Abstract
Currently, there is a considerable need to develop new treatments for osteosarcoma (OS), a very aggressive bone cancer. The activation of STAT3 signaling is positively associated with poor prognosis and aggressive progression in OS patients. Our previous study reported that the FDA-approved antipsychotic drug pimozide had anti-tumor activity against hepatocellular carcinoma and prostate cancer cells by suppressing STAT3 activity. Therefore, the aim of this study was to investigate the specific effect of pimozide on OS cells and the underlying molecular mechanism. Pimozide inhibited cell proliferation, colony formation, and sphere formation capacities of the OS cells in a dose-dependent manner, inducing G0/G1 phase cell cycle arrest. Pimozide reduced the percentage of side population cells representing cancer stem-like cells and enhanced the sensitivity of OS cells to 5-FU induced proliferative inhibition. In addition, pimozide induced apoptosis of U2OS cells, which showed increased expression of cleaved-PARP, a marker of programmed cell death. Moreover, pimozide suppressed Erk signaling in OS cells. Importantly, pimozide induced ROS generation by downregulating the expression of the antioxidant enzyme catalase (CAT). NAC treatment partially reversed the ROS generation and cytotoxic effects induced by pimozide. CAT treatment attenuated the pimozide-induced proliferation inhibition. The decrease of CAT expression induced by pimozide was potentially mediated through the suppression of cellular STAT3 activity in OS cells. Thus, pimozide may be a novel STAT3 inhibitor that suppresses cellular STAT3 activity to inhibit OS cells or stem-like cells and is a novel potential anti-cancer agent in OS treatment.
Keywords: Pimozide, osteosarcoma, STAT3 inhibitor, ROS, translational medicine
Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor, predominantly affecting children and young adults [1,2]. It is a very aggressive cancer and is universally fatal if left untreated. The 5-year survival rate of OS patients is 60%-70%, with no significant improvements in prognosis since the advent of multi-agent chemotherapy [3]. Many patients develop metastases that are difficult to treat and confer a poor prognosis [4]. The most frequent metastases sites are lung and bone [5]. The current available adjuvant chemotherapy provides no survival advantage for patients with pulmonary metastases [6]. Therefore, the development of novel strategies is critical to overcoming the poor prognosis of OS.
Importantly, the Signal Transducer and Activator of Transcription 3 (STAT3) signaling is constitutively active in various human cancers, including osteosarcoma [7,8]. The activation of STAT3 signaling is positively associated with poor prognosis and aggressive progression in OS patients [9]. STAT3 signaling is involved in tumor initiation, proliferation, and metastasis of cancer cells [10]. Several studies have suggested that inhibiting STAT3 signaling by an antagonist or small interfering RNA induced apoptosis and prevented from metastasis of OS cells [11]. STAT3 signaling activity was blocked by an IL-6-neutralizing antibody, leading to reduced proliferation, migration, and invasion of osteosarcoma cells through the downregulation of Survivin and Bcl-xL and the upregulation of pro-apoptotic genes [12]. Thus, it is indicated that targeting STAT3 activation appears to be an effective therapy for the treatment of OS.
Increasing numbers of FDA-approved clinical drugs have been re-purposed to treat various cancers, leading to increased choices and improved effectiveness of anti-tumor therapies [13]. Pimozide, an FDA-approved compound used to clinically treat psychotic diseases, has anti-cancer effects on various tumors and leukemia, such as breast cancer [14], melanoma [15] and myelogenous leukemia [16]. Pimozide mechanically inhibited the activation of STAT5 signaling to decrease the cell viability of leukemia cells [16]. Moreover, our previous study showed that the antipsychotic drug pimozide inhibited the proliferation and self-renewal abilities of cancer cells, including hepatocellular carcinoma and prostate cancer cells, by suppressing STAT3 activity [17,18]. Furthermore, pimozide was reported to inhibit Wnt/β-catenin signaling and reduce the expression of the stem cell marker EpCAM in hepatocellular carcinoma cells [19]. However, neither the anti-tumor effect of pimozide nor the inhibition of STAT3 activity by pimozide treatment in OS cells has been fully demonstrated.
The aim of this study was to investigate whether the neuroleptic drug pimozide has anti-cancer effects on osteosarcoma cells through the suppression of STAT3 signaling activity. The results showed that pimozide inhibited U2OS cancer cell proliferation, colony formation, and sphere formation and induced G0/G1 phase cell cycle arrest. In addition, pimozide reduced the percentage of side population and induced the apoptosis of cancer cells. Importantly, pimozide induced the reactive oxygen species (ROS) generation by suppressing catalase expression. Therefore, pimozide may be a novel potential anti-tumor agent in treating OS cells or stem-like cells.
Materials and methods
Cell line and cell culture
Human osteosarcoma U2OS cell lines (provided by Medical Centre of Shenzhen University, Shenzhen, China) was cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; Sigma, St. Louis, MO, USA) in incubator with 5% CO2 at 37°C.
Cell proliferation assay using WST-8
Cell proliferation was assessed by WST-8 colorimetric assay (Dojindo Laboratories, Gaithersburg, MD). Human osteosarcoma cells were plated in 96-well plates with 2,500 cells per well and exposed to the treatment of different concentrations of pimozide for various time intervals (24 h, 48 h, and 72 h). The WST-8 solution was added to each well after indicated time. After incubated at 37°C for another 4 hours, the absorbance was measured at 450 nm using a multi-well plate reader (BioTek, Vermont, USA).
Cell cycle assay
Cell cycle was determined by propidium iodide (PI, BD Biosciences Clontech, USA) staining. Briefly, equal amounts of cells were seeded in 6-well plates and treated with pimozide at different concentration for 48 h. The cells were harvested, washed with PBS containing 0.1% BSA, and then, cold 70% ethanol was added while mixing the cells. PI buffer (40 μg/mL, containing 100 μg/mL RNase) was added, and the cells were analyzed by flow cytometry.
Colony formation assay
Cells with different concentrations of pimozide were plated in 10% FBS medium for 7 days. Cells were stained with 0.5% crystal violet and photographed. The morphology and the number of colonies were observed using the microscope.
Sphere formation assay
Sphere formation assay was performed as previously described [20]. To establish sphere cultures, single cells were cultured in 200 μl of serum-free DMEM/F12 medium (Gibco) supplemented with 20 ng/ml human recombinant epidermal growth factor (EGF, PeproTech), 20 ng/ml human recombinant basic fibroblast growth factor (bFGF, PeproTech), and B27 (1:50; Gibco). Cells were cultured in ultra-low attachment plates with or without pimozide treatment. After plating for 14 days, all spheres in each well were photographed.
Apoptosis assay using Annexin V/PI staining
The percentage of apoptotic cells was measured using an Annexin V-FITC/PI Apoptosis Detection Kit according to the manufacturer’s instructions (Keygen, Nanjing, China). Briefly, cells were treated with the indicated concentrations of pimozide. The cells were then collected and used for Annexin V/PI staining. The stained cells were analyzed by flow cytometry (Calibur, BD Biosciences, San Diego, CA, USA).
