Abstract
Purpose
Tools for survival prediction for non-small cell lung cancer (NSCLC) patients treated with chemoradiation or radiation therapy are of limited quality. In this work, we developed a predictive model of survival at 2 years. The model is based on a large volume of historical patient data and serves as a proof of concept to demonstrate the distributed learning approach.
Methods and Materials
Clinical data from 698 lung cancer patients, treated with curative intent with chemoradiation or radiation therapy alone, were collected and stored at 2 different cancer institutes (559 patients at Maastro clinic (Netherlands) and 139 at Michigan university [United States]). The model was further validated on 196 patients originating from The Christie (United Kingdon). A Bayesian network model was adapted for distributed learning (the animation can be viewed at https://www.youtube.com/watch?v=ZDJFOxpwqEA). Two-year posttreatment survival was chosen as the endpoint. The Maastro clinic cohort data are publicly available at https://www.cancerdata.org/publication/developing-and-validating-survival-prediction-model-nsclc-patients-through-distributed, and the developed models can be found at www.predictcancer.org.
Results
Variables included in the final model were T and N category, age, performance status, and total tumor dose. The model has an area under the curve (AUC) of 0.66 on the external validation set and an AUC of 0.62 on a 5-fold cross validation. A model based on the T and N category performed with an AUC of 0.47 on the validation set, significantly worse than our model (P<.001). Learning the model in a centralized or distributed fashion yields a minor difference on the probabilities of the conditional probability tables (0.6%); the discriminative performance of the models on the validation set is similar (P=.26).
Conclusions
Distributed learning from federated databases allows learning of predictive models on data originating from multiple institutions while avoiding many of the data-sharing barriers. We believe that distributed learning is the future of sharing data in health care.
Summary.
Tools for survival prediction for non-small cell lung cancer patients treated with chemoradiation or radiation therapy are of limited quality. The gold standard (TNM staging) was originally developed for patients undergoing surgery. We developed a predictive model for survival in non-small cell lung cancer patients following chemoradiation or radiation therapy. The model was trained on a large volume of patients from multiple institutes by use of the distributed learning approach. The model outperforms the gold standard (TNM staging).
Introduction
Learning from large volumes of patient data can greatly increase our capacity to generate and test hypotheses about health care (1). To capture and use the knowledge contained in large volumes of patient data, predictive models are essential for clinical decision making 1, 2, 3, 4, 5, 6, 7, 8. Predictive models can be trained on large volumes of data—from patients who have been treated in the past—to make predictions about survival, disease control, and side effects of treatment for a patient who has yet to be treated 9, 10.
Radiation therapy (RT), alone or in combination with chemotherapy, is a common choice of treatment in non-small cell lung cancer (NSCLC) patients whose tumors are inoperable because of metastases to mediastinal lymph node stations and/or because of patients' physical condition (11). The TNM staging system for survival risk stratification of this group of patients is, however, inaccurate (12). The TNM classification has initially been made to look at operability rather than prognosis after chemoradiation therapy (CRT). Other prognostic factors have been identified, such as performance status 13, 14, 15, weight loss 13, 14, presence of comorbidity (15), chemotherapy use in combination with RT 13, 16, radiation dose 13, 17, tumor size 12, 18, 19, 20, 21, and image features, the so-called radiomics approach 22, 23, 24, 25, 26. For other factors such as age and sex, the results have been inconclusive (14).
Currently, the TNM staging system is the gold standard for risk stratification. Studies have indicated the TNM system performs poorly for patients receiving CRT or RT, creating an increasing need for more reliable prediction models 27, 28.
In this study, we conducted a level III and IV modeling effort on historical data, as a proof of concept, conforming with the TRIPOD (Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis) statement, in which validation is performed at an independent site that has not seen the data (29). Our hypothesis is 2-fold: Learning from different centers without moving the data is possible, and by learning from a large volume of patient data, we can develop a better model for 2-year survival prediction that is more robust than the existing prognostic tools.
Methods and Materials
Data
Clinical data from 698 lung cancer patients, treated with curative intent with CRT or RT alone, were collected and stored at 2 different medical institutes (559 patients at Maastro clinic [Netherlands] and 139 at Michigan University [United States]). The model was validated on 196 patients originating from The Christie (United Kingdon). None of the patients received stereotactic body RT, and all patients had inoperable stage I through IIIB NSCLC. Patients were treated for their primary lung tumor and did not receive a diagnosis of another tumor in the 5 years before treatment. The patient details are shown in Table 1. Two-year survival, taken from the start of RT, was used as the outcome of this study. The Maastro clinic cohort data are publicly available at https://www.cancerdata.org/publication/developing-and-validating-survival-prediction-model-nsclc-patients-through-distributed.
Table 1.
