure 3A, miR-204 levels were remarkably higher in the mimic-treated group and lower in the inhibitor-treated group than those in the corresponding NC group (P<0.05 for all treatments). We next determined the effects of ectopic miR-204 expression on cell chemosensitivity using MTT assay. Upon treatment with miR-204 mimics, PC-3-R cells showed significantly higher sensitivity to docetaxel than the NC group (Figure 3B). On the other hand, PC-3-R cells showed increased docetaxel resistance than the NC group upon treatment with miR-204 inhibitors (Figure 3C). Subsequent apoptotic analysis revealed that miR-204 overexpression significantly promoted cell apoptosis, and miR-204 downregulation effectively inhibited cell apoptosis (Figure 3D, 3E).
Figure 3.
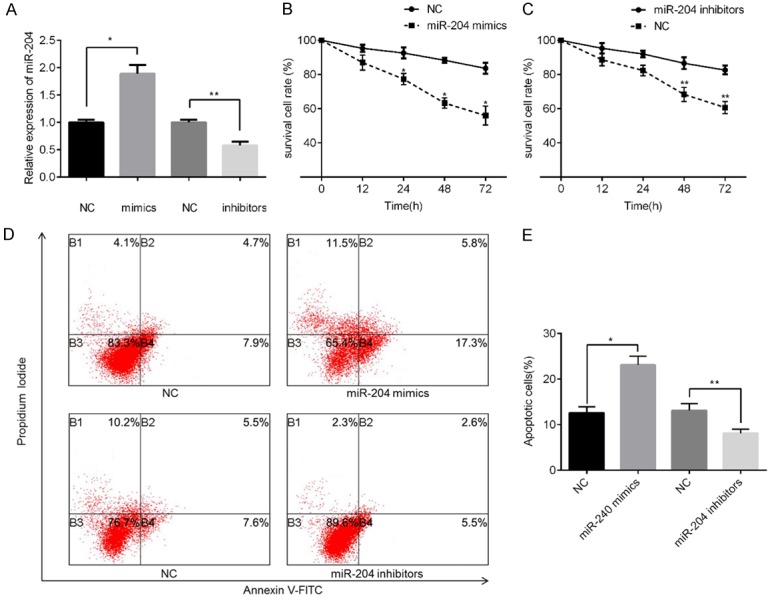
miR-204 modulates chemosensitivity and cell apoptosis of PC cells. miR-204 expression levels in PC-3-R cells transfected with miRNA mimics, miRNA inhibitors, or corresponding negative-controls (NC) (A). Cell viability was compared between cells transfected with miR-204 or control mimics (B) and between cells transfected with miR-204 inhibitors or control inhibitors (C) in PC-3-R cells incubated with 20 nM docetaxel at varying incubation periods. Cell apoptosis rates were detected in PC-3-R cells transfected with miRNA mimics, miRNA inhibitors, or NC, followed by incubation with 20 nM docetaxel for 24 h using flow cytometry (D and E). *P<0.05 vs. mimic NC group; **P<0.05 vs. inhibitors NC group.
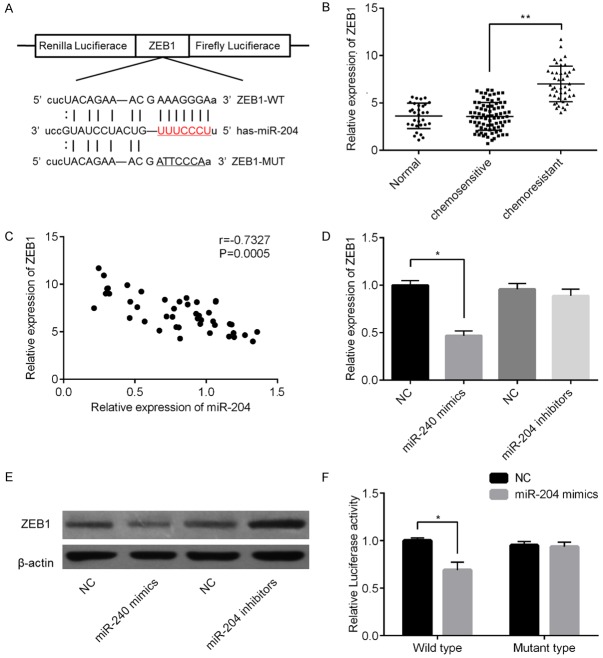
miR-204 directly targets ZEB1 in PC cells
Potential targets of miR-204 were predicted using the public database microRNA.org (http://www.microrna.org/microrna/home.do), and the putative complementary sequence of miR-204 was identified in the 3’-UTR of ZEB1 mRNA, as illustrated in Figure 4A. ZEB1 mRNA expression levels were determined; results in Figure 4B showed that ZEB1 expression was distinctly higher in the tissues of patients with chemoresistant PC than in those of chemosensitive tissues or non-neoplastic tissues. Subsequently, Pearson correlation analysis revealed that ZEB1 levels were negatively correlated with miR-204 levels in PC tissues (r=-0.7327, P=0.0005, Figure 4C). In addition, ZEB1 levels were assessed in PC-3-R cells transfected with miR-204 mimics or corresponding NC mimics and miR-204 inhibitors or NC inhibitors via qPCR and western blotting. Samples in the mimic-treated group showed higher ZEB1 expression levels than those of the NC group; however, miR-204 inhibitor-treated groups did not show significant differences in