Reactive oxygen species assay
ROS generation was detected using the fluorescent dyes 2,7-dichlorofluorescein diacetate (DCFH-DA) and dihydroethidium (DHE) (Beyotime, Jiangsu, China). Briefly, after pimozide treatment for 48 h, cells were washed with medium and were incubated with DCFH-DA (10 μM) or DHE (10 μM) in DMEM for 30 min at 37°C in the dark. Then, cells were washed twice with DMEM and analyzed using a flow cytometer (FACSCalibur, BD Biosciences, San Diego, CA, USA) at an excitation/emission wavelength of 488/525 nm and 488/610 nm, respectively [21].
Total RNA extraction and real-time PCR assay
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s protocol and reversely transcribed into cDNA using a Revert Aid First-Strand cDNA Synthesis Kit (Thermo Scientific, USA). The primers are provided in Table 1. HPRT1 expression was used as an internal control [22].
Table 1.
The primers for qPCR
| Name | Genbank accession | Sense sequence (5’-3’) | Antisense sequence (5’-3’) |
|---|---|---|---|
| HPRT1 | NM_000194 | GCGTCGTGATTAGTGATGATGA | GCACACAGAGGGCTACAATG |
| MCL1 | NM_021960 | GGCAGGATTGTGACTCTC | CTCCTACTCCAGCAACAC |
| BCL-xL | NM_138578 | GCTGGTGGTTGACTTTCTCTC | GGTCTCCATCTCCGATTCAGT |
| MYC | NM_002467 | AGGAACAAGAAGATGAGGAAGA | CTGCGTAGTTGTGCTGATG |
| STAT3 | NM_003150 | GGTCTGGCTGGACAATATCATT | GAGGCTTAGTGCTCAAGATGG |
| SOD1 | NM_000454 | GGTCCTCACTTTAATCCTCTAT | CATCTTTGTCAGCAGTCACATT |
| SOD2 | NM_000636 | TGACAAGTTTAAGGAGAAGC | GAATAAGGCCTGTTGTTCC |
| CAT | NM_001752 | TTAATCCATTCGATCTCACC | GGCGGTGAGTGTCAGGATAG |
| GPX1 | NM_000581 | CGCCACCGCGCTTATGACCG | GCAGCACTGCAACTGCCAAGCAG |
| CHiP FOS | / | ATTGAACCAGGTGCGAATGT | GGAGGGATTGACGGGAACT |
| CHiP CAT-1 | / | GGGTGCTAAAGATCAATTTGTG | CCTCAGGTGCTAGGATTTATCT |
| CHiP CAT-2 | / | ACCAGTCTTGTTTCTCCATTTC | TCATCTCCAAAGTCCACAGTTT |
Abbreviations: HPRT1, hypoxanthine phosphoribosyltransferase 1; MCL1, myeloid cell leukemia 1; STAT3, signal transducer and activator of transcription 3; SOD1, cytosolic superoxide dismutase; SOD2, mitochondrial superoxide dismutase; CAT, catalase; GPX1, glutathione peroxidase 1.
Western blotting assay
Equal amounts of protein from cells sample harvested with RIPA Lysis Buffer were subjected to electrophoresis in SDS-polyacrylamide gel and transferred to nitrocellulose membrane (Merck Millipore, Billerica MA, USA). Blots were detected using primary antibodies against GAPDH (Ambion, Austin, TX, USA), p21, p27, Nanog, SOX2, phospho-STAT3 (Tyr705) (p-STAT3), STAT3, cleaved poly ADP-ribose polymerase (Asp214) (cleave-PARP), Erk, phosphorylated Erk (p-Erk) (Cell Signaling Technology, Beverly, MA, USA), CAT (Beyotime, Jiangsu, China), Cyclin D1, c-Myc and β-actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Antibody binding was detected with an enhanced chemiluminescence (ECL) kit (Thermo Fisher Scientific, Rockford, USA) [23].
STAT3 luciferase reporter assay
Transient transfection was conducted using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions. For the luciferase reporter assay, U2OS cells were seeded in 24-well plates and transfected with the STAT3 luciferase reporter plasmid STAT3-Luc (pGL3.0, Promega, USA). The cells were collected 48 h after transfection, and the luciferase activities in the cell lysates were determined using the Dual Luciferase Reporter Assay System (Promega, WI, USA). Each transfection was performed in triplicate and repeated at least three times.
Chromatin immunoprecipitation assay
Chromatin Immunoprecipitation (ChIP) assay was performed by the Chromatin Immunoprecipitation Kits (Millipore, Billerica, MA, USA) according to manufacturer’s instructions. Fold enrichment method as described was used to analyze the ChIP-qPCR data.The primers are provided in Table 1.
Statistical analysis
The data were presented as the mean ± SD and analyzed using GraphPad Prism 6.0 (GraphPad Software, Inc.). The Student’s two-tailed t-test was used to assess statistically significant differences between experimental and control groups. Analysis of variance (ANOVA) was used when more than two data sets were analyzed, followed by an appropriate post hoc test. The level of significance was set at P < 0.05. The statistical results were shown, *P < 0.05, **P < 0.01.
Results
The antipsychotic agent pimozide inhibits STAT3 signaling activation and reduces cell proliferation of osteosarcoma cells
Initially, we examined whether the antipsychotic agent pimozide inhibited STAT3 signaling activity. A Western blot analysis was performed to determine the phosphorylation of STAT3 at tyrosine 705 (pY-STAT3), which reflects STAT3 signaling activation. Figure 1A shows that pimozide reduced the basal expression of pY-STAT3 in U2OS cells in a dose-dependent manner. Moreover, pimozide inhibited STAT3-dependent luciferase activity (Figure 1B). RT-qPCR results demonstrated that pimozide treatment inhibited the transcriptional levels of the STAT3 signaling downstream genes MYC, BCL-xL and MCL1 (Figure 1C). Thus, it indicated that U2OS cells showed decreased STAT3 activity after pimozide treatment.
Figure 1.
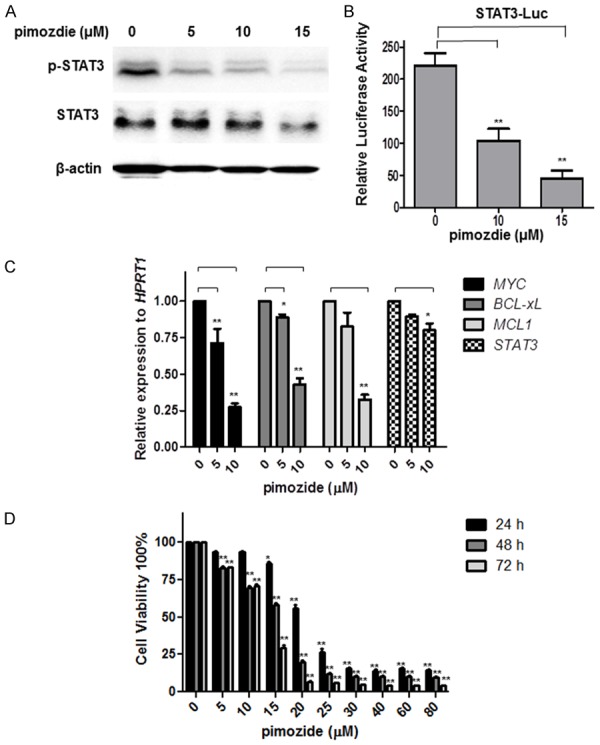
The antipsychotic agent pimozide inhibits STAT3 signaling activation and reduced cell proliferation of osteosarcoma cells. U2OS cells were incubated with the indicated doses of pimozide for 48 h before being subjected to Western blot or qPCR assays to analyze cellular STAT3 signaling activity. A. Western blot analysis of the protein expression of p-STAT3 and STAT3. B. U2OS cells were transfected with STAT3 reporter and pRL-TK renilla luciferase reporter plasmids. After 24 h of transfection, the cells were treated with different doses of pimozide for another 48 h. A dual luciferase assay was performed to detect the relative luciferase activity. C. An RT-qPCR assay was performed to analyze the expression of the STAT3 downstream genes MYC, BCL-xL, MCL1, and STAT3. D. U2OS cells were treated with various concentrations of pimozide for various times, and cell viability was determined by WST-8 colorimetric assay. The results are shown as the mean values ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, compared with the control.
