Abstract
Esophageal cancer-related gene 4 (ECRG4) is a tumor suppressor gene associated with the prognosis of esophageal squamous-cell carcinoma (ESCC). Studies have reported that ECRG4 effectively inhibits the proliferation, migration and invasion of ESCC cells. In the current study, ectopic expression of ECRG4 significantly induced ESCC cell apoptosis. To further understand the molecular profile of ECRG4 overexpression in ESCC cells, tandem mass tag (TMT) labeling followed by LC-MS/MS analysis was applied on samples from ECRG4 overexpressed cells and control cells. Among the identified differentially expressed proteins, four up-regulated proteins (PLK1, CDK4, PLOD1 and PLOD2) associated with cell apoptosis, cell cycle and metastasis were chosen for the further investigation. The effects of ECRG4 protein levels on the expression of these four proteins were validated by manipulating the expression of ECRG4 in ESCC cells followed by Western blotting analysis. The immunohistochemical staining results showed a significant decrease in ECRG4 levels and a notable increase in the four proteins in ESCC samples as compared to matched esophageal tissues (n=75). More importantly, the protein levels of ECRG4 had a negative association with those of PLK1, CDK4, PLOD1 and PLOD2. Thus, our data suggested that the tumor-repressor functions of ECRG4 may be associated with PLK1, CDK4, PLOD1 and PLOD2, providing important insights into the molecular mechanisms of esophageal carcinogenesis.
Keywords: ECRG4, esophageal squamous-cell carcinoma, quantitative proteomics
Introduction
Esophageal cancer, one of the most common human malignancies, is the sixth leading cause of cancer-related mortality worldwide [1]. Esophageal cancer is classified as two main subtypes, esophageal adenocarcinoma (EAC) and esophageal squamous-cell carcinoma (ESCC). China is a high incidence area of esophageal cancer. ESCC is the predominant type in China and other Asian countries [2,3]. Although the advances in the earlier detection and multimodal therapies have moderately reduced the mortality rates, the 5-year survival rate of ESCC is still less than 30% and ESCC poses a serious threat to human health [4]. Therefore, it is of great clinical importance to identify biological markers for the diagnosis and prognosis of ESCC, and elucidate the molecular mechanisms involved in esophageal carcinogenesis.
Esophageal cancer related gene 4 (ECGR4, also called C2ORF40) was firstly identified and cloned from human normal esophageal epithelium [5,6]. ECRG4 gene was widely expressed in normal adult human tissues [7]. The expression of ECRG4 was down-regulated in several cancers including ESCC [8,9], glioma [10,11], prostate cancer [12], colorectal cancer [10,13] and breast cancer [14] and closely related with the development and prognosis of these malignancies. Our previous studies have found that ECRG4 can impede ESCC cell growth by inducing cell cycle arrest [9], and inhibit the migration and invasion of ESCC cells as well [15]. However, the biological pathways regulated by ECRG4 are far from being understood.
Initially, we investigated the global effects of ECRG4 expression on ESCC by bioinformatics analysis on public available expression data of ESCC tissues. Then we identified differentially expressed proteins between ECRG4 overexpressed cells and control cells by quantitative proteomics analysis. The expression of 4 differentially expressed proteins, polo-like kinase 1 (PLK1), CDK4, PLOD1 and PLOD2, were validated in ESCC cells by Western blotting. Further, ECRG4 expression showed a negative correlation with PLK1, CDK4, PLOD1 and PLOD2 in ESCC specimens as indicated by immunohistochemical staining.
Materials and methods
Bioinformatics analysis
The gene expression profiling data of ESCC tissues were downloaded from The Cancer Genome Atlas (TCGA) website. In order to investigate the global effect of ECRG4 on ESCC, we used Gene Set Enrichment Analysis (GSEA) (http://www.broad.mit.edu/gsea) [16] to perform the pathway analysis on the TCGA data. The pathways of fewer than 10 genes were excluded. The pathways with a P-value of <0.01 and a false discovery rate (FDR) of <0.05 were considered significantly related with ECRG4 expression.