ZEB1 levels with samples in the NC-treated group (Figure 4D). In contrast, ZEB1 protein levels were higher in the miR-204 mimic-treated group and lower in miR-204 inhibitor-treated group than in their corresponding NC groups (Figure 4E). To further validate our hypothesis, we performed a luciferase reporter assay. As shown in Figure 4F, cells transfected with ZEB1-WT-3’-UTR vector containing a miR-204 mimic showed significantly lower luciferase activity than cells transfected with control RNAs (P<0.01). In contrast, no changes in relative luciferase activity were observed when the miR-204 binding site was mutated. Based on these findings, we suggest that ZEB1 is a potential a direct target of miR-204 in PC cells.
Figure 4.
miR-204 targets ZEB1 by binding to its 3’-UTR in PC. Binding of miR-204 to the 3’-UTR of ZEB1 was predicted using microRNA.org (A). Varying ZEB1 expression levels in different treatment groups (B). Relationship between miR-204 mRNA and ZEB1 mRNA expression levels were analysis using Pearson correlation (C). Changes in ZEB1 mRNA levels in PC-3-R cells after transfection with miRNA mimics, miRNA inhibitors, or NC based on qPCR (D) and western blotting (E). Results of luciferase reporter assays using PC-3-R cells co-transfected with the constructed reporter gene harboring miR-204 or control mimic (F). *P<0.05 vs. mimic NC group; **P<0.05 vs. chemosensitive group.
ZEB1 is involved in miR-204-mediated chemoresistance in PC cells
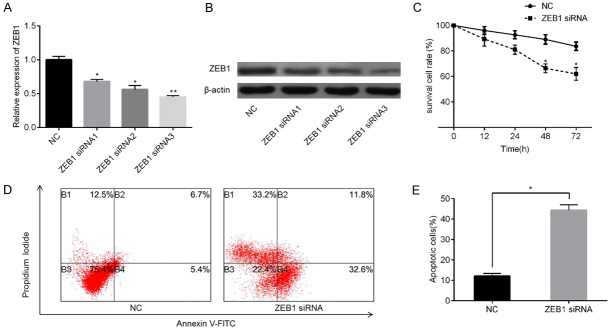
Given that miR-204 directly targets ZEB1, we further investigated whether miR-204 exerts its function in PC by regulating ZEB1. ZEB1 knockdown was performed via treatment with specific siRNAs, and results were validated via qPCR (Figure 5A) and western blotting (Figure 5B). Next, PC-3-R cells transfected with ZEB1 siRNA or NC siRNA, as well as PC-3-R cells supplemented with miR-204 mimics, were subjected to MTT assay and apoptotic analysis. Results showed that ZEB1 downregulation enhanced docetaxel sensitivity (Figure 5C) and promoted cell apoptosis (Figure 5D, 5E) in PC-3-R cells than in cells from the NC group. In addition, we overexpressed ZEB1 via transgenic overexpression and verified expression both at mRNA (Figure 6A) and protein levels (Figure 6B). As expected, ZEB1 upregulation promoted docetaxel-resistance (Figure 6C) and inhibited cell apoptosis (Figure 6D, 6E) to a greater extent in PC-3-R cells than in cells in the NC group. These results suggest that miR-204 can induce docetaxel sensitivity and apoptosis in PC cells by negatively regulating the ZEB1 pathway.