Overview of patient characteristics per hospital
| Maastro clinic (n=559) | Michigan University (n=139) | The Christie (n=196) | |
|---|---|---|---|
| Age | |||
| Mean, y | 68 | 66 | 66 |
| SD, y | 10 | 10 | 10 |
| Missing, n | 0 (0%) | 0 (0%) | 2 (1%) |
| Sex, n | |||
| Male | 370 (62%) | 107 (77%) | 89 (45%) |
| Female | 189 (32%) | 32 (23%) | 117 (60%) |
| Missing | 0 (0%) | 0 (0%) | 1 (1%) |
| ECOG performance status, n | |||
| 0 | 102 (17%) | 16 (12%) | 40 (20%) |
| 1 | 301 (50%) | 100 (72%) | 103 (53%) |
| 2 | 11 (2%) | 21 (15%) | 45 (23%) |
| 3 | 21 (4%) | 1 (1%) | 4 (2%) |
| 4 | 4 (1%) | 0 (0%) | 0 (0%) |
| Missing | 120 (20%) | 1 (1%) | 4 (2%) |
| T stage, n | |||
| 0 | 83 (14%) | 25 (18%) | 1 (1%) |
| 1 | 154 (26%) | 33 (24%) | 13 (7%) |
| 2 | 89 (15%) | 40 (29%) | 54 (28%) |
| 3 | 198 (33%) | 40 (29%) | 51 (26%) |
| 4 | 0 (0%) | 0 (0%) | 70 (36%) |
| Missing | 35 (6%) | 1 (1%) | 7 (4%) |
| N stage, n | |||
| 0 | 150 (25%) | 33 (24%) | 51 (26%) |
| 1 | 32 (5%) | 17 (12%) | 14 (7%) |
| 2 | 214 (36%) | 58 (42%) | 86 (44%) |
| 3 | 136 (23%) | 31 (22%) | 40 (20%) |
| Missing | 27 (5%) | 0 (0%) | 5 (3%) |
| M stage, n | |||
| 0 | 505 (84%) | 139 (100%) | 191 (97%) |
| 1 | 0 (0%) | 0 (0%) | 0 (0%) |
| Missing | 54 (9%) | 0 (0%) | 5 (3%) |
| Chemotherapy timing, n | |||
| No chemotherapy | 119 (20%) | 25 (18%) | 73 (37%) |
| Sequential | 53 (9%) | 0 (0%) | 62 (32%) |
| Concurrent | 279 (47%) | 114 (82%) | 60 (31%) |
| Missing | 108 (18%) | 0 (0%) | 0 (0%) |
| Stage group, n | |||
| IA | 44 (8%) | 9 (6%) | 7 (4%) |
| IB | 26 (5%) | 8 (6%) | 12 (6%) |
| IIA | 18 (3%) | 0 (0%) | 0 (0%) |
| IIB | 53 (9%) | 16 (11%) | 14 (7%) |
| IIIA | 350 (63%) | 105 (76%) | 153 (78%) |
| IIIB | 0 (0%) | 0 (0%) | 0 (0%) |
| Missing | 68 (12%) | 1 (1%) | 10 (5%) |
| 2-y survival, n | |||
| No | 339 (57%) | 72 (52%) | 151 (77%) |
| Yes | 220 (37%) | 67 (48%) | 45 (23%) |
| Missing | 0 (0%) | 0 (0%) | 0 (0%) |
Abbreviation: ECOG = Eastern Cooperative Oncology Group.
Maastro clinic cohort
The institute 1 data were collected under an institutional review board (IRB)–approved and registered clinical trial (NCT01949259). Patients were treated between 2007 and 2014. Three different protocol types were administered to the patients in this study:
-
1.
One hundred eighty-nine NSCLC patients were treated according to the new protocol for sequential CRT, which was introduced in August 2005 30, 31, 32. The individualized radiation dose ranged from 54.0 to 79.2 Gy, delivered in fractions of 1.8 Gy, twice daily, until the mean lung dose or maximum dose to the spinal cord was reached. The minimum interval between the fractions was 8 hours.
-
2.
Two hundred eighty-three NSCLC patients received concurrent CRT. A radiation dose of 45 Gy was delivered in fractions of 1.5 Gy, twice daily. The minimum interval between the fractions was 8 to 10 hours. This treatment was followed by an individualized dose ranging from 8 to 24 Gy, delivered in fractions of 2.0 Gy, once daily, until the normal tissue dose constraints were reached. Cisplatin and etoposide were given concurrently on days 2, 9, 23, and 30.
-
3.
Thirty-six NSCLC patients received accelerated high-dose conformal RT: 66 Gy in 24 fractions (2.75 Gy/fraction). Some of these patients received chemotherapy. The concurrent chemotherapy used consisted of intravenous administration of 80 mg/m2 of cisplatin and 100 mg/m2 of etoposide on days 1 through 3. The first cycle was administered before RT, and cycles 2 and 3 were given during RT. In total, 3 cycles of chemotherapy were given (33). For patients receiving sequential CRT, 3 courses of etoposide (100 mg/m2 on days 1 and 8) and cisplatin (75 mg/m2 on day 2) were given (34).
The remaining 51 patients received a treatment regimen tailored specifically to the patients.
Michigan University cohort
The Michigan University data were collected from prospective protocols under IRB approval (UMCC 2006.040 and UMCC 2007.123). All patients were treated with curative intent between May 2007 and July 2014.
-
1.
The first study treated patients to standard doses (60-66 Gy) with once-daily fractions of 2 Gy.
-
2.
The second study was a dose-escalation study intensifying doses to persistent positron emission tomography-avid target volumes during treatment with 2.1 to 2.85 Gy/fraction up to a total dose of 85.5 Gy in 30 fractions.
The Christie cohort
One external validation set was used in this study. The Christie cohort consisted of 196 anonymized lung cancer patients with stage I through IIIB NSCLC. The study was conducted under IRB approval. All patients were treated with curative intent between December 2008 and May 2013. Two different protocols were used for treating patients in this dataset.
-
1.
One hundred twenty-one NSCLC patients received 55 Gy in 20 daily fractions (2.75 Gy/fraction), either without chemotherapy or with sequential chemotherapy.
-
2.
Seventy-three NSCLC patients received 60 to 66 Gy in 30 to 33 daily fractions (2 Gy/fraction) with concurrent chemotherapy.
The remaining 2 patients received a treatment regimen tailored specifically to the patients.