Cell culture, transfection, lentiviral infection and RNA interference
ESCC cell lines, EC9706 [17], EC-18 [18] and TE-1 [19] were cultured in RPMI-1640 medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS, Hyclone) 100 U/ml penicillin and 100 μg/ml streptomycin. All cells were maintained in 5% CO2 at 37°C.
Human ECRG4 cDNA was inserted into the Xho I and BamH I sites of pLVX-Puro vector (Clontech). HEK293T cells were co-transfected with pLVX-Puro-ECRG4 with helper plasmids (psPAX2 and pMD2.G) by using lipofectamine 2000 (Invitrogen) per the manufacturer’s instructions. pLVX-Puro vector was served as negative control. The lentiviral supernatant was harvested 48 h later. EC9706 and EC-18 cells were transduced with lentiviral particles in the presence of 10 μg/mL polybrene (Sigma). The expression of ECRG4 was detected by Western blot analysis at 48 h after viral transduction.
ECRG4 siRNA (siECRG4, CCAGGUGGCAUAAGUGGAA) and negative control siRNA (siNC) were designed and synthesized by Genepharma. TE-1 cells were transfected with siECRG4 or siNC with Lipofectamine 2000. The expression of ECRG4 was assessed by Western blot analysis at 48 h after siRNA transfection.
Apoptosis analysis by flow cytometry
Cells in a 6-well culture plates were treated with ECRG4 expressing virus or Vector virus. At 48 h after transduction, cells were gently harvested by trypsin digestion and washed with ice-cold PBS. Then the cells were stained with Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kits (Beyotime). After incubation for 30 min at 37°C in the dark, cell apoptosis was analyzed by flow cytometry (BD Biosciences).
Tandem mass tag (TMT) labeling and LC-MS/MS analysis
EC9706 cells were transduced with ECRG4 expressing lentivirus or control vector virus. At 48 h after viral transduction, protein was extracted from Control (C1-C3) and ECRG4-overexpressed cells (E1-E3) by using lysis buffer (7 M urea and 4% SDS) with 1% proteinase inhibitor cocktail (Sigma), and quantified by BCA method. Equal amounts of protein (100 μg) were reduced, alkylated and then precipitated with acetone. The precipitates were then re-suspended in 200 mM Tetraethylammonium Bromide (TEAB) and digested with trypsin. Peptides from C1-C3 was labeled with TMT-126, TMT-127N and TMT-128N, respectively. Peptides from E1-E3 were labeled with was labeled with TMT-129N, TMT-130N and TMT-131N, respectively. The obtained samples were then combined, separated by high-performance liquid chromatography (HPLC), and analyzed by LC-MS/MS. MS raw data were analyzed with Proteome Discoverer Software version 2.1 (Thermo Fisher Scientific).
Western blot analysis
For western blotting analysis, lysate containing 15 μg protein was separated on a 10% or 5% SDS-PAGE gel under denaturing conditions and subsequently transferred to a nitrocellulose membrane by electroblotting. The membranes were blocked in 5% non-fat milk for 1 h at room temperature, and then incubated overnight at 4°C with the following primary antibodies: ECRG4, PLK1, PLOD1 and PLOD2 (Abcam); CDK4 and GAPDH (Cell signaling Technology). After three washing steps in TBST, the membranes were incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (Beyotime) for 1 h at room temperature and then analyzed using Enhanced Chemiluminescence kit (BioRad).
Tissue specimens
The study protocol was approved by the Institutional Review Board of Zhengzhou University People’s Hospital (Henan Provincial People’s Hospital). A total of 75 pairs of formalin-fixed paraffin-embedded ESCC samples and adjacent histological normal esophageal tissues were collected from the Zhengzhou University People’s Hospital after obtaining patients’ written informed consents. About 70% of the patients were men, and the mean age was 63 years (ranging from 41 to 80 years). Representative sections of each specimen were stained with hematoxylin and eosin to confirm the diagnosis of ESCC.