Figure 5.
Effect ZEB1 inhibition on docetaxel sensitivity and cell apoptosis. To knock down ZEB1, PC-3-R cells overexpressing miR-204 were transfected with three synthetic ZEB1 siRNAs. ZEB1 levels were then measured via qPCR (A) and western blotting (B). Cell viability was measured after ZEB1 knockdown in PC-3-R cells overexpressing miR-204 and incubated with 20 nM docetaxel at varying incubation periods (C). Cell apoptosis rate were determined in PC-3-R cells overexpressing miR-204 transfected with ZEB1 siRNA or NC siRNA, followed by incubation with 20 nM docetaxel for 24 h using flow cytometry (D and E). *P<0.05, **P<0.01 vs. NC siRNA group.
Figure 6.
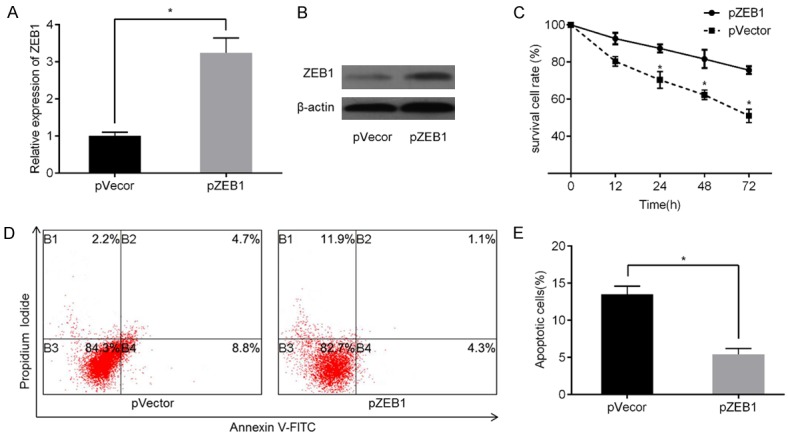
Effect of ZEB1 overexpression on docetaxel sensitivity and cell apoptosis. To induce ZEB1 overexpression, PC-3-R cells overexpressing miR-204 were transfected with pcDNA3.1+ZEB1 or negative control pVector. ZEB1 expression levels were then evaluated via qPCR (A) and western blotting (B). Cell viability was measured after inducing ZEB1 overexpression using MTT assay (C). Flow cytometry was used to analyze cell apoptosis rates in PC-3-R cells overexpressing miR-204 in pZEB1 group or pVector group (D and E). *P<0.05 vs. pVector group.
Discussion
Previous docetaxel-based clinical studies have demonstrated the potential benefits of chemotherapy to prolong the survival and improve the quality of life of patients with PC [22,23]. However, the drug-resistant nature of prostate cancer (PC) still challenges the effectiveness of such therapies. To date, the roles of miRNAs in PC progression have been increasingly studied [17,24], and miRNAs that exert regulatory effects on chemoresistance remain the focus of research in recent years. miRNAs that are involved in regulating chemoresistance have been considered promising targets for PC management. Here, we found that chemoresistant PC samples exhibit miR-204 downregulation when compared to control samples. Similarly, Lin et al. showed that miR-204 is downregulated in different PC cell lines with respect to expression in normal prostate epithelial cells and up-regulation of miR-204 promotes cell apoptosis and reduces cell viability by targeting BCL2 [9]. Another study showed that miR-204 levels are elevated in human prostate tumor samples and that miR-204 enhances cellular growth, migration, and invasion of PC cells by inhibiting the expression of prostate-derived epithelial factor [25]. However, so far, few studies have determined the effects of ectopic expression of miR-204 on chemoresistance during cancer therapy. In 2012, Sacconi et al. found that decreased miR-204 expression has prognostic value and correlates with increased Bcl-2 protein expression in gastric cancer specimens. Further experiments indicated that overexpressing miR-204 enhances the response of gastric cancer cells to 5-fluorouracil (5-FU) by targeting the 3’-UTR of BCL [26]. In a recent study, Wang et al. investigated the role of miR-204 in docetaxel resistance in PC cells and suggested that the long noncoding RNA urothelial carcinoma-associated 1 (UCA1) can modulate docetaxel resistance of PC cells by targeting the miR-204/Sirt1 axis [27]. Consistent with previous studies, we found that forced overexpression of miR-204 reduced docetaxel resistance of PC-3-R cells. Furthermore, miR-204 upregulation was observed to promote cell apoptosis. These results provide evidence that miR-204 could serve as a crucial post-transcriptional regulator during the pathogenesis of chemoresistance in PC.