Bayesian network
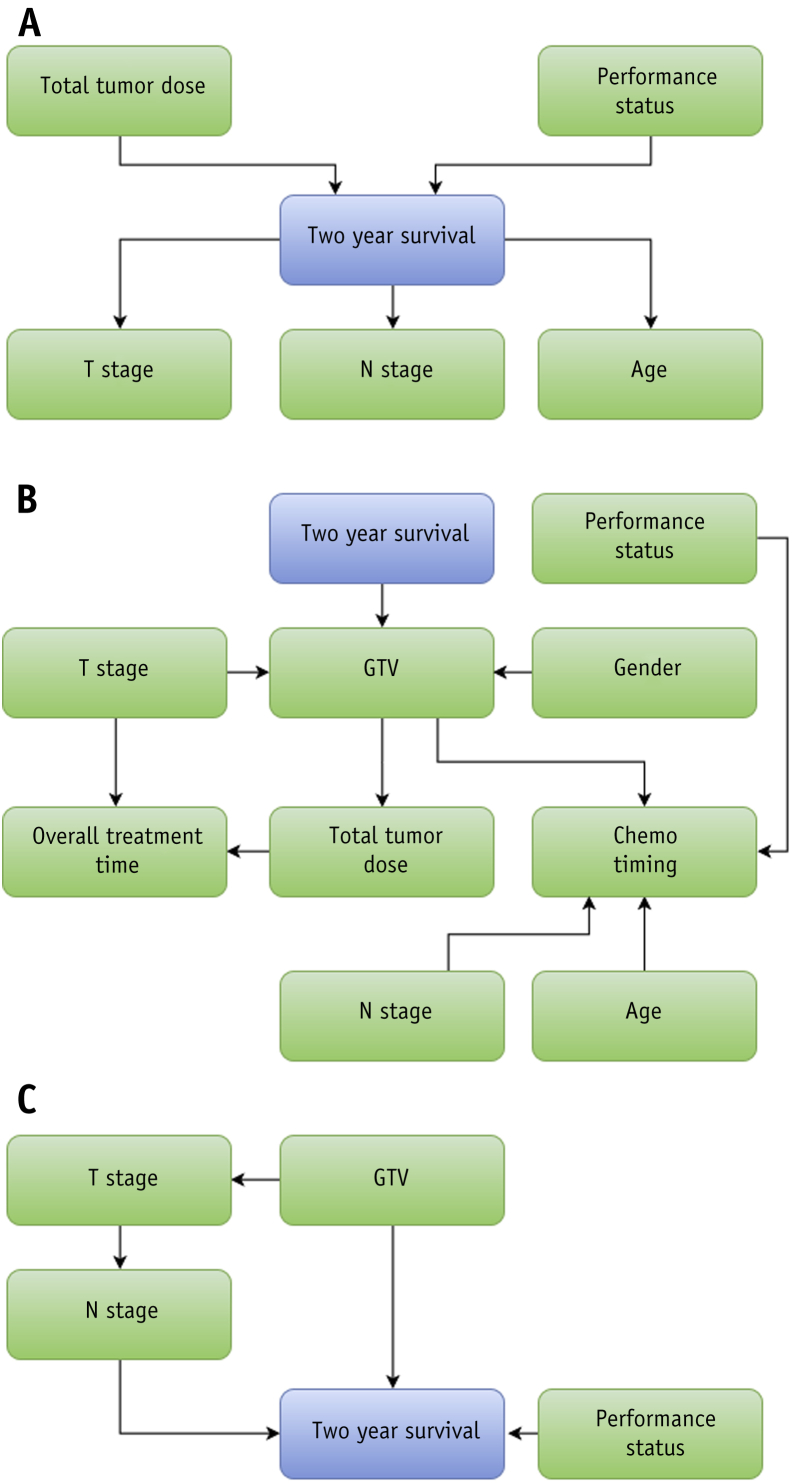
A Bayesian network model was developed to predict survival at 2 years after RT start. The model used T category, N category, age, total tumor dose (defined as the prescribed dose to a reference point according to the International Commission on Radiation Units & Measurements), and World Health Organization performance status to make predictions. (The target dose definition for patients who received a boost to [positron emission tomography-avid] tumor subvolumes can be found in the publication of van Elmpt et al (35).) The network structure of this model was prespecified by experts and is a balance between the desire for causal links and the desire for a limit on the number of incoming links to the survival endpoint. It is shown in Figure 1. A further elaboration on the method used to determine the network structure is presented in Appendix E1 (available online at www.redjournal.org).
Fig. 1.
Bayesian network structures. The blue nodes represent outcome. (A) Network structure based on expert opinion. (B) Network structure built using algorithmic approach. (C) Network structure adapted from Jayasurya et al (9). Abbreviations: Chemo = chemotherapy; GTV = gross tumor volume.
The model's performance is expressed as the area under the curve (AUC) of the receiver operating characteristic (ROC) curve. The maximum value of the AUC is 1.0, indicating a perfect prediction model. A value of 0.5 indicates that patients are only correctly classified in 50% of the cases, that is, as good as chance.
A Bayesian network is a probabilistic graphical model that represents a set of variables and their dependencies in a directed acyclic graph (DAG). Within the DAG, variables are depicted as nodes and statistical dependencies are represented as directed edges.
We have determined the DAG dependencies based on expert knowledge. The conditional probability tables (CPTs) associated with each variable have been computed by use of a maximum likelihood technique based on the expectation maximization algorithm (36). All continuous variables underwent discretization into either 2 or 3 bins by use of a method described by Kuschner et al (37). The developed models can be found at www.predictcancer.org.
Distributed learning
Distributed learning is defined by learning from data from multiple hospitals without the data leaving the hospitals. To realize distributed learning for this study, we have used a formerly adapted method 38, 39.
Statistical analysis
The Bayesian network model was programmed in Java (Oracle, Redwood Shores, California) by use of the JSMILE (Structural Modeling, Inference, and Learning Engine) framework developed by the Dynamic Systems Laboratory of Pittsburg University (40) and made freely available for academic purposes by BayesFusion (Pittsburgh, PA; http://www.bayesfusion.com/).
Analysis of ROC curves was performed in R (version 3.0.0; R Foundation for Statistical Computing, Vienna, Austria) by use of the pROC package (41). Comparison of ROC curves and computation of confidence intervals (CIs) of AUC values were performed with the method described by DeLong et al (42). Kaplan-Meier curves were made in R using the survival package (43). The log-rank test with a ρ value of 0 was used to evaluate differences in Kaplan-Meier curves (44). CI estimation of Kaplan-Meier curves was performed with the method described by Dorey and Korn (45). Tests for significant differences between groups were performed with the Wilcoxon rank sum test 46, 47.
Comparison with previous models
Imputation of missing variables was done by taking the mean for the corresponding variable in the training set. This mean was used for imputation in the training data and validation sets. As Bayesian networks rely on Bayesian inference to do imputation, imputation by taking the mean was omitted for validating and training these models. The number of positive lymph node stations (PLNSs) was used as a variable in the older models, which is unavailable in our current dataset. Nodal stage was used to impute the number of PLNSs. In subsequent analyses of the data, we partitioned the validation set into a young patients' cohort and old patients' cohort. We set the cutoff point at 67 years, as this most closely partitioned the data into 2 sets of equal size. To investigate the performance of the TNM staging variables alone, a logistic regression was performed with these variables.