Next, the anti-proliferative effect of pimozide in U2OS cells was detected using a WST-8 colorimetric assay. U2OS cells were exposed to a series of concentrations of pimozide for 24, 48, and 72 h. As shown in Figure 1D, pimozide inhibited the proliferation of U2OS cells in both a dose- and time-dependent manner. The IC50 value at 24, 48, and 72 h was 22.16 ± 2.54, 17.49 ± 1.14 and 13.78 ± 0.34 μΜ, respectively. Similar results were observed in two other OS cell lines, MG-63 and SW1353 (Supplementary Figure 1). Pimozide, might, therefore have therapeutic potential in treating OS cells.
Pimozide inhibits the colony- and sphere-forming abilities of osteosarcoma cells
We next examined whether pimozide inhibited the colony- and sphere-forming abilities of U2OS cells. Colony and sphere formation assays showed that pimozide treatment suppressed the numbers of colonies or spheres in a dose-dependent manner (Figure 2A and 2B). After treatment with 10 μΜ pimozide for 7days, U2OS cells showed a 95.34% ± 0.577% decrease in the number of colonies. A sphere formation assay showed that the inhibition rate was 56.24% ± 5.08%, 87.12% ± 2.22% and 98.44% ± 0.22% in U2OS cells treated with 5, 10, and 15 μM pimozide, respectively. Similar results were observed in the two other OS cell lines, MG-63 and SW1353 (Supplementary Figure 2). In addition, a Western blot assay showed that U2OS cells had significantly reduced expression of the stemness genes Nanog, Sox2, and c-Myc after treatment with pimozide for 48 h (Figure 2C).
Figure 2.
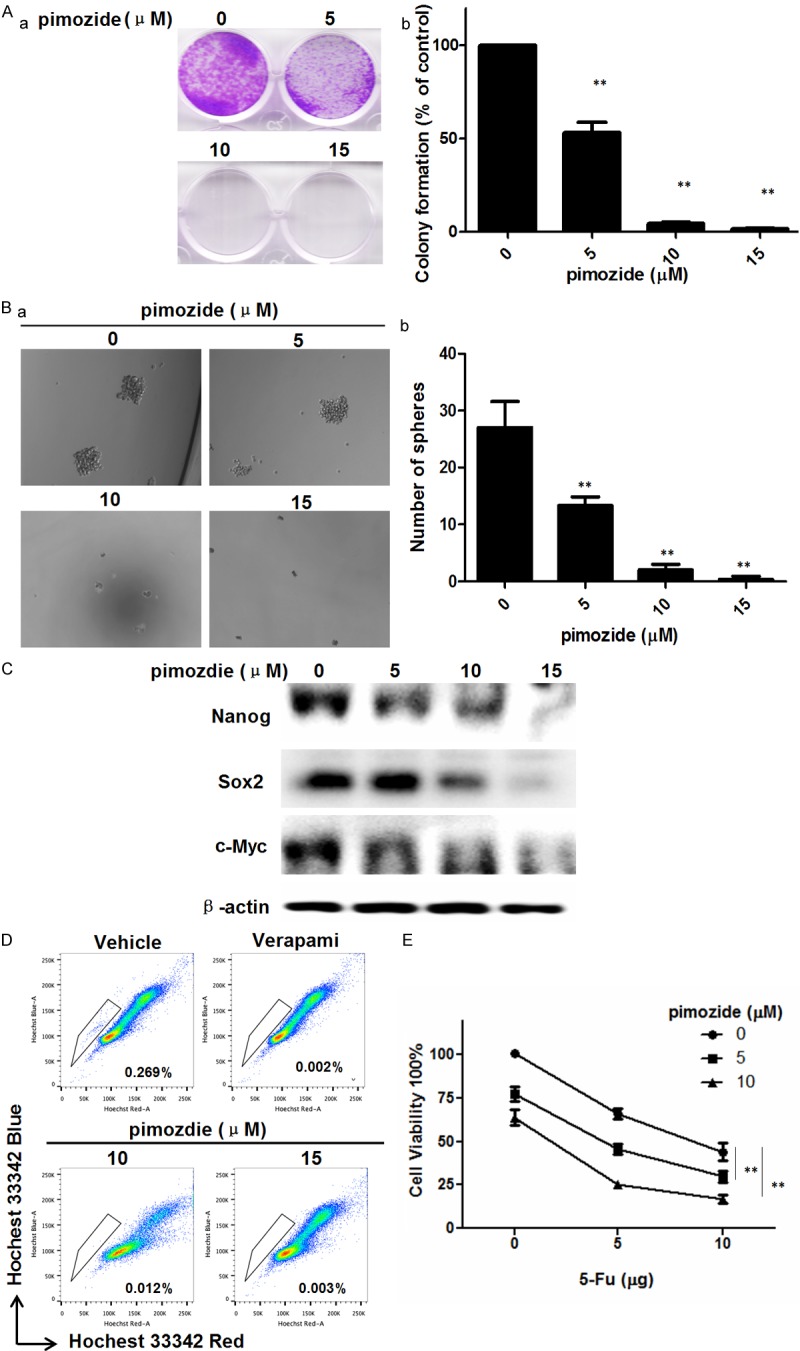
Pimozide inhibits the colony- and sphere-forming abilities of osteosarcoma cells.U2OS cells were treated with indicated concentrations of pimozide for the indicated times. A. Colony formation assay of U2OS cells treated with pimozide. The numbers of colonies were counted after staining with crystal violet. The graph indicates the number of colonies (a) and the statistical results are displayed (b). B. Sphere formation assay of U2OS cells treated with pimozide. The spheres were imaged under a light microscope (magnification, 100×) (a), and the statistical results are shown (b). C. Western blot analysis of the expression of stemness factors in U2OS cells. Cells were treated with the indicated concentrations of pimozide for 48 h. The cell lysates were subjected to SDS-PAGE and immunoblotted with antibodies against Nanog, Sox2, and c-Myc. β-actin was used as a loading control. D. Side population analysis of U2OS with indicated treatment. The population rate was shown. Relative representative figures are shown from 1 of 3 independent experiments. E. The cell viability assay was performed to show whether pimozide enhanced the sensitivity of U2OS cells to 5-FU induced proliferative inhibition. The results are from 3 independent experiments. The statistical results are shown, *P < 0.05, **P < 0.01.
