Abstract
Objectives
Serum protein electrophoresis is a commonly used test in the diagnosis and follow-up of patients with monoclonal gammopathies. The practice of documenting the location of the peak may serve as delta check flags in SPEP samples.
Methods
We report on the inconsistent finding on two tests performed about three months apart. The inconsistency was discovered due to recognition of the change in the location of the monoclonal immunoglobulin on protein electrophoresis.
Results
Repeat testing with a third specimen revealed that the first test was run on a wrong specimen.
Conclusion
Recording the location of the monoclonal spike is recommended to serve as an additional “delta” check.
Keywords: Monoclonal gammopathies, M-peak location, Delta check, Specimen Mislabeling
Highlights
-
•
Delta checks compare earlier results and flag data for further investigation.
-
•
The monoclononal immunoglobulin peak can serve as a delta check.
-
•
Recording the location of the peak of a monoclonal immunoglobulin is proposed.
1. Introduction
Specimen mislabeling is one of the reasons for pre-analytical laboratory errors [1]. Laboratories have introduced measures to curtail these errors, including barcode labeling, repeat blood group testing for specimens used in blood transfusion, and delta checks [2], [3], [4]. Delta checks are generally programmed into the analyzers so that the results of the current specimen are compared to earlier results and significant departures are flagged for further investigation. Only a limited number of analytes are suitable candidates for delta check program due to rapid and marked variation in results in acutely ill patients [4].
Plasma cell proliferative disorders are commonly associated with the synthesis and secretion of a monoclonal immunoglobulin. These abnormal proteins are usually diagnosed and monitored by serum protein electrophoresis (SPEP), serum protein immunofixation electrophoresis (SIFE), urine protein electrophoresis (UPEP), urine protein immunofixation electrophoresis (UIFE) [5], [6], [7]. Electrophoretic methods depend upon the visual identification of the monoclonal immunoglobulin and pathologists are increasingly using the electronic medical record information to add value to the results of these assays [8]. One item in reporting the results is documentation of the location of the peak, or M spike. The location of the peak may be noted as extreme cathodic end of SPEP, cathodal region of SPEP, mid-gamma region, anodal region, anodal end, point of application, beta region, co-migrating with C3, peak between C3 and transferrin bands, co-migrating with transferrin and uncommonly in the alpha-2 region.
We present a patient in whom a specimen misidentification occurred in the laboratory, and documentation of the location of the monoclonal peak was one of the factors that allowed detection of the error.
2. Materials and methods
The patient was a 67-year-old Caucasian male who presented to clinic for anemia work-up, on referral from his primary care physician. On presentation, he complained of gum bleeding while brushing his teeth, but denied any night sweats, fatigue, or unintentional weight loss.
In addition to other studies for the work-up of anemia, serum protein electrophoresis was performed with agarose gel electrophoresis using the Helena SPIFE 3000 system Manufacturer protocols were followed for all portions of the SPEP and SIFE as well as urine protein electrophoresis (UPEP) and urine protein immunofixation electrophoresis (UIFE).
3. Results