A Bayesian network model previously developed by Jayasurya et al (9) was compared with the model in this study. PLNS was imputed by nodal stage. The nodal stage node was removed from the structure because it is obsolete as a result of the imputation of PLNS. The resulting network structure is shown in Figure 1C.
Results
The Bayesian network model was learned in a distributed setting on the Maastro clinic and Michigan University cohorts. The CPTs of the learned model are shown in Tables E1-E6 (available online at www.redjournal.org). The AUC of the model was 0.62 (95% CI, 0.57-0.66) on a 5-fold cross validation.
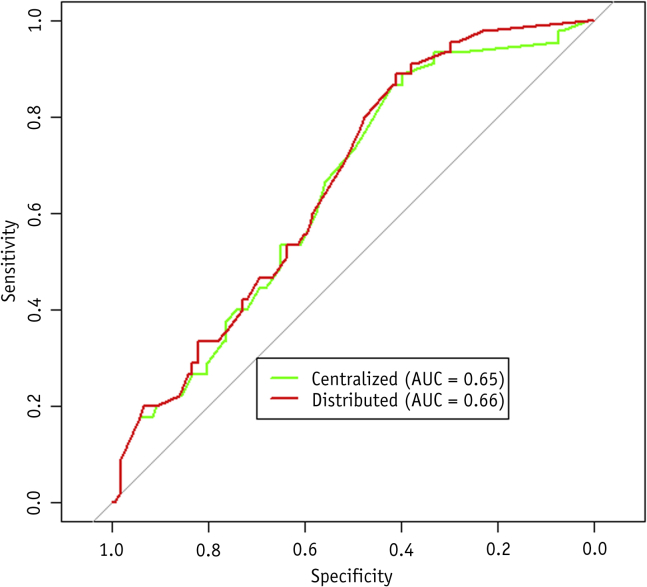
To investigate the effectivity of the distributed learning methodology used, we compared the CPTs of a model learned on the training data of institutes 1 and 2 in a centralized manner versus the CPTs of the model learned on the same datasets in a distributed manner. The average difference in probability per CPT was 0.6%. The results of using both models on the validation set of The Christie are shown in Figure 2. The discriminative performance of both models on the validation set was similar (P=.26).
Fig. 2.
Receiver operating characteristic curves for the Bayesian network model learned using centralized learning and distributed learning. Validation was done on the institute 3 cohort. Abbreviation: AUC = area under curve.
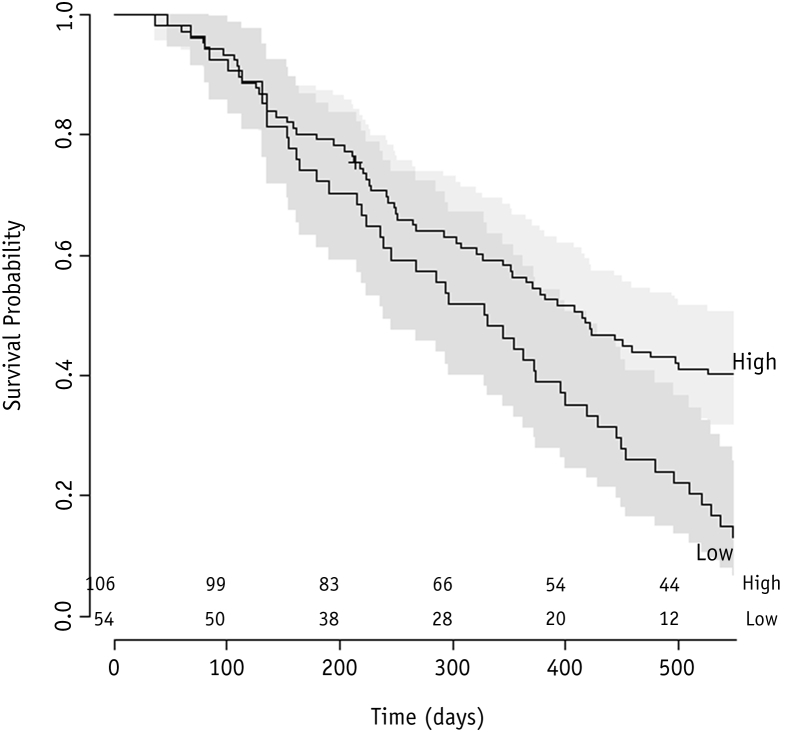
Splitting the The Christie cohort into 2 subgroups, according to the mean of the predicted survival probability by the model for all patients in the training cohort, resulted in the identification of a group with a high chance of survival and a group with a low chance of survival. The 2-year survival rate was 33% (95% CI, 25%-42%) for the high–survival chance group and 0% for the low–survival chance group (Fig. 3). A log-rank test on these curves indicated that they were significantly different (P<.01).
Fig. 3.
Kaplan-Meier curves for risk group stratification for model. A log-rank test indicated that these curves were significantly different (P<.01).