Immunohistochemiscal staining
For immunohistochemiscal staining, sections were deparaffinized with xylem, rehydrated in a graded series of ethanol and treated with methanol containing 0.3% hydrogen peroxide to remove endogenous peroxidase activity. Antigen retrieval was performed by heating the slides for 10 min in 0.01 M citrate-buffer (pH 6.0) in a pressure cooker. After three washing steps in PBS, the slides were incubated with anti-ECRG4, anti-PLK1, anti-CDK4, anti-PLOD1 or anti-PLOD2 (Abcam) at 4°C overnight. After washing again, the slides were processed with HRP conjugated secondary antibody for 1 h at room temperature followed by 3,3-diaminobenzidine (DAB) solution (Vector Laboratories). After counterstaining with hematoxylin, the slides were dehydrated and sealed with neutral resin. The staining was evaluated by two observers according to the percentage of positive cells: 0-25% (low positive), 25-50% (medium positive), and more than 50% (high positive).
Statistical analysis
The correlations between proteins expression was analyzed using Chi-square tests. The differences among groups were compared using one-way analysis of variance, and data were expressed as mean ± SD. Statistical difference was set at P<0.05.
Results
ECRG4-associated pathways in ESCC
Our previous study has demonstrated that ECRG4 protein was downregulated in most ESCC samples [20]. In order to investigate the global effect of ECRG4 on ESCC, we performed GSEA on gene expression profiling data of ESCC tissues obtained from TCGA. Our data implied that high ECRG4 expression was negatively correlated with the processes of metastasis, apoptosis and cell cycle in ESCC tissue samples (Figure 1).
Figure 1.
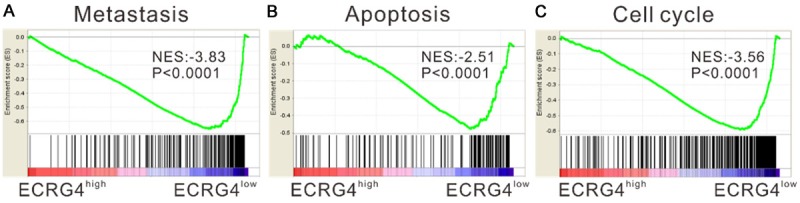
Gene Set Enrichment Analysis (GSEA). GSEA analysis in ESCC tissues with higher ECRG4 expression versus lower ECRG4 expression based on TCGA dataset. NES, normalized enrichment score. Metastasis, apoptosis and cell cycle processes showed negative association with ECRG4-higher expression.
Effects of ECRG4 overexpression on cell apoptosis
The regulatory roles of ECRG4 in the metastasis, proliferation and cell cycle have been previously studied [20]. We then measured the effects of ECRG4 overexpression on ESCC cell apoptosis. EC9706 and EC-18 cells, which had relative lower ECRG4 level (Figure 2A), were transduced with ECRG4 expressing virus or vector virus. At 48 h after ECRG4 viral transduction, ECRG4 expression was significantly increased (Figure 2B). ECRG4 overexpression remarkably increased the percentages of cells undergoing early apoptosis compared to cells transduced with vector (P<0.0001, Figure 2C).
Figure 2.