We then examined the percentage of side population cells in U2OS cells treated with pimozide using flow cytometry. Pimozide treatment decreased the percentage of side population cells from 0.269% to 0.012% (Figure 2D). Moreover, pimozide treatment enhanced the sensitivity of U2OS cells to 5-Fluorouracil (5-FU) induced proliferative inhibition (Figure 2E). Therefore, these results suggested that pimozide decreased the stemness of OS cells, leading to inhibit OS cells or stem-like cells.
Pimozide induces G0/G1 phase cell cycle arrest in osteosarcoma cells
To determine whether pimozide could induce cell cycle arrest to inhibit cell growth, we analyzed the effect of pimozide on cell cycle distribution using PI staining. After U2OS cells were treated with different concentrations of pimozide for 24 h, the percentage of cells in G0/G1 phase increased significantly in a dose-dependent manner (Figure 3A and 3B). Following treatment with 15 μΜ pimozide, U2OS cells exhibited a significant increase in the percentage of G0/G1 phase cells, from 48.23 ± 2.34% to 64.95 ± 3.18%. In addition, we assessed whether pimozide inhibited the expression of molecular markers associated with G0/G1 phase arrest. A Western blot assay indicated that the p21 and p27 levels were remarkably increased and the Cyclin D1 level was decreased in U2OS cells treated with pimozide compared with control cells (Figure 3C). Besides, the expression of Ki67, a proliferation marker, was decreased from 83.2% to 45.2% in U2OS cells treated with 15 μΜ pimozide for 48 h (Figure 3D). Thus, these data indicated that pimozide decreased the viability of OS cells in positive association with G0/G1 phase cell cycle arrest.
Figure 3.
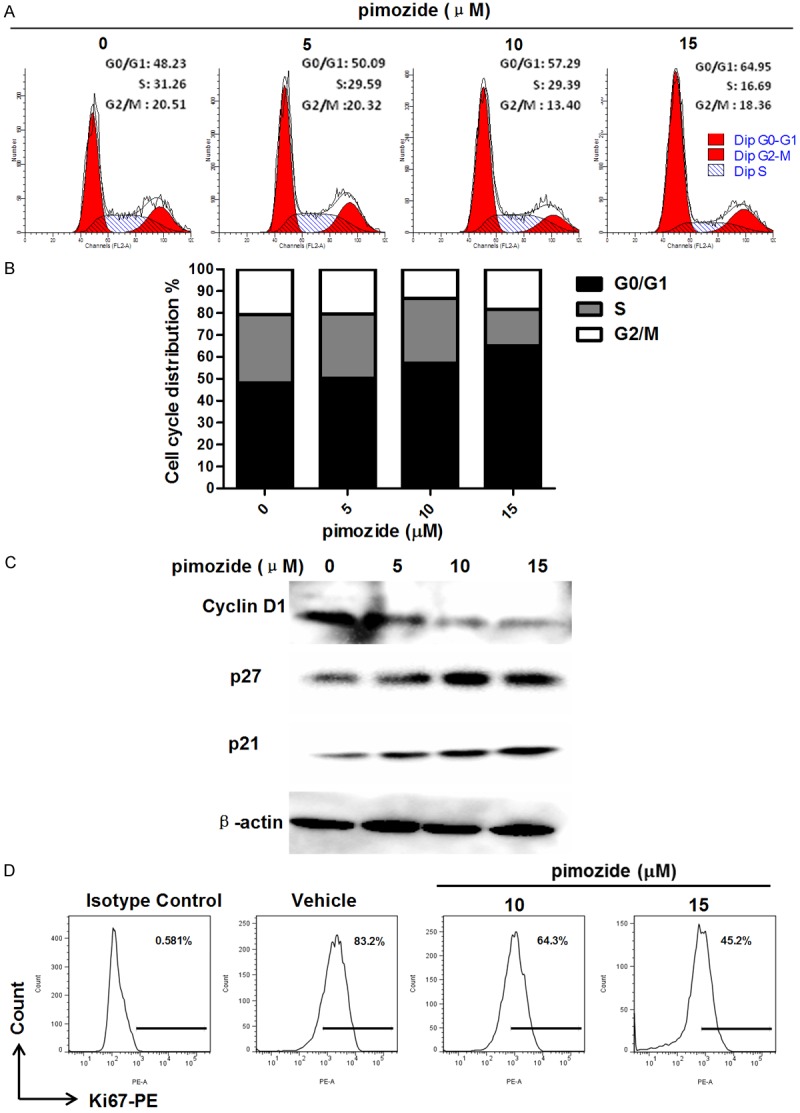
Pimozide induces G0/G1 phase cell cycle arrest in osteosarcoma cells. A. U2OS cells were stained with PI and subjected to flow cytometry to determine the cell distributions at each phase of the cell cycle. B. The statistical results are shown from above data. C. Western blot analysis of the expression of cell cycle-related genes. Cell extracts were probed with antibodies against p21, p27 and Cyclin D1. β-actinwas used as an internal control. D. Analysis of Ki67 in U2OS cells treated as indicated. The population rate was shown.Relative representative figures are shown from 1 of 3 independent experiments.
Pimozide promotes apoptosis in U2OS osteosarcoma cells
An Annexin-V-FITC/PI double staining assay showed that pimozide promoted the apoptosis of U2OS cells in a dose-dependent manner (Figure 4A and 4B). Flow cytometry showed that the total apoptosis rate was 20.22% ± 2.04%, 25.63% ± 2.16% and 38.09% ± 2.18%, respectively, in U2OS cells treated for 24 h with 5, 10 and 15 μM pimozide, while the apoptosis rate of the control cells was 8.12% ± 1.08%. Moreover, the expression levels of apoptosis-related proteins were measured by a Western blot assay (Figure 4C). Cells treated with pimozide for 24 h exhibited significantly increased expression of cleaved PARP, which is produced by caspase-3 cleavage during programmed cell death. These results indicated that pimozide promoted apoptosis in U2OS cells.
Figure 4.
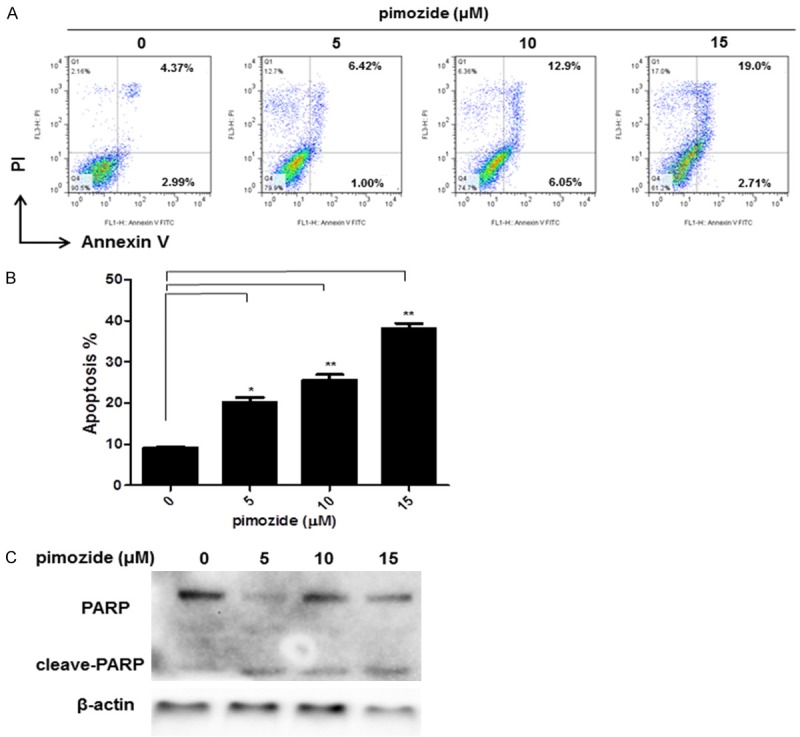
Pimozide promotes apoptosis in osteosarcoma cells. U2OS cells were treated with pimozide at different concentrations for 48 h. A. Apoptotic cells were measured by Annexin V/PI staining. Relative representative figures are shown from 1 of 3 independent experiments. B. Data summarized three independent experiments, *P < 0.05, **P < 0.01, compared to control. C. Western blot analysis of the expression of the apoptotic marker cleaved-PARP in U2OS after pimozide treatment. β-actin was used as an internal control.