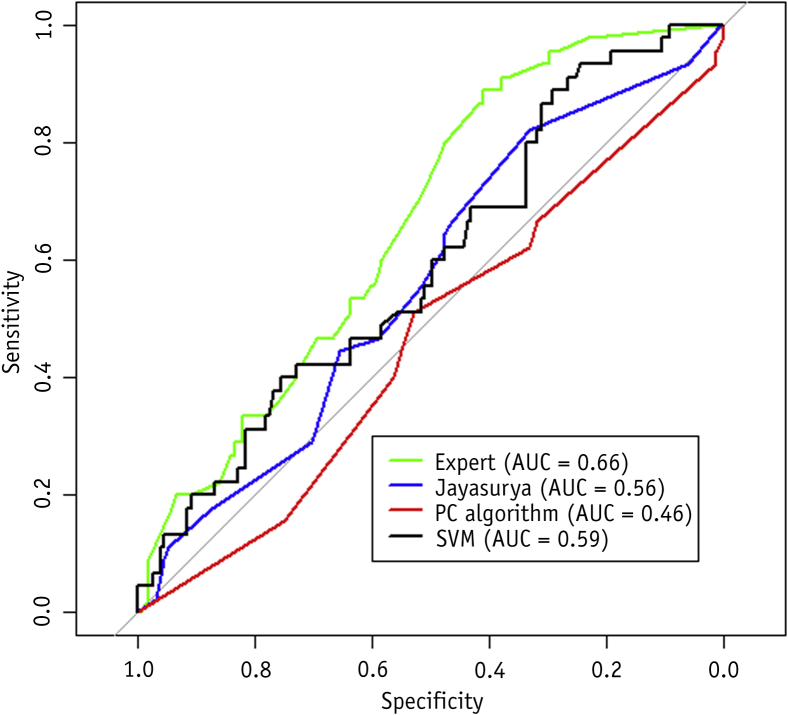
We conducted an external validation on the The Christie cohort (n=196). The AUC of the model was 0.66 (95% CI, 0.57-0.74). The support vector machine (SVM) model developed by Oberije et al was also validated on this cohort. The AUC of the SVM model developed by Oberije et al was 0.59 (95% CI, 0.49-0.68). Comparison of the 2 ROC curves indicated they were not significantly different (P=.19). The 2 ROC curves are shown in Figure 4. Performance of a model based on the T and N category alone was compared with the Bayesian network. The T and N model performed with an AUC of 0.47 (95% CI, 0.37-0.57) on the external validation set, significantly lower than the Bayesian network model (P<.001). The T and N model performed with an AUC of 0.53 on a 5-fold cross validation on the training set. A model that used stage groups, rather than T and N category separately, performed with an AUC of 0.56 on the validation set (95% CI, 0.49-0.63) and an AUC of 0.54 on a 5-fold cross validation on the training set.
Fig. 4.
Receiver operating characteristic curves of models compared in this study. The expert Bayesian network structure is shown in Figure 1A. The Bayesian network structure of Jayasurya et al (9) is shown in Figure 1C. The Patch Condition (PC) algorithm using the Bayesian network structure is shown in Figure 1B. Validation was done on the institute 3 cohort. Abbreviations: AUC = area under curve; SVM = support vector machine.
To investigate the effect of using different network structures, we have compared the performance of the model using the network structure presented in the work by Jayasurya et al (9). In addition, we have learned a network structure on the data using an algorithmic approach (48). Network structures are shown in Figure 1B and C. Results are shown in Figure 4. Neither network structure equaled the performance of the network structure defined by experts (P<.05 for Jayasurya et al, P<.001 for the algorithmic approach.
To further evaluate all models tested in this study, we present their performances in terms of discrimination (expressed as AUC), calibration (expressed as the coefficient of determination, r2), and actual chance (Brier score) in Tables E7 and E8 (available online at www.redjournal.org). Comparison of the ROC curves for the Bayesian network model on the 5-fold cross validation and external validation indicated that they were not significantly different (P=.33).
Discussion
In this study, we used a distributed learning approach for Bayesian networks to develop a model using data from 3 hospitals in 3 countries without the need for any data to leave the individual hospital. Previous work exists on survival prediction for NSCLC patients receiving CRT 9, 12, 49. Oberije et al found that sex, performance status, forced expiratory volume in 1 second, number of PLNSs, and gross tumor volume were relevant predictive factors and used these in their model (12). Their model was based on a smaller cohort of patients (n=377), and we found that some of our observations are in contrast to theirs. Age was included in our model as it was significantly correlated with 2-year survival (P<.01). Oberije et al found that there was no significant trend between survival and age (12). Sex was found to correlate significantly with survival in the cohort of Oberije et al. In our study, sex did not significantly correlate with outcome (P=.07). Contradictory results regarding both sex and age have been reported previously 50, 51.
Dose was included as a parameter in our model. A clinical trial showed that higher dose decreased overall survival (52). A meta-analysis suggested there may be an effect of hyperfractionated and/or accelerated non-concurrent CRT (53). As most of the patients included in the model were treated with the highest dose achievable without exceeding normal tissue constraints, it is possible that RT dose in our model was a surrogate for low volumetric disease burden. To provide conclusive evidence of whether total tumor dose affects overall survival for specific patient groups, large multicenter studies are required.
Model comparison
We have compared our model with an SVM model that was developed previously (49). Although the Bayesian network did not perform significantly better on the validation set as a whole, we observed a significant improvement in survival prediction on the older patients of the validation cohort (Fig. E1; available online at www.redjournal.org). This could be because of the larger volumes of patient data on which we have based our model. As more data are used, a larger variety of patients are included in the model. This in turn makes the model more robust. Variables and trends previously not significant may turn out to be of high value when larger sample sizes are used, as was the case in this study for the age variable. Other weakly significant variables, such as sex, may ultimately be found to be irrelevant.
Another advantage of our model over the previously developed model is that it handles missing values better, as has been shown in previous work (9). Furthermore, the T and N model performs poorly on the validation set, whereas both the SVM model of Oberije et al and the current model perform above the chance level, indicating added value of this work (12).
Our model outperformed the previous model of Jayasurya et al (9) on this validation set. One possible explanation for this finding is that the network structure used by Jayasurya et al has too many arcs pointing directly to the outcome node. This in turn means that the CPT for this node becomes very large. Such a large CPT will be undersampled, and therefore the model may be subject to overfitting. Every node pointing toward the outcome will interact with all the other variables pointing to the outcome. This is analogous to introducing interaction terms in conventional regression models. For all arrows pointing from the outcome to a variable, independence is assumed among these variables. These are the only kinds of connections observed in naive Bayes classifiers (NBCs). A weakness of NBCs is that they assume independence among variables. An advantage of NBCs is that they require fewer data, because of this independence assumption. In the Bayesian network presented in this study, we used a partially NBC structure and combined it with 2 arcs pointing directly toward the outcome. We thereby avoided overfitting while still enabling enough complexity to allow the model to perform with high discriminative power.