Emerging evidence has shown that docetaxel resistance is driven by the epithelial-mesenchymal transition. Intriguingly, ZEB1, one of the master genes that initiate the epithelial-mesenchymal transition, is considered as a key transcriptional factor in cancer progression [28,29]. Previous studies suggested that ZEB1 plays a vital role in inhibiting cell apoptosis [30,31]. Importantly, Wang et al. suggested that overexpression of ZEB1 in androgen-dependent PC cells leads to increased resistance to bicalutamide, whereas ZEB1 knockdown results in bicalutamide sensitization [32]. In the present work, tissues of patients with chemoresistant PC showed increased ZEB1 levels than chemosensitive tissues or non-tumor tissues. Subsequent analysis revealed that downregulation of ZEB1 sensitized docetaxel-resistant PC cells and promoted cell apoptosis in PC cells by increasing sensitivity to docetaxel-induced cy-totoxicity. Previous studies indicated that ZEB1 function is regulated by other important genes. For instance, Leshem et al. demonstrated that the TMPRSS2 and ERG directly bind to the promoter of ZEB1, thereby promoting PC progression [33]. Jung et al. demonstrated that nesfatin-1/nucleobindin-2 enhances cell migration and invasion through ZEB1 in colon cancer [34]. However, considering that miRNAs can regulate protein expression in subtle levels, it is intriguing whether certain miRNAs can function as mediators that determine the functional mode of ZEB1. Over the past decade, an increasing number of researchers have focused on miRNAs, which could function as important gene regulators of up to 60% of human protein-coding genes [35]. We used the comprehensive resource in microRNA.org and used microRNA target predictions to identify potential binding sequences between miR-204 and ZEB1. To validate this prediction, we analyzed the relationship between the ZEB1 and miR-204 expression levels in tissues of patients with chemoresistant PC and found a negative correlation between them. miR-204 could regulate ZEB1 expression both at protein and mRNA levels. Results of the luciferase reporter assay indicated that miR-204 targets ZEB1 by directly binding to its 3’-UTR. In addition, after transfection of docetaxel-resistance PC cells with miR-204 mimics, we demonstrate that treatment with ZEB1 siRNA resulted in sensitization of docetaxel-resistance PC cells; on the other hand, forced ZEB1 expression inhibited miR-204-induced apoptosis in response to docetaxel treatment. Taken together, our data suggest that miR-204 plays important role in regulating ZEB1 expression. Furthermore, miR-204 potentiates docetaxel sensitivity and promotes apoptosis of prostate cancer cells by targeting ZEB1.
In summary, we demonstrate that miR-204 is downregulated in chemoresistant PC samples and cells. miR-204 sensitizes PC cells to docetaxel and promotes cell apoptosis at least partly by regulating ZEB1. Our findings indicate that the miR-204/ZEB1 axis is important in modulating the in vitro docetaxel sensitivity of PC cells and could serve as a novel therapeutic target for chemoresistant PC.