Pimozide suppresses the extracellular signal-regulated kinase (Erk) signaling to inhibit cell viability of osteosarcoma cells
Previous studies have found cross talk between the STAT3 and Erk pathways [24]. To examine whether pimozide inhibited Erk signaling, a Western blot analysis was performed to evaluate the expression of Erk phosphorylation (p-Erk). The results showed that pimozide reduced the expression of p-Erk of U2OS cells in a dose-dependent manner (Figure 5A). In addition, a proliferation curve showed that the rate of cell viability was 39.63% ± 1.44% in U2OS cells treated with 15 μΜ pimozide alone and 38.23% ± 1.39% in cells treated with 15 μΜ pimozide plus 10 μΜ U0126, an indirect Erk signaling inhibitor (Figure 5B). There was no statistically significant difference in the anti-proliferative effect of pimozide between the groups treated with and without U0126. Similar results were found after treatment with 20 μΜ pimozide and in U2OS cells treated with pimozide alone or with pimozide plus the direct Erk signaling inhibitor FR180204 (Figure 5D).
Figure 5.
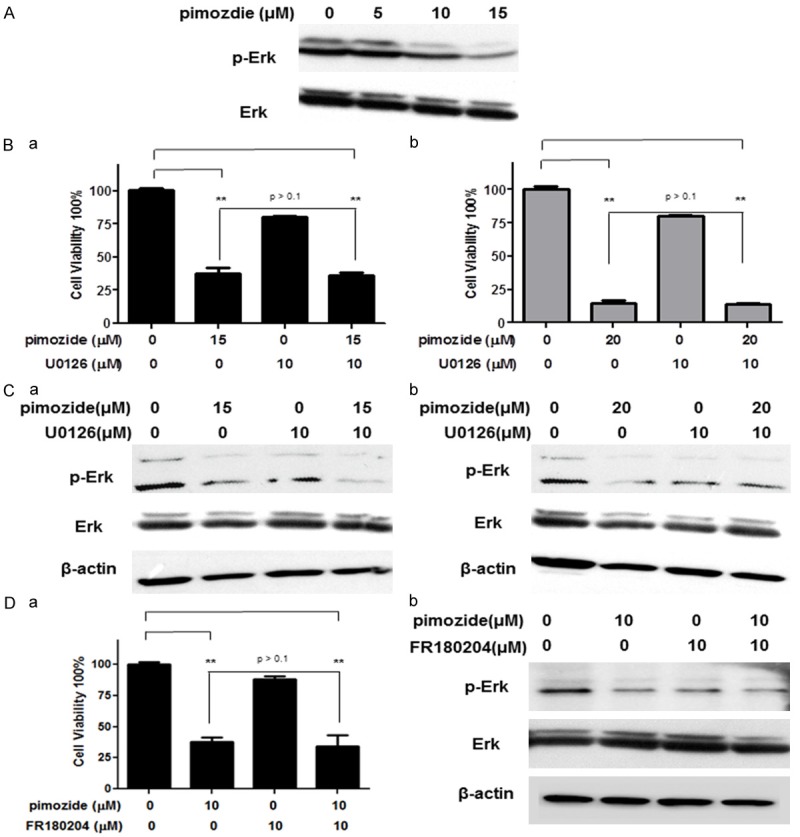
Pimozide suppresses the extracellular signal-regulated kinase (Erk) signaling to inhibit cell viability of osteosarcoma cells. U2OS cells were incubated with the indicated doses of pimozide for 48 h before being subjected to a Western blot assay to analyze cellular Erk signaling. A. Western blot analysis of the level of p-Erk and Erk expression. U2OS cells were treated with the indicated doses of pimozide alone or with the Erk signaling inhibitor U0126 or FR180204 for 72 h before being subjected to a cell viability assay or Western blot assay. B, Da. Cell proliferative analysis of U2OS cells after specific treatments. The results are shown as the mean values ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, compared with the control. C, Db. Western blot analysis of the level of p-Erk and Erk expression. β-actin was used as a loading control.
Erk and p-Erk expression were further examined using a Western blot assay to detect Erk signaling activation. As shown in Figure 5C, U2OS cells did not exhibit significantly decreased p-Erk expression after treatment with pimozide alone or with pimozide plus U0126. Similar data were observed in the FR180204 group (Figure 5D). According to these results, pimozide could suppress Erk signaling to inhibit the viability of OS cells.
Pimozide produces ROS generation in osteosarcoma cells through inhibiting antioxidant enzyme gene catalase expression
Reports have indicated that ROS production could partially contribute to cell apoptosis and affect cell proliferation. We next evaluated the effect of pimozide on the ROS levels in U2OS cells using the fluorescent dyes DCFH-DA and DHE. U2OS cells were incubated with 15 μΜ pimozide for 48 h, and the cells were then collected, stained with DCFH-DA or DHE and subjected to flow cytometry. As expected, the ROS levels increased in U2OS cells after treatment with pimozide (Figure 6A). Moreover, the ROS production-related genes SOD1, SOD2, CAT and GPX1 were analyzed by qPCR to demonstrate the decreased ROS levels induced by pimozide. The results showed that pimozide treatment inhibited the transcription levels of the CAT gene but had little effect on the SOD1, SOD2 and GPX1 genes (Figure 6B). Presumably, the pimozide-induced ROS generation was associated with the expression of the antioxidant enzyme gene CAT in OS cells.
Figure 6.