Limitations
This study has a number of limitations. First, a Bayesian network structure has been chosen based on expert opinion. An alternative option is to take an algorithmic approach and let the data determine the best Bayesian network structure. Previous work by Sesen et al (54) has shown that structure learning algorithms outperform structures selected by experts. However, applying an algorithmic approach proved ineffective in this work (Fig. E2; available online at www.redjournal.org). This conflicting finding may be because of the difference in datasets and variables used. Further research is necessary to identify the optimal structure given a certain modeling scenario.
Another weakness of this study is related to the comparison of our model with the SVM (49). The SVM model developed previously used the number of PLNSs to make predictions (12). This variable was unavailable in our dataset, making a fair comparison more difficult.
An additional weakness of our study is that the model was largely trained on historical patients. Such models have less added value to clinical practice today as RT practice is subject to continual innovation. Phase III clinical trials provide high-grade evidence; however, they have a downside, as such studies take a relatively long time to complete. Distributed learning can fill the temporal gap by learning from large volumes of patient data from different hospitals as soon as the data are entered into the electronic health record system. A model trained on historical patients, as was used in this study, may serve to generate hypotheses for future experiments. In addition, once the system is in place, distributed learning can be repeated to include more recent patients and thus update the model using the latest practice insights with the ultimate goal to realize a rapid learning health care system.
Although distributed learning is a major stride towards easier data sharing and learning models on larger volumes of data, some hurdles remain. It takes several months to set up the infrastructure in an institution. Furthermore, retrieving the data from the electronic medical records is still challenging, as data are often scattered across many databases and applications. Once the data are retrieved and the infrastructure is set up, however, distributed learning of any number of models can be done.
Future work
Our future vision is summarized in an animation at https://www.youtube.com/watch?v=ZDJFOxpwqEA. As our methodology permits learning from features of all kinds, we intend to include variables that could potentially have higher predictive value, such as radiomics and genomics features, to learn more sophisticated models in a distributed manner 22, 23. We intend to include these models in customized patient decision aids and use them for patient stratification in clinical trials 3, 7, 55.
Acknowledgments
The authors would like to thank Varian for providing the distributed learning manager and Wolfgang Wiessler for his dedicated support. They would also like to thank Grant Weyburne for his help in collecting and curating the data used in this study.
Footnotes
This work was supported by the Interreg grant euroCAT and the Dutch Technology Foundation STW (DuCAT, grant No. 10696; Radiomics STRaTegy, grant No. P14-19), which is the applied science division of Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO); the Technology Programme of the Ministry of Economic Affairs; and a Manchester Cancer Research UK major center grant. Financial support was also provided by the following: EU Seventh Framework program (ARTFORCE, grant No. 257144; REQUITE, grant No. 601826), CTMM-TraIT, EUROSTARS (CloudAtlas), Kankeronderzoekfonds Limburg from the Health Foundation Limburg, Alpe d'HuZes-KWF (DESIGN), Dutch Cancer Society, NIH P01 CA059827, European Program H2020-2015-17 (ImmunoSABR, grant No. 733008), an European Research Council (ERC) advanced grant (ERC-ADG-2015, grant No. 694812; Hypoximmuno), and SME Phase 2 (European Union (EU) proposal 673780, RAIL). This publication was supported by the Dutch national program COMMIT (Prana Data project).
Arthur Jochems and Timo M. Deist contributed equally to this publication.
Conflict of interest: Andre Dekker is funded by Varian Medical Systems for projects other than that described in this study.
Supplementary material for this article can be found at www.redjournal.org.
Supplementary Data
References
- 1.Etheredge L.M. A rapid-learning health system. Health Aff (Millwood) 2007;26:w107–w118. doi: 10.1377/hlthaff.26.2.w107. [DOI] [PubMed] [Google Scholar]
- 2.Abernethy A.P., Etheredge L.M., Ganz P.A. Rapid-learning system for cancer care. J Clin Oncol. 2010;28:4268–4274. doi: 10.1200/JCO.2010.28.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambin P., van Stiphout R.G.P.M., Starmans M.H.W. Predicting outcomes in radiation oncology–multifactorial decision support systems. Nat Rev Clin Oncol. 2013;10:27–40. doi: 10.1038/nrclinonc.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambin P., Roelofs E., Reymen B. “Rapid Learning health care in oncology” - an approach towards decision support systems enabling customised radiotherapy’. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2013;109:159–164. doi: 10.1016/j.radonc.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Lambin P., Petit S.F., Aerts H.J.W.L. The ESTRO Breur Lecture 2009. From population to voxel-based radiotherapy: Exploiting intra-tumour and intra-organ heterogeneity for advanced treatment of non-small cell lung cancer. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2010;96:145–152. doi: 10.1016/j.radonc.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Lambin P., Zindler J., Vanneste B. Modern clinical research: How rapid learning health care and cohort multiple randomised clinical trials complement traditional evidence based medicine. Acta Oncol Stockh Swed. 2015;54:1289–1300. doi: 10.3109/0284186X.2015.1062136. [DOI] [PubMed] [Google Scholar]