Acknowledgements
This study was supported by the Natural Science Foundation of China (No. 80170407); the Science and Technology Plan Project of Beijing (No. 2013A13207).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Pang C, Guan Y, Li H, Chen W, Zhu G. Urologic cancer in China. Jpn J Clin Oncol. 2016;46:497–501. doi: 10.1093/jjco/hyw034. [DOI] [PubMed] [Google Scholar]
- 3.Cookson MS, Roth BJ, Dahm P, Engstrom C, Freedland SJ, Hussain M, Lin DW, Lowrance WT, Murad MH, Oh WK, Penson DF, Kibel AS. Castration-resistant prostate cancer: AUA guideline. J Urol. 2013;190:429–438. doi: 10.1016/j.juro.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Berthold DR, Sternberg CN, Tannock IF. Management of advanced prostate cancer after first-line chemotherapy. J. Clin. Oncol. 2005;23:8247–8252. doi: 10.1200/JCO.2005.03.1435. [DOI] [PubMed] [Google Scholar]
- 5.Feng B, Wang R, Chen LB. Review of miR-200b and cancer chemosensitivity. Biomed Pharmacother. 2012;66:397–402. doi: 10.1016/j.biopha.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Bhatnagar N, Li X, Padi SK, Zhang Q, Tang MS, Guo B. Downregulation of miR-205 and miR-31 confers resistance to chemotherapyinduced apoptosis in prostate cancer cells. Cell Death Dis. 2010;1:e105. doi: 10.1038/cddis.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corcoran C, Friel AM, Duffy MJ, Crown J, O’Driscoll L. Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 2011;57:18–32. doi: 10.1373/clinchem.2010.150730. [DOI] [PubMed] [Google Scholar]
- 8.Kopczynska E. Role of microRNAs in the resistance of prostate cancer to docetaxel and paclitaxel. Contemp Oncol (Pozn) 2015;19:423–427. doi: 10.5114/wo.2015.56648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin YC, Lin JF, Tsai TF, Chou KY, Chen HE, Hwang TI. Tumor suppressor miRNA-204-5p promotes apoptosis by targeting BCL2 in prostate cancer cells. Asian J Surg. 2016 doi: 10.1016/j.asjsur.2016.07.001. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Li F, Zhou X. miR-204-5p regulates cell proliferation and metastasis through inhibiting CXCR4 expression in OSCC. Biomed Pharmacother. 2016;82:202–207. doi: 10.1016/j.biopha.2016.04.060. [DOI] [PubMed] [Google Scholar]
- 11.Zhang S, Gao L, Thakur A, Shi P, Liu F, Feng J, Wang T, Liang Y, Liu JJ, Chen M, Ren H. miRNA-204 suppresses human non-small cell lung cancer by targeting ATF2. Tumour Biol. 2016;37:11177–11186. doi: 10.1007/s13277-016-4906-4. [DOI] [PubMed] [Google Scholar]
- 12.Song S, Fajol A, Tu X, Ren B, Shi S. miR-204 suppresses the development and progression of human glioblastoma by targeting ATF2. Oncotarget. 2016;7:70058–70065. doi: 10.18632/oncotarget.11732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, Liang Y, Shen L, Shen L. MicroRNA-204 modulates colorectal cancer cell sensitivity in response to 5-fluorouracil-based treatment by targeting high mobility group protein A2. Biol Open. 2016;5:563–570. doi: 10.1242/bio.015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Peng X, Tan J, Darling DS, Kaplan HJ, Dean DC. Zeb1 mutant mice as a model of posterior corneal dystrophy. Invest Ophthalmol Vis Sci. 2008;49:1843–1849. doi: 10.1167/iovs.07-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 16.Hanrahan K, O’Neill A, Prencipe M, Bugler J, Murphy L, Fabre A, Puhr M, Culig Z, Murphy K, Watson RW. The role of epithelial-mesenchymal transition drivers ZEB1 and ZEB2 in mediating docetaxel-resistant prostate cancer. Mol Oncol. 2017;11:251–265. doi: 10.1002/1878-0261.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun X, Li Y, Yu J, Pei H, Luo P, Zhang J. miR-128 modulates chemosensitivity and invasion of prostate cancer cells through targeting ZEB1. Jpn J Clin Oncol. 2015;45:474–482. doi: 10.1093/jjco/hyv027. [DOI] [PubMed] [Google Scholar]