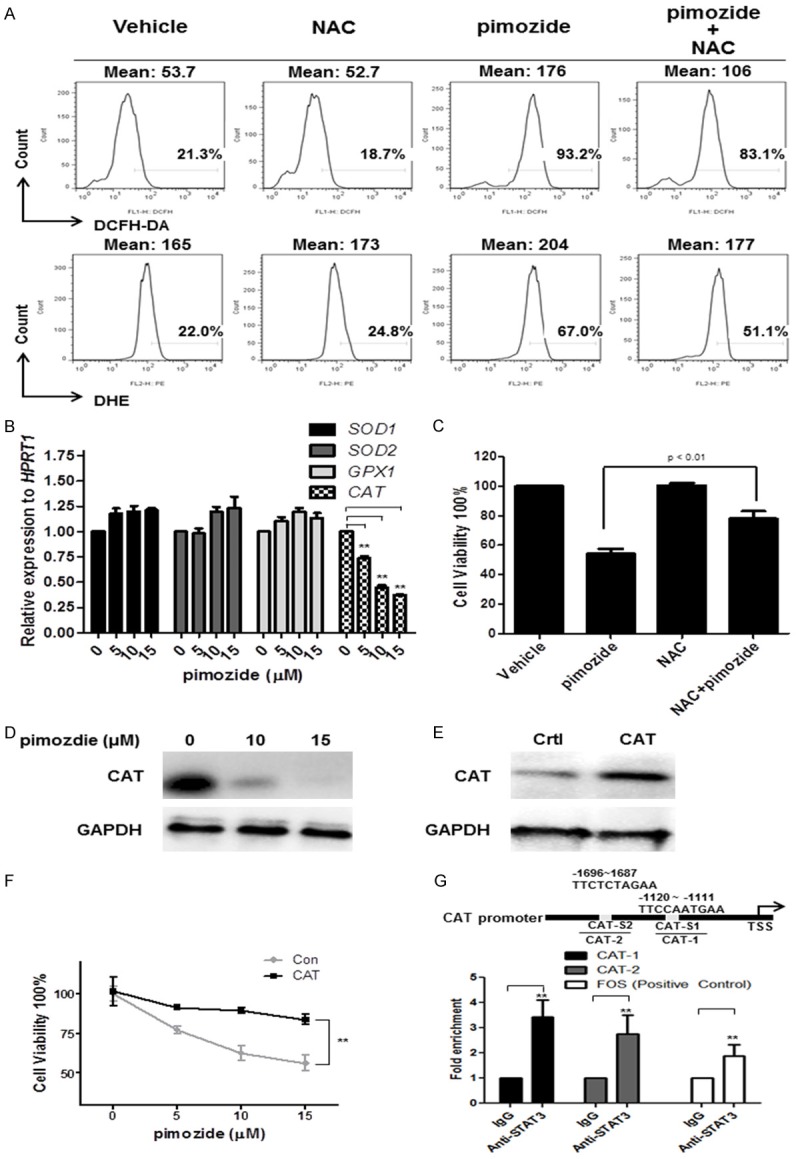
Pimozide produces ROS generation in osteosarcoma cells through inhibiting antioxidant enzyme gene catalase expression. U2OS cells were treated with 15 μΜ pimozide alone, 2 mM NAC alone or a combination of the two for 48 h, and the cells were then collected, stained with DCFH-DA or DHE and subjected to flow cytometry. A. ROS analysis of U2OS cells treated as indicated. The intensity or the population rate is shown. Relative representative figures are shown from 1 of 3 independent experiments. B. The expression of the ROS production-related genes SOD1, SOD2, CAT, and GPX1 were analyzed by RT-qPCR. C. Cell viability was determined by a WST-8 colorimetric assay. D. Western blot analysis of the expression of the CAT protein in U2OS cells treated as indicated. E. Western blot analysis of the expression of the CAT protein in U2OS cells transfected with a CAT expression plasmid. F. Cell viability was determined by a WST-8 colorimetric assay. G. The conserved STAT3 binding sequence “TTCN3~4 GAA” was searched in the promoter of the CAT gene, and two putative STAT3-binding sites were found. A ChIP assay was performed with an antibody against STAT3 in U2OS cells. Real-time PCR was then used to measure the enrichment of the putative STAT3-binding sites in the CAT gene. The results are shown as the mean values ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, compared with the control.
To determine whether the pimozide-induced ROS generation was affected by the presence of antioxidant compounds, we analyzed the pimozide-induced effects in the presence of NAC. NAC treatment partially reversed the level of ROS generation induced by pimozide in U2OS cells (Figure 6A). The cytotoxic effects observed in U2OS cells treated with pimozide were decreased in the presence of NAC (Figure 6C). In addition, pimozide reduced the expression levels of the CAT protein (Figure 6D). Moreover, we examined whether increased CAT expression reversed the pimozide-induced inhibitory effect on OS cells. A Western blot analysis revealed increased expression of the CAT protein in U2OS cells transfected with CAT overexpression plasmid (Figure 6E). CAT treatment attenuated the pimozide-induced proliferation inhibition (Figure 6F). These results suggested that pimozide induced ROS generation in OS cells by inhibiting the expression of the antioxidant enzyme gene CAT.
To determine whether CAT is a direct target of STAT3 signaling, we searched for STAT3-binding sites in the regulatory regions of the CAT gene and found two putative STAT3-binding sites (Figure 6G). We then performed a ChIP analysis of STAT3 binding to the promoter of the CAT gene in OS cells and found that STAT3 was able to bind the CAT promoter. These data indicated that the decrease in CAT expression induced by pimozide was potentially mediated through the suppression of cellular STAT3 activity in OS cells.
Discussion
Drug discovery and development for the clinical treatment of OS has been taken seriously. Drug repurposing, new applications for existing or abandoned pharmacotherapies, is one of the most important strategies used to treat cancer cells [25]. For example, metformin, an anti-diabetic drug, can inhibit cancer cell growth in vitro and in vivo, functioning as an anti-cancer drug [26]. Clinically approved drugs used to treat a new disease typically have two important advantages. One is that the drug often has low toxicity to cells in vivo. The other is that the drug has clear molecular targets or treatment for specific diseases [27]. Drug repurposing brings new advances in drug discovery and development, making pharmaceutical research more predictable and reliable and less costly. In addition, since the common chemotherapeutics cannot obviously improve the disease conditions of OS patients, it is meaningful that an FDA approved drug is repurposed for the treatment of OS. In this study, we found that pimozide, a clinical drug approved by the FDA to treat neuroleptic disorders, has anti-cancer effects in OS cells, reducing the proliferation ability and increasing the apoptosis of the osteosarcoma cells in vitro. The antipsychotic agent pimozide may, therefore, be a potential and novel curative for patients with osteosarcoma.
Although pimozide can induce cardiac toxicity, it has not been reported to have adverse effects on other normal functional cells, such as hepatic or hematopoietic cells. A previous study showed that pimozide has almost no effect on hematopoietic progenitors derived from healthy donors [16]. Pimozide treatment was well tolerated in a mouse model with no significant effects on body weight [16]. Similar results were observed in our previous study [16]. The dose (25 mg/kg) of pimozide used to treat cancer in-vivo is relatively low compared to the commonly used dose for treating CNS diseases. Additionally, according to the previous study, the precise lethal dose of pimozide in humans is unknown. The oral LD50 is 228 mg/kg in mice, 5120 mg/kg in rats, 188 mg/kg in guinea pigs, and 40 mg/kg in dogs (DrugBank: pimozide (DB01100)). Therefore, pimozide may also be a safe drug for treating OS cells or stem-like cells.