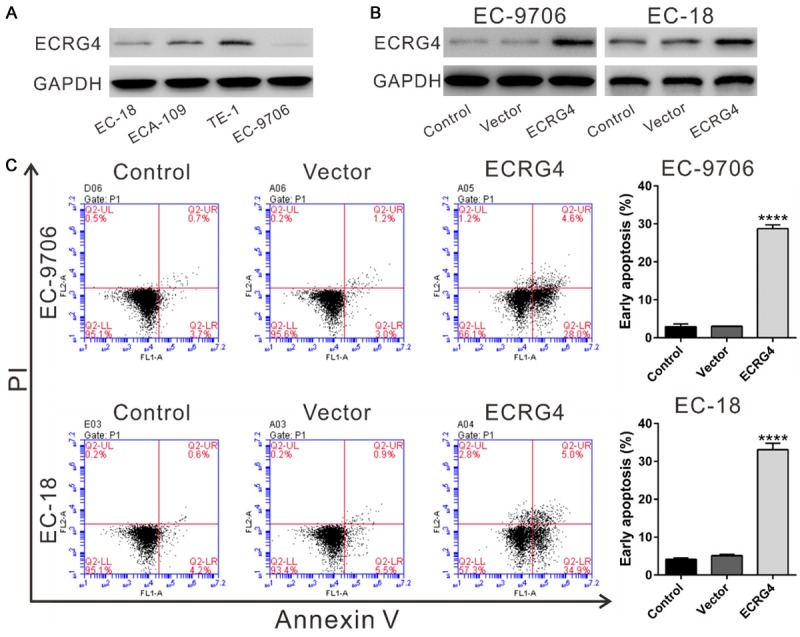
ECRG4 overexpression induced ESCC cell apoptosis. A: ECRG4 protein levels in 4 ESCC cell lines were analyzed by Western blotting. Experiments were performed three times and representative blots were presented. B: EC9706 and EC-18 cells were transduced with ECRG4 expressing virus (ECRG4) or vector virus (Vector), and 48 h later, ECRG4 expression was determined by Western blotting. C: Early apoptotic cells were evaluated by Annexin V-PI staining and flow cytometry analysis at 48 h after transduction. The cells without any treatment served as Control. ****P<0.0001 versus Control and Vector cells.
Identification of differentially expressed proteins between ECRG4 overexpressed cells and control cells
To further understand the effects of ECRG4 overexpression on ESCC cells, TMT labeling and LC-MS/MS analysis was applied to identify the differentially expressed proteins between EC9706-ECRG4 cells and Control cells. The experiments were conducted in triplicate and 5897 proteins were identified. Based on the criteria of P<0.05 and fold change ≥1.2, 485 abnormally expressed proteins were identified in ECRG4 overexpressed cells, including 175 up-regulated proteins (Table S1, ECRG4/Control ≥1.2) and 310 down-regulated proteins (Table S2, ECRG4/Control ≤0.83). The differentially expressed proteins were then subjected to Gene ontology (GO) analysis and categorized into three GO groups, biological processes, molecular function and cellular components (Figure S1).
Among the down-regulated proteins, cell cycle-, apoptosis- and invasion-related proteins, PLK1, CDK4, PLOD1 and PLOD2, attracted our attention because their mRNA levels in ESCC samples showed an increase trend comparing to those in normal tissues based on expression data from TCGA (Figure 3).
Figure 3.
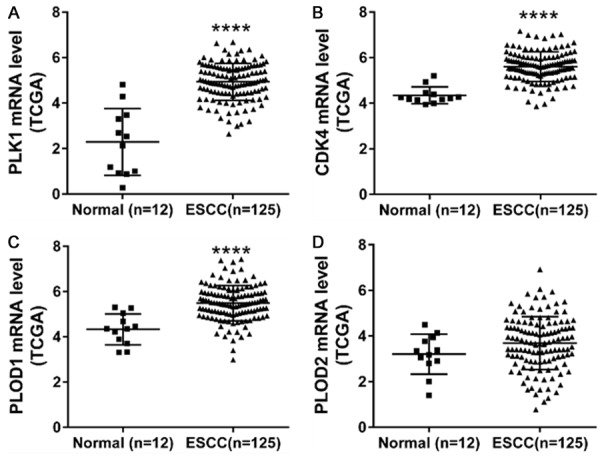
The mRNA expression of PLK1, CDK4, PLOD1 and PLOD2 in ESCC tissues (n=125) and the non-tumorous tissues (n=12) from TCGA dataset. ****P<0.0001.
To further validate the proteomics data, the changes of the above four proteins were detected by Western blotting after manipulating the expression of ECRG4 in ESCC cells. At 48 h after viral transduction, the protein expression levels of PLK1, CDK4, PLOD1 and PLOD2 were up-regulated in EC9706-ECRG4 cells and EC-18-ECRG4 cells as comparing to control cells (Figure 4A). TE-1 cells, which had relative higher ECRG4 level were transfected with siECRG4 or control siNC. ECRG4 knockdown significantly increased the expression of the above four proteins in TE-1 cells, which were complementary to the results from overexpression experiment (Figure 4B).