- 18.Yu L, Su YS, Zhao J, Wang H, Li W. Repression of NR4A1 by a chromatin modifier promotes docetaxel resistance in PC-3 human prostate cancer cells. FEBS Lett. 2013;587:2542–2551. doi: 10.1016/j.febslet.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 19.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 20.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: a toolkit for illustrating heatmaps. PLoS One. 2014;9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Neill AJ, Prencipe M, Dowling C, Fan Y, Mulrane L, Gallagher WM, O’Connor D, O’Connor R, Devery A, Corcoran C, Rani S, O’Driscoll L, Fitzpatrick JM, Watson RW. Characterisation and manipulation of docetaxel resistant prostate cancer cell lines. Mol Cancer. 2011;10:126. doi: 10.1186/1476-4598-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, Oudard S, Theodore C, James ND, Turesson I, Rosenthal MA, Eisenberger MA. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 23.Petrylak DP, Tangen CM, Hussain MH, Lara PN Jr, Jones JA, Taplin ME, Burch PA, Berry D, Moinpour C, Kohli M, Benson MC, Small EJ, Raghavan D, Crawford ED. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351:1513–1520. doi: 10.1056/NEJMoa041318. [DOI] [PubMed] [Google Scholar]
- 24.Corcoran C, Rani S, O’Driscoll L. miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate. 2014;74:1320–1334. doi: 10.1002/pros.22848. [DOI] [PubMed] [Google Scholar]
- 25.Turner DP, Findlay VJ, Moussa O, Semenchenko VI, Watson PM, LaRue AC, Desouki MM, Fraig M, Watson DK. Mechanisms and functional consequences of PDEF protein expression loss during prostate cancer progression. Prostate. 2011;71:1723–1735. doi: 10.1002/pros.21389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sacconi A, Biagioni F, Canu V, Mori F, Di Benedetto A, Lorenzon L, Ercolani C, Di Agostino S, Cambria AM, Germoni S, Grasso G, Blandino R, Panebianco V, Ziparo V, Federici O, Muti P, Strano S, Carboni F, Mottolese M, Diodoro M, Pescarmona E, Garofalo A, Blandino G. miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis. 2012;3:e423. doi: 10.1038/cddis.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Yang B, Ma B. The UCA1/miR-204/Sirt1 axis modulates docetaxel sensitivity of prostate cancer cells. Cancer Chemother Pharmacol. 2016;78:1025–1031. doi: 10.1007/s00280-016-3158-8. [DOI] [PubMed] [Google Scholar]
- 28.Aigner K, Descovich L, Mikula M, Sultan A, Dampier B, Bonne S, van Roy F, Mikulits W, Schreiber M, Brabletz T, Sommergruber W, Schweifer N, Wernitznig A, Beug H, Foisner R, Eger A. The transcription factor ZEB1 (deltaEF1) represses Plakophilin 3 during human cancer progression. FEBS Lett. 2007;581:1617–1624. doi: 10.1016/j.febslet.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selth LA, Das R, Townley SL, Coutinho I, Hanson AR, Centenera MM, Stylianou N, Sweeney K, Soekmadji C, Jovanovic L, Nelson CC, Zoubeidi A, Butler LM, Goodall GJ, Hollier BG, Gregory PA, Tilley WD. A ZEB1-miR-375-YAP1 pathway regulates epithelial plasticity in prostate cancer. Oncogene. 2017;36:24–34. doi: 10.1038/onc.2016.185. [DOI] [PubMed] [Google Scholar]
- 30.Gu Y, Zhao Y, Zhou Y, Xie Y, Ju P, Long Y, Liu J, Ni D, Cao F, Lyu Z, Mao Z, Hao J, Li Y, Wan Q, Kanyomse Q, Liu Y, Ren D, Ning Y, Li X, Zhou Q, Li B. Zeb1 Is a potential regulator of Six2 in the proliferation, apoptosis and migration of metanephric mesenchyme cells. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17081283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiu LY, Hsin IL, Yang TY, Sung WW, Chi JY, Chang JT, Ko JL, Sheu GT. The ERK-ZEB1 pathway mediates epithelial-mesenchymal transition in pemetrexed resistant lung cancer cells with suppression by vinca alkaloids. Oncogene. 2017;36:242–253. doi: 10.1038/onc.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P, Wang J, Chu M, Zhang K, Yang R, Gao WQ. Zeb1 promotes androgen independence of prostate cancer via induction of stem celllike properties. Exp Biol Med (Maywood) 2014;239:813–822. doi: 10.1177/1535370214538727. [DOI] [PubMed] [Google Scholar]
- 33.Leshem O, Madar S, Kogan-Sakin I, Kamer I, Goldstein I, Brosh R, Cohen Y, Jacob-Hirsch J, Ehrlich M, Ben-Sasson S, Goldfinger N, Loewenthal R, Gazit E, Rotter V, Berger R. TMPRSS2/ERG promotes epithelial to mesenchymal transition through the ZEB1/ZEB2 axis in a prostate cancer model. PLoS One. 2011;6:e21650. doi: 10.1371/journal.pone.0021650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kan JY, Yen MC, Wang JY, Wu DC, Chiu YJ, Ho YW, Kuo PL. Nesfatin-1/Nucleobindin-2 enhances cell migration, invasion, and epithelial-mesenchymal transition via LKB1/AMPK/TORC1/ZEB1 pathways in colon cancer. Oncotarget. 2016;7:31336–31349. doi: 10.18632/oncotarget.9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]