In our previous study, we reported that the neuroleptic drug pimozide had anti-tumor activity against hepatocellular carcinoma and prostate cancer cells through the suppression of STAT3 activity [17,18]. Numerous studies have demonstrated constitutive activation of STAT3 in a wide variety of human malignancies, including osteosarcoma [9,12]. Aberrantly STAT3 activation contributes to oncogenesis by preventing apoptosis, inducing cell proliferation, angiogenesis, invasion, and metastasis as well as suppressing anti-tumor immune responses [28,29]. Since STAT3 signaling is important for OS cell proliferation, we hypothesized that pimozide may inhibit the proliferation of OS cells and reduce STAT3 activity. Similarly, pimozide also reduced the basal expression of pY-STAT3 in U2OS cells in a dose-dependent manner, inhibited the transcription levels of the STAT3 signaling downstream genes MYC, BCL-xL and MCL1 and weakened cellular STAT3 reporter luciferase activity. Pimozide also inhibited U2OS cell proliferation, colony formation, and sphere formation and induced G0/G1 phase cell cycle arrest. In addition, pimozide reduced the percentage of side population cells. Our present study showed that pimozide, as a STAT3 inhibitor, inhibited the proliferation of OS cells or stem-like cells. Thus, these results further suggest that pimozide may be a novel STAT3 inhibitor that can be used to suppress cellular STAT3 activation for anti-cancer treatment.
In addition, our results showed that pimozide could suppress Erk signaling to inhibit the viability of osteosarcoma cells, reducing the expression of p-Erk in a dose-dependent manner, while no statistically significant difference in the anti-proliferative effect of pimozide was observed between cells treated with and without U0126. Because there is cross talk between the STAT3 and Erk pathways, it is likely that pimozide is a potential STAT3 activity inhibitor, leading to its anti-cancer effects in cancer cells. The mechanism will further demonstrate why pimozide has an anti-cancer effect on cancer cells, paving the way for the translational application of pimozide in cancer therapy.
Previous studies show that pimozide is a well-known antagonist of serotonin 5HT7 receptors (Ki = 0.5 nM) for anti-depressant effect and of dopamine receptor D2 (D2R) (Ki = 0.33 nM) for treating schizophrenia [30]. Pharmacologic antagonists pimozide reduced proliferation of pancreatic cancer cells by inhibiting DRD2 expression [30]. No reports have been demonstrated how 5HT7 receptor or DRD2 to activate STAT3 signaling in OS cells. In future, it needs to determine whether 5HT7 or DRD2 expression is associated with STAT3 activation of OS cells.
Our current study is to provide novel insight that pimozide produced ROS generation in cancer cells through inhibiting antioxidant enzyme gene catalase expression. ROS are closely related to carcinogenesis and play an important role in cancer [31,32]. Previous studies have shown that ROS may be involved in multiple tumorigenesis processes, including tumor initiation and transformation, tumor progression, tumor promotion, tumor angiogenesis, and tumor metastasis [33,34]. ROS are generated by both mitochondria and NADPH oxidases and scavenged by antioxidant enzyme [35]. By screening the expression of antioxidant enzyme genes, we found that pimozide treatment inhibited CAT gene expression but had little effect on the expression of SOD1, SOD2, and GPX1. Moreover, CAT treatment attenuated the pimozide-induced proliferation inhibition. Presumably, the pimozide-induced ROS generation was associated with the expression of the antioxidant enzyme gene CAT in cancer cells. Mechanically, pimozide inhibited cellular STAT3 activity to reduce CAT expression in OS cells.
In conclusion, this study illustrated that the neuroleptic drug pimozide has anti-tumor activity against OS cells or stem-like cells. Moreover, pimozide as a STAT3 inhibitor can suppress Erk signaling and promote ROS production possibly through reducing the expression of the antioxidant enzyme gene CAT. Thus, the antipsychotic agent pimozide may be a potential and novel therapeutic for OS patients. Of note, the development of new pimozide derivatives with stronger anti-cancer effects but lower side effects is urgently needed.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81602595 to J.-J Chen), the PhD Start-up Fund of National Natural Science Foundation of Guangdong Province of China (2015A030310191 to J.-J Chen), the Science and Technology Planning Project of Guangdong Province of China (2016A020214020 to G. Chang; 2017A020215069 to J.-J Chen), the Science Foundation of Guangzhou Education Bureau of China (1201630358 to W. Zhou) and the Natural Science Foundation of Shenzhen University (2016088 to J.-J Chen).
Disclosure of conflict of interest
None.
Authors’ contribution
JJ Chen and G Chang made substantial contributions to conception and design of the study and to data interpretation. N Cai and W Zhou performed the real-time PCR analysis, western blot analysis and preformed statistical analysis. LL Ye and J Chen carried out the experiments of cell culture and cellular proliferation. QN Liang made AnnexinV/PI staining using FASC. N Cai, W Zhou, JJ Chen and G Chang wrote the manuscript. All authors reviewed and approved the manuscript.
Abbreviations
- OS
osteosarcoma
- STAT3
the Signal Transducer and Activator of Transcription 3
- ROS
reactive oxygen species
- PI
propidium iodide
- EGF
epidermal growth factor
- bFGF
basic fibroblast growth factor
- DCFH-DA
2,7-dichlorofluorescein diacetate
- DHE
dihydroethidium
- SP
side population
- 5-FU
5-fluorouracil
- ChIP
chromatin immunoprecipitation
- pY-STAT3
phosphorylation of STAT3 at tyrosine 705
- Erk
extracellular signal regulated kinase
- p-Erk
Erk phosphorylation
- HPRT1
hypoxanthine phosphoribosyltransferase 1
- MCL1
myeloid cell leukemia 1
- STAT3
signal transducer and activator of transcription 3
- SOD1
cytosolic superoxide dismutase
- SOD2
mitochondrial superoxide dismutase
- CAT
catalase
- GPX1
glutathione peroxidase 1
Supporting Information
References
- 1.Dai X, Ma W, He X, Jha RK. Review of therapeutic strategies for osteosarcoma, chondrosarcoma, and Ewing’s sarcoma. Med Sci Monit. 2011;17:Ra177–190. doi: 10.12659/MSM.881893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biermann JS, Adkins DR, Agulnik M, Benjamin RS, Brigman B, Butrynski JE, Cheong D, Chow W, Curry WT, Frassica DA, Frassica FJ, Hande KR, Hornicek FJ, Jones RL, Mayerson J, McGarry SV, McGrath B, Morris CD, O’Donnell RJ, Randall RL, Santana VM, Satcher RL, Siegel HJ, von Mehren M, Bergman MA, Sundar H. Bone cancer. J Natl Compr Canc Netw. 2013;11:688–723. doi: 10.6004/jnccn.2013.0088. [DOI] [PubMed] [Google Scholar]