Figure 4.
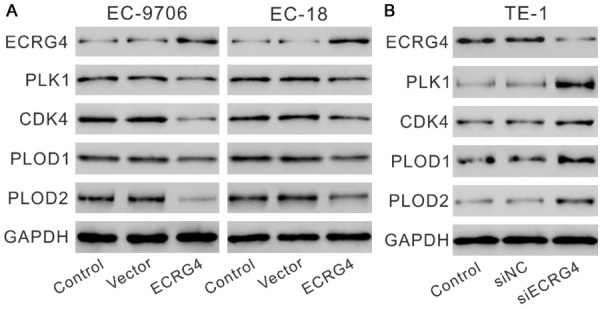
ECRG4 expression levels affected the expression of PLK1, CDK4, PLOD1 and PLOD2 in ESCC cells. EC9706 and EC-18 cells (A) were transduced with ECRG4 expressing virus (ECRG4) or vector virus (Vector), while TE-1 cells (B) were transfected with siECRG4 or control siNC. The cells without any treatment served as Control. At 48 h after treatment, the protein levels of ECRG4, PLK1, CDK4, PLOD1 and PLOD2 were detected by Western blotting. Experiments were performed three times and representative blots were presented.
ECRG4 expression was negatively correlated with PLK1, CDK4, PLOD1 and PLOD2
Immunohistochemical staining was then performed on 75 pairs of ESCC and normal specimens with antibodies against ECRG4, PLK1, CDK4, PLOD1 or PLOD2. Comparing to the matched normal specimens, ECRG4 was down-regulated in 73.3% (55/75) of ESCC samples, while PLK1, CDK4, PLOD1 and PLOD2 was up-regulated in 89.3% (67/75), 86.7% (65/75), 81.3% (61/75) and 81.3% (61/75) of ESCC samples, respectively. Interestingly, ECRG4 expression was negatively correlated with PLK1, CDK4, PLOD1 and PLOD2 (P<0.0001, Tables 1, 2, 3 and 4 and Figure 5).
Table 1.
Correlation between ECRG4 and PLK1 expression in 150 ESCC patients
| ECRG4 | PLK4 | ||
|---|---|---|---|
|
| |||
| Medium positive (%) | High positive (%) | Total (%) | |
| Low positive (%) | 8 (10.7%) | 23 (30.7%) | 31 (41.3%) |
| Medium positive (%) | 32 (42.7%) | 8 (10.7%) | 40 (53.3%) |
| High positive (%) | 4 (5.3%) | 0 (0%) | 4 (5.3%) |
| Total (%) | 44 (58.7%) | 31 (41.3%) | 75 (100%) |
X 2 test P<0.0001.
Table 2.
Correlation between ECRG4 and CDK4 expression in 150 ESCC patients
| ECRG4 | CDK4 | ||
|---|---|---|---|
|
| |||
| Medium positive (%) | High positive (%) | Total (%) | |
| Low positive (%) | 11 (14.7%) | 20 (26.7%) | 31 (41.3%) |
| Medium positive (%) | 35 (46.7%) | 5 (6.7%) | 40 (53.3%) |
| High positive (%) | 4 (5.3%) | 0 (0%) | 4 (5.3%) |
| Total (%) | 50 (66.7%) | 26 (33.3%) | 75 (100%) |
X 2 test P<0.0001.
Table 3.
Correlation between ECRG4 and PLOD1 expression in 150 ESCC patients
| ECRG4 | PLOD1 | ||
|---|---|---|---|
|
| |||
| Medium positive (%) | High positive (%) | Total (%) | |
| Low positive (%) | 10 (13.3%) | 21 (28%) | 31 (41.3%) |
| Medium positive (%) | 38 (50.7%) | 2 (2.7%) | 40 (53.3%) |
| High positive (%) | 4 (5.3%) | 0 (0%) | 4 (5.3%) |
| Total (%) | 52 (69.3%) | 25 (30.7%) | 75 (100%) |
X 2 test P<0.0001.