- 7.Lambin P., Zindler J., Vanneste B.G.L. Decision support systems for personalized and participative radiation oncology. Adv Drug Deliv Rev. 2016 doi: 10.1016/j.addr.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Cheng Q., Roelofs E., Ramaekers B.L.T. Development and evaluation of an online three-level proton vs photon decision support prototype for head and neck cancer - Comparison of dose, toxicity and cost-effectiveness. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2016;118:281–285. doi: 10.1016/j.radonc.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 9.Jayasurya K., Fung G., Yu S. Comparison of Bayesian network and support vector machine models for two-year survival prediction in lung cancer patients treated with radiotherapy. Med Phys. 2010;37:1401–1407. doi: 10.1118/1.3352709. [DOI] [PubMed] [Google Scholar]
- 10.Dehing-Oberije C., De Ruysscher D., Petit S. Development, external validation and clinical usefulness of a practical prediction model for radiation-induced dysphagia in lung cancer patients. Radiother Oncol. 2010;97:455–461. doi: 10.1016/j.radonc.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 11.Vulto A., Louwman M., Rodrigus P. Referral rates and trends in radiotherapy as part of primary treatment of cancer in South Netherlands, 1988-2002. Radiother Oncol. 2006;78:131–137. doi: 10.1016/j.radonc.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Dehing-Oberije C., De Ruysscher D., van der Weide H. Tumor volume combined with number of positive lymph node stations is a more important prognostic factor than TNM stage for survival of non-small-cell lung cancer patients treated with (chemo)radiotherapy. Int J Radiat Oncol Biol Phys. 2008;70:1039–1044. doi: 10.1016/j.ijrobp.2007.07.2323. [DOI] [PubMed] [Google Scholar]
- 13.Pfister D.G., Johnson D.H., Azzoli C.G. American Society of Clinical Oncology treatment of unresectable non-small-cell lung cancer guideline: Update 2003. J Clin Oncol. 2004;22:330–353. doi: 10.1200/JCO.2004.09.053. [DOI] [PubMed] [Google Scholar]
- 14.Brundage M.D., Davies D., Mackillop W.J. Prognostic factors in non-small cell lung cancer: A decade of progress. Chest. 2002;122:1037–1057. doi: 10.1378/chest.122.3.1037. [DOI] [PubMed] [Google Scholar]
- 15.Firat S., Byhardt R.W., Gore E. Comorbidity and Karnofksy performance score are independent prognostic factors in stage III non-small-cell lung cancer: An institutional analysis of patients treated on four RTOG studies. Radiation Therapy Oncology Group. Int J Radiat Oncol Biol Phys. 2002;54:357–364. doi: 10.1016/s0360-3016(02)02939-5. [DOI] [PubMed] [Google Scholar]
- 16.Non-Small Cell Lung Cancer Collaborative Group Chemotherapy for non-small cell lung cancer. Cochrane Database Syst Rev. 2000;(2):CD002139. doi: 10.1002/14651858.CD002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solan M.J., Werner-Wasik M. Prognostic factors in non-small cell lung cancer. Semin Surg Oncol. 2003;21:64–73. doi: 10.1002/ssu.10023. [DOI] [PubMed] [Google Scholar]
- 18.Zhao L., West B.T., Hayman J.A. High radiation dose may reduce the negative effect of large gross tumor volume in patients with medically inoperable early-stage non-small cell lung cancer. Int J Radiat Oncol Biol Phys. 2007;68:103–110. doi: 10.1016/j.ijrobp.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 19.Werner-Wasik M., Swann R.S., Bradley J. Increasing tumor volume is predictive of poor overall and progression-free survival: Secondary analysis of the Radiation Therapy Oncology Group 93-11 phase I-II radiation dose-escalation study in patients with inoperable non-small-cell lung cancer. Int J Radiat Oncol Biol Phys. 2008;70:385–390. doi: 10.1016/j.ijrobp.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 20.Bradley J.D., Ieumwananonthachai N., Purdy J.A. Gross tumor volume, critical prognostic factor in patients treated with three-dimensional conformal radiation therapy for non-small-cell lung carcinoma. Int J Radiat Oncol Biol Phys. 2002;52:49–57. doi: 10.1016/s0360-3016(01)01772-2. [DOI] [PubMed] [Google Scholar]
- 21.Basaki K., Abe Y., Aoki M. Prognostic factors for survival in stage III non-small-cell lung cancer treated with definitive radiation therapy: Impact of tumor volume. Int J Radiat Oncol Biol Phys. 2006;64:449–454. doi: 10.1016/j.ijrobp.2005.07.967. [DOI] [PubMed] [Google Scholar]
- 22.Lambin P., Rios-Velazquez E., Leijenaar R. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer Oxf Engl. 1990;2012(48):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anon. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. - PubMed - NCBI. [DOI] [PMC free article] [PubMed]
- 24.Leijenaar R.T.H., Carvalho S., Hoebers F.J.P. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol Stockh Swed. 2015;54:1423–1429. doi: 10.3109/0284186X.2015.1061214. [DOI] [PubMed] [Google Scholar]
- 25.Leijenaar R.T.H., Nalbantov G., Carvalho S. The effect of SUV discretization in quantitative FDG-PET Radiomics: The need for standardized methodology in tumor texture analysis. Sci Rep. 2015;5:11075. doi: 10.1038/srep11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panth K.M., Leijenaar R.T.H., Carvalho S. Is there a causal relationship between genetic changes and radiomics-based image features? An in vivo preclinical experiment with doxycycline inducible GADD34 tumor cells. Radiother Oncol. 2015;116:462–466. doi: 10.1016/j.radonc.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Langendijk H., de Jong J., Wanders R. The relevance of the revised version of the international lung cancer staging system for non-small-cell lung cancer treated with radiotherapy. Clin Lung Cancer. 2001;3:33–36. doi: 10.3816/clc.2001.n.015. [DOI] [PubMed] [Google Scholar]
- 28.Berghmans T., Lafitte J.J., Thiriaux J. Survival is better predicted with a new classification of stage III unresectable non-small cell lung carcinoma treated by chemotherapy and radiotherapy. Lung Cancer. 2004;45:339–348. doi: 10.1016/j.lungcan.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Collins G.S., Reitsma J.B., Altman D.G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. BMC Med. 2015;13:1. doi: 10.1186/s12916-014-0241-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Baardwijk A., Wanders S., Boersma L. Mature results of an individualized radiation dose prescription study based on normal tissue constraints in stages I to III non-small-cell lung cancer. J Clin Oncol. 2010;28:1380–1386. doi: 10.1200/JCO.2009.24.7221. [DOI] [PubMed] [Google Scholar]