- 3.Broadhead ML, Clark JC, Myers DE, Dass CR, Choong PF. The molecular pathogenesis of osteosarcoma: a review. Sarcoma. 2011;2011:959248. doi: 10.1155/2011/959248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerrand C, Athanasou N, Brennan B, Grimer R, Judson I, Morland B, Peake D, Seddon B, Whelan J. UK guidelines for the management of bone sarcomas. Clin Sarcoma Res. 2016;6:7. doi: 10.1186/s13569-016-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs B, Pritchard DJ. Etiology of osteosarcoma. Clin Orthop Relat Res. 2002:40–52. doi: 10.1097/00003086-200204000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Zhou W, Hao M, Du X, Chen K, Wang G, Yang J. Advances in targeted therapy for osteosarcoma. Discov Med. 2014;17:301–307. [PubMed] [Google Scholar]
- 7.Kirk R. Targeted therapies: STAT3 and EGFR target resistance. Nat Rev Clin Oncol. 2012;9:489. doi: 10.1038/nrclinonc.2012.140. [DOI] [PubMed] [Google Scholar]
- 8.Wu P, Wu D, Zhao L, Huang L, Shen G, Huang J, Chai Y. Prognostic role of STAT3 in solid tumors: a systematic review and meta-analysis. Oncotarget. 2016;7:19863–19883. doi: 10.18632/oncotarget.7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fossey SL, Liao AT, McCleese JK, Bear MD, Lin J, Li PK, Kisseberth WC, London CA. Characterization of STAT3 activation and expression in canine and human osteosarcoma. BMC Cancer. 2009;9:81. doi: 10.1186/1471-2407-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Zhang W. New molecular insights into osteosarcoma targeted therapy. Curr Opin Oncol. 2013;25:398–406. doi: 10.1097/CCO.0b013e3283622c1b. [DOI] [PubMed] [Google Scholar]
- 11.Yan J, Wang Q, Zou K, Wang L, Schwartz EB, Fuchs JR, Zheng Z, Wu J. Inhibition of the JAK2/STAT3 signaling pathway exerts a therapeutic effect on osteosarcoma. Mol Med Rep. 2015;12:498–502. doi: 10.3892/mmr.2015.3439. [DOI] [PubMed] [Google Scholar]
- 12.Tu B, Du L, Fan QM, Tang Z, Tang TT. STAT3 activation by IL-6 from mesenchymal stem cells promotes the proliferation and metastasis of osteosarcoma. Cancer Lett. 2012;325:80–88. doi: 10.1016/j.canlet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Boguski MS, Mandl KD, Sukhatme VP. Drug discovery. Repurposing with a difference. Science. 2009;324:1394–1395. doi: 10.1126/science.1169920. [DOI] [PubMed] [Google Scholar]
- 14.Strobl JS, Kirkwood KL, Lantz TK, Lewine MA, Peterson VA, Worley JF 3rd. Inhibition of human breast cancer cell proliferation in tissue culture by the neuroleptic agents pimozide and thioridazine. Cancer Res. 1990;50:5399–5405. [PubMed] [Google Scholar]
- 15.Taub RN, Baker MA. Treatment of metastatic malignant melanoma with pimozide. Lancet. 1979;1:605. doi: 10.1016/s0140-6736(79)91025-0. [DOI] [PubMed] [Google Scholar]
- 16.Nelson EA, Walker SR, Weisberg E, Bar-Natan M, Barrett R, Gashin LB, Terrell S, Klitgaard JL, Santo L, Addorio MR, Ebert BL, Griffin JD, Frank DA. The STAT5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors. Blood. 2011;117:3421–3429. doi: 10.1182/blood-2009-11-255232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JJ, Cai N, Chen GZ, Jia CC, Qiu DB, Du C, Liu W, Yang Y, Long ZJ, Zhang Q. The neuroleptic drug pimozide inhibits stem-like cell maintenance and tumorigenicity in hepatocellular carcinoma. Oncotarget. 2017;8:17593–17609. doi: 10.18632/oncotarget.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou W, Chen MK, Yu HT, Zhong ZH, Cai N, Chen GZ, Zhang P, Chen JJ. The antipsychotic drug pimozide inhibits cell growth in prostate cancer through suppression of STAT3 activation. Int J Oncol. 2016;48:322–328. doi: 10.3892/ijo.2015.3229. [DOI] [PubMed] [Google Scholar]
- 19.Fako V, Yu Z, Henrich CJ, Ransom T, Budhu AS, Wang XW. Inhibition of wnt/beta-catenin signaling in hepatocellular carcinoma by an antipsychotic drug pimozide. Int J Biol Sci. 2016;12:768–775. doi: 10.7150/ijbs.14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levina V, Marrangoni AM, DeMarco R, Gorelik E, Lokshin AE. Drug-selected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS One. 2008;3:e3077. doi: 10.1371/journal.pone.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen P, Luo X, Nie P, Wu B, Xu W, Shi X, Chang H, Li B, Yu X, Zou Z. CQ synergistically sensitizes human colorectal cancer cells to SN-38/CPT-11 through lysosomal and mitochondrial apoptotic pathway via p53-ROS cross-talk. Free Radic Biol Med. 2017;104:280–297. doi: 10.1016/j.freeradbiomed.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 22.Ao X, Nie P, Wu B, Xu W, Zhang T, Wang S, Chang H, Zou Z. Decreased expression of microRNA-17 and microRNA-20b promotes breast cancer resistance to taxol therapy by upregulation of NCOA3. Cell Death Dis. 2016;7:e2463. doi: 10.1038/cddis.2016.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou Z, Yuan Z, Zhang Q, Long Z, Chen J, Tang Z, Zhu Y, Chen S, Xu J, Yan M, Wang J, Liu Q. Aurora kinase A inhibition-induced autophagy triggers drug resistance in breast cancer cells. Autophagy. 2012;8:1798–1810. doi: 10.4161/auto.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neiva KG, Zhang Z, Miyazawa M, Warner KA, Karl E, Nor JE. Cross talk initiated by endothelial cells enhances migration and inhibits anoikis of squamous cell carcinoma cells through STAT3/Akt/ERK signaling. Neoplasia. 2009;11:583–593. doi: 10.1593/neo.09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 26.Kasznicki J, Sliwinska A, Drzewoski J. Metformin in cancer prevention and therapy. Ann Transl Med. 2014;2:57. doi: 10.3978/j.issn.2305-5839.2014.06.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fishman MC, Porter JA. Pharmaceuticals: a new grammar for drug discovery. Nature. 2005;437:491–493. doi: 10.1038/437491a. [DOI] [PubMed] [Google Scholar]
- 28.Zhao M, Jiang B, Gao FH. Small molecule inhibitors of STAT3 for cancer therapy. Curr Med Chem. 2011;18:4012–4018. doi: 10.2174/092986711796957284. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Crowe PJ, Goldstein D, Yang JL. STAT3 inhibition, a novel approach to enhancing targeted therapy in human cancers (review) Int J Oncol. 2012;41:1181–1191. doi: 10.3892/ijo.2012.1568. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Ji ZL, Chen YZ. TTD: therapeutic target database. Nucleic Acids Res. 2002;30:412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Zhang H, Zhou HJ, Ji W, Min W. Mitochondrial redox signaling and tumor progression. Cancers (Basel) 2016;8 doi: 10.3390/cancers8040040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manda G, Isvoranu G, Comanescu MV, Manea A, Debelec Butuner B, Korkmaz KS. The redox biology network in cancer pathophysiology and therapeutics. Redox Biol. 2015;5:347–357. doi: 10.1016/j.redox.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa A, Scholer-Dahirel A, Mechta-Grigoriou F. The role of reactive oxygen species and metabolism on cancer cells and their microenvironment. Semin Cancer Biol. 2014;25:23–32. doi: 10.1016/j.semcancer.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 34.Poillet-Perez L, Despouy G, Delage-Mourroux R, Boyer-Guittaut M. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox Biol. 2015;4:184–192. doi: 10.1016/j.redox.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buldak RJ, Buldak L, Kukla M, Gabriel A, Zwirska-Korczala K. Significance of selected antioxidant enzymes in cancer cell progression. Pol J Pathol. 2014;65:167–175. doi: 10.5114/pjp.2014.45779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