Table 4.
Correlation between ECRG4 and PLOD2 expression in 150 ESCC patients
| ECRG4 | PLOD2 | ||
|---|---|---|---|
|
| |||
| Medium positive (%) | High positive (%) | Total (%) | |
| Low positive (%) | 11 (14.7%) | 20 (26.7%) | 31 (41.3%) |
| Medium positive (%) | 34 (45.3%) | 6 (8%) | 40 (53.3%) |
| High positive (%) | 4 (5.3%) | 0 (0%) | 4 (5.3%) |
| Total (%) | 67 (65.3%) | 29 (34.7%) | 75 (100%) |
X 2 test P<0.0001.
Figure 5.
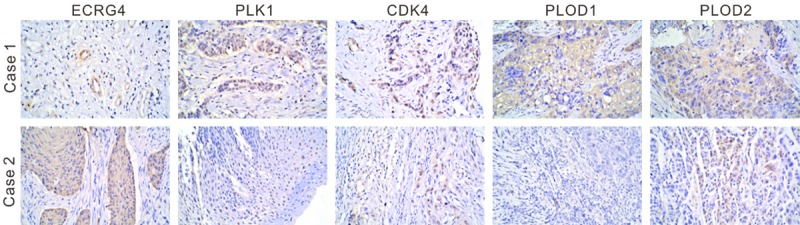
Correlation between ECRG4 and PLK1, CDK4, PLOD1 and PLOD2 expression in human ESCC. The tissues were incubated with indicated primary antibodies and visualized by DAB. Magnification: ×200. Case 1 exhibited low positive expression of ECRG4 and high positive expression of PLK1, CDK4, PLOD1 and PLOD2. Case 2 showed the contrary results.
Discussion
ECRG4 was a putative tumor suppressor gene in several cancers. ECRG4 overexpression can impede cancer cell growth by inducing cell cycle arrest and cell apoptosis, as well as inhibit the migration and invasion of cancer cells [11,13,15,20-22]. In the present study, by analyzing expression data of ESCC tissues from TCGA, we found a negatively correlation between ECRG4 and the processes of metastasis, apoptosis and cell cycle, which was consistent with the previous findings. The functions of ECRG4 overexpression on ESCC cell cycle progression, migration and invasion have been studied [15,20]. The current study revealed the induction effects of ECRG4 overexpression on ESCC cell apoptosis, which further indicated the tumor-suppressor function of ECRG4 in ESCC.
To further understand the effects of ECRG4 overexpression on ESCC cells, we carried out proteomic analysis to identify differentially expressed proteins between EC9706 ECRG4 overexpressed and control cells. Among the 485 abnormally expressed proteins, the expression of PLK1, CDK4, PLOD1 and PLOD2 were down-regulated, which may contribute to the functions of ECRG4 on the cell cycle progression, apoptosis and metastasis of ESCC cells. PLK1 protein expression was frequently increased in ESCC, and its overexpression was an independently prognosis factor for ESCC. Knockdown of PLK1 can induce cell apoptosis via mitochondrial pathway [23,24]. CDK4 is a key cell cycle regulator, and its inhibitors have anti-tumor activity in a wide range of cancers [25,26]. PLOD1 and PLOD2 encode procollagen hydroxylases, which are responsible for collagen remodeling [27]. PLOD2 is activity involved in cancer cell migration and invasion [28]. In the current study, up-regulation or down-regulation of ECRG expression in ESCC cells decreased or increased the expression of PLK1, CDK4, PLOD1 and PLOD2. As indicated by immunohistochemical staining, ECRG4 expression was frequently decreased in ESCC, while the expression of PLK1, CDK4, PLOD1 and PLOD2 was increased in ESCC. Most importantly, statistical analysis showed that ECRG4 expression negatively correlated with the expression of PLK1, CDK4, PLOD1 and PLOD2. The detailed mechanisms how ECRG4 affected their expression remain to be explored in the future study.