- 31.Dehing-Oberije C., Aerts H., Yu S. Development and Validation of a Prognostic Model Using Blood Biomarker Information for Prediction of Survival of Non–Small-Cell Lung Cancer Patients Treated With Combined Chemotherapy and Radiation or Radiotherapy Alone (NCT00181519, NCT00573040, and NCT00572325) Int J Radiat Oncol. 2011;81:360–368. doi: 10.1016/j.ijrobp.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 32.van Baardwijk A., Bosmans G., Boersma L. Individualized radical radiotherapy of non–small-cell lung cancer based on normal tissue dose constraints: A feasibility study. Int J Radiat Oncol Biol Phys. 2008;71:1394–1401. doi: 10.1016/j.ijrobp.2007.11.070. [DOI] [PubMed] [Google Scholar]
- 33.Belderbos J., Uitterhoeve L., van Zandwijk N. Randomised trial of sequential versus concurrent chemo-radiotherapy in patients with inoperable non-small cell lung cancer (EORTC 08972-22973) Eur J Cancer. 2007;43:114–121. doi: 10.1016/j.ejca.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Vansteenkiste J., Crino L., Dooms C. 2nd ESMO Consensus Conference on Lung Cancer: Early-stage non-small-cell lung cancer consensus on diagnosis, treatment and follow-up. Ann Oncol. 2014;25:1462–1474. doi: 10.1093/annonc/mdu089. [DOI] [PubMed] [Google Scholar]
- 35.van Elmpt W., Ruysscher D.D., van der Salm A. The PET-boost randomised phase II dose-escalation trial in non-small cell lung cancer. Radiother Oncol. 2012;104:67–71. doi: 10.1016/j.radonc.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Lauritzen S.L. The EM algorithm for graphical association models with missing data. Comput Stat Data Anal. 1995;19:191–201. [Google Scholar]
- 37.Kuschner K.W., Malyarenko D.I., Cooke W.E. A Bayesian network approach to feature selection in mass spectrometry data. BMC Bioinformatics. 2010;11:177. doi: 10.1186/1471-2105-11-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jochems A., Deist T.M., van Soest J. Distributed learning: Developing a predictive model based on data from multiple hospitals without data leaving the hospital - A real life proof of concept. Radiother. Radiother Oncol. 2016;121:459–467. doi: 10.1016/j.radonc.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Deist T.M., Jochems A., van Soest J. Infrastructure and distributed learning methodology for privacy-preserving multi-centric rapid learning health care: euroCAT. Clin Transl Radiat Oncol. 2017;4:24–31. doi: 10.1016/j.ctro.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Druzdzel MJ. SMILE: Structural Modeling, Inference, and Learning Engine and GeNIe: A development environment for graphical decision-theoretic models. Proceeding AAAI ‘99/IAAI '99 Proceedings of the sixteenth national conference on Artificial intelligence and the eleventh Innovative applications of artificial intelligence conference innovative applications of artificial intelligence Orlando, Florida, USA — July 18 - 22, 1999. American Association for Artificial Intelligence Menlo Park, CA, USA; 1999:902-903.
- 41.Robin X., Turck N., Hainard A. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 43.Therneau T.M., Grambsch P.M. Springer Science+Business Media; Berlin, Germany: 2000. Modeling Survival Data: Extending the Cox Model. [Google Scholar]
- 44.Harrington D.P., Fleming T.R. A class of rank test procedures for censored survival data. Biometrika. 1982;69:553–566. [Google Scholar]
- 45.Dorey F.J., Korn E.L. Effective sample sizes for confidence intervals for survival probabilities. Stat Med. 1987;6:679–687. doi: 10.1002/sim.4780060605. [DOI] [PubMed] [Google Scholar]
- 46.Bauer D.F. Constructing confidence sets using rank statistics. J Am Stat Assoc. 1972;67:687–690. [Google Scholar]
- 47.Hollander M., Wolfe D.A., Chicken E. John Wiley & Sons; Hoboken, NJ: 2013. Nonparametric Statistical Methods. [Google Scholar]
- 48.Spirtes P., Glymour C. An algorithm for fast recovery of sparse causal graphs. Soc Sci Comput Rev. 1991;9:62–72. [Google Scholar]
- 49.Dehing-Oberije C., Yu S., De Ruysscher D. Development and external validation of prognostic model for 2-year survival of non-small-cell lung cancer patients treated with chemoradiotherapy. Int J Radiat Oncol Biol Phys. 2009;74:355–362. doi: 10.1016/j.ijrobp.2008.08.052. [DOI] [PubMed] [Google Scholar]
- 50.Werner-Wasik M., Scott C., Cox J.D. Recursive partitioning analysis of 1999 Radiation Therapy Oncology Group (RTOG) patients with locally-advanced non-small-cell lung cancer (LA-NSCLC): Identification of five groups with different survival. Int J Radiat Oncol Biol Phys. 2000;48:1475–1482. doi: 10.1016/s0360-3016(00)00801-4. [DOI] [PubMed] [Google Scholar]
- 51.Komaki R., Scott C.B., Byhardt R. Failure patterns by prognostic group determined by recursive partitioning analysis (RPA) of 1547 patients on four radiation therapy oncology group (RTOG) studies in inoperable nonsmall-cell lung cancer (NSCLC) Int J Radiat Oncol Biol Phys. 1998;42:263–267. doi: 10.1016/s0360-3016(98)00213-2. [DOI] [PubMed] [Google Scholar]
- 52.Bradley J.D., Paulus R., Komaki R. Standard-dose versus high-dose conformal radiotherapy with concurrent and consolidation carboplatin plus paclitaxel with or without cetuximab for patients with stage IIIA or IIIB non-small-cell lung cancer (RTOG 0617): A randomised, two-by-two factorial phase 3 study. Lancet Oncol. 2015;16:187–199. doi: 10.1016/S1470-2045(14)71207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mauguen A., Le Péchoux C., Saunders M.I. Hyperfractionated or accelerated radiotherapy in lung cancer: An individual patient data meta-analysis. J Clin Oncol. 2012;30:2788–2797. doi: 10.1200/JCO.2012.41.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sesen M.B., Nicholson A.E., Banares-Alcantara R. Bayesian networks for clinical decision support in lung cancer care. PLoS One. 2013;8:e82349. doi: 10.1371/journal.pone.0082349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stacey D., Légaré F., Col N.F. Decision aids for people facing health treatment or screening decisions. Cochrane Database Syst Rev. 2014;(1):CD001431. doi: 10.1002/14651858.CD001431.pub4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.