In summary, ectopic expression of ECRG4 induced ESCC cell apoptosis. Altered expression of ECRG4 affected the expression of PLK1, CDK4, PLOD1 and PLOD2 in ESCC cells. ECRG protein expression in ESCC tissues had a negative association with the expression of PLK1, CDK4, PLOD1 and PLOD2. Our study provided possible mechanism of the tumor-repressor functions of ECRG4.
Acknowledgements
This work was supported by the Chinese National Natural Science Foundation (U1304817).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stewart B, Wild CP. World Cancer Report 2014. 2014 [Google Scholar]
- 3.Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y, Ueda J, Wei W, Inoue M, Tanaka H. Epidemiology of esophageal cancer in Japan and China. J Epidemiol. 2013;23:233–242. doi: 10.2188/jea.JE20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambert R, Hainaut P. The multidisciplinary management of gastrointestinal cancer. Epidemiology of oesophagogastric cancer. Best Pract Res Clin Gastroenterol. 2007;21:921–945. doi: 10.1016/j.bpg.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Su T, Liu H, Lu S. Cloning and identification of cDNA fragments related to human esophageal cancer. Zhonghua Zhong Liu Za Zhi. 1998;20:254–257. [PubMed] [Google Scholar]
- 6.Bi MX, Han WD, Lu SX. Using lab on-line to clone and identify the esophageal cancer related gene 4. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue. 2000;33:257–261. [PubMed] [Google Scholar]
- 7.Matsuzaki J, Torigoe T, Hirohashi Y, Tamura Y, Asanuma H, Nakazawa E, Saka E, Yasuda K, Takahashi S, Sato N. Expression of ECRG4 is associated with lower proliferative potential of esophageal cancer cells. Pathol Int. 2013;63:391–397. doi: 10.1111/pin.12079. [DOI] [PubMed] [Google Scholar]
- 8.Mori Y, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Kurehara H, Mori R, Tomoda K, Ogawa R, Katada T, Harata K, Fujii Y. Expression of ECRG4 is an independent prognostic factor for poor survival in patients with esophageal squamous cell carcinoma. Oncol Rep. 2007;18:981–985. [PubMed] [Google Scholar]
- 9.Li LW, Yu XY, Yang Y, Zhang CP, Guo LP, Lu SH. Expression of esophageal cancer related gene 4 (ECRG4), a novel tumor suppressor gene, in esophageal cancer and its inhibitory effect on the tumor growth in vitro and in vivo. Int J Cancer. 2009;125:1505–1513. doi: 10.1002/ijc.24513. [DOI] [PubMed] [Google Scholar]
- 10.Gotze S, Feldhaus V, Traska T, Wolter M, Reifenberger G, Tannapfel A, Kuhnen C, Martin D, Muller O, Sievers S. ECRG4 is a candidate tumor suppressor gene frequently hypermethylated in colorectal carcinoma and glioma. BMC Cancer. 2009;9:447. doi: 10.1186/1471-2407-9-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Liu X, Zhang B, Qi D, Zhang L, Jin Y, Yang H. Overexpression of candidate tumor suppressor ECRG4 inhibits glioma proliferation and invasion. J Exp Clin Cancer Res. 2010;29:89. doi: 10.1186/1756-9966-29-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanaja DK, Ehrich M, Van den Boom D, Cheville JC, Karnes RJ, Tindall DJ, Cantor CR, Young CY. Hypermethylation of genes for diagnosis and risk stratification of prostate cancer. Cancer Invest. 2009;27:549–560. doi: 10.1080/07357900802620794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Z, Liang P, Xuan J, Wan J, Guo H. ECRG4 as a novel tumor suppressor gene inhibits colorectal cancer cell growth in vitro and in vivo. Tumor Biol. 2016;37:9111–9120. doi: 10.1007/s13277-015-4775-2. [DOI] [PubMed] [Google Scholar]
- 14.Sabatier R, Finetti P, Adelaide J, Guille A, Borg JP, Chaffanet M, Lane L, Birnbaum D, Bertucci F. Down-regulation of ECRG4, a candidate tumor suppressor gene, in human breast cancer. PloS One. 2011;6:e27656. doi: 10.1371/journal.pone.0027656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Zhang C, Li X, Lu S, Zhou Y. The candidate tumor suppressor gene ECRG4 inhibits cancer cells migration and invasion in esophageal carcinoma. J Exp Clin Cancer Res. 2010;29:133. doi: 10.1186/1756-9966-29-133. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han Y, Wei F, Xu X, Cai Y, Chen B, Wang J, Xia S, Hu H, Huang X, Wu M. Establishment and comparative genomic hybridization analysis of human esophageal carcinomas cell line EC9706. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2002;19:455–457. [PubMed] [Google Scholar]
- 18.Pan QQ. Studies on esophageal cancer cells in vitro. Proc Chin Acad Med Sci Peking Union Med Coll. 1988;4:52–57. [PubMed] [Google Scholar]
- 19.Miyazaki T, Kato H, Nakajima M, Sohda M, Fukai Y, Masuda N, Manda R, Fukuchi M, Tsukada K, Kuwano H. FAK overexpression is correlated with tumour invasiveness and lymph node metastasis in oesophageal squamous cell carcinoma. Br J Cancer. 2003;89:140–145. doi: 10.1038/sj.bjc.6601050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li LW, Yu XY, Yang Y, Zhang CP, Guo LP, Lu SH. Expression of esophageal cancer related gene 4 (ECRG4), a novel tumor suppressor gene, in esophageal cancer and its inhibitory effect on the tumor growth in vitro and in vivo. Int J cancer. 2009;125:1505–1513. doi: 10.1002/ijc.24513. [DOI] [PubMed] [Google Scholar]
- 21.Jiang CP, Wu BH, Wang BQ, Fu MY, Yang M, Zhou Y, Liu F. Overexpression of ECRG4 enhances chemosensitivity to 5-fluorouracil in the human gastric cancer SGC-7901 cell line. Tumour Biol. 2013;34:2269–2273. doi: 10.1007/s13277-013-0768-1. [DOI] [PubMed] [Google Scholar]
- 22.Xu T, Xiao D, Zhang X. ECRG4 inhibits growth and invasiveness of squamous cell carcinoma of the head and neck in vitro and in vivo. Oncol Lett. 2013;5:1921–1926. doi: 10.3892/ol.2013.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng YB, Lin DC, Shi ZZ, Wang XC, Shen XM, Zhang Y, Du XL, Luo ML, Xu X, Han YL, Cai Y, Zhang ZQ, Zhan QM, Wang MR. Overexpression of PLK1 is associated with poor survival by inhibiting apoptosis via enhancement of survivin level in esophageal squamous cell carcinoma. Int J Cancer. 2009;124:578–588. doi: 10.1002/ijc.23990. [DOI] [PubMed] [Google Scholar]
- 24.Bu Y, Yang Z, Li Q, Song F. Silencing of pololike kinase (Plk) 1 via siRNA causes inhibition of growth and induction of apoptosis in human esophageal cancer cells. Oncology. 2008;74:198–206. doi: 10.1159/000151367. [DOI] [PubMed] [Google Scholar]
- 25.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 26.Dickson MA. Molecular pathways: CDK4 inhibitors for cancer therapy. Clin Cancer Res. 2014;20:3379–3383. doi: 10.1158/1078-0432.CCR-13-1551. [DOI] [PubMed] [Google Scholar]
- 27.van der Slot AJ, Zuurmond AM, Bardoel AF, Wijmenga C, Pruijs HE, Sillence DO, Brinckmann J, Abraham DJ, Black CM, Verzijl N. Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J Biol Chem. 2003;278:40967–40972. doi: 10.1074/jbc.M307380200. [DOI] [PubMed] [Google Scholar]
- 28.Kurozumi A, Kato M, Goto Y, Matsushita R, Nishikawa R, Okato A, Fukumoto I, Ichikawa T, Seki N. Regulation of the collagen crosslinking enzymes LOXL2 and PLOD2 by tumorsuppressive microRNA-26a/b in renal cell carcinoma. Int J Oncol. 2016;48:1837–1846. doi: 10.3892/ijo.2016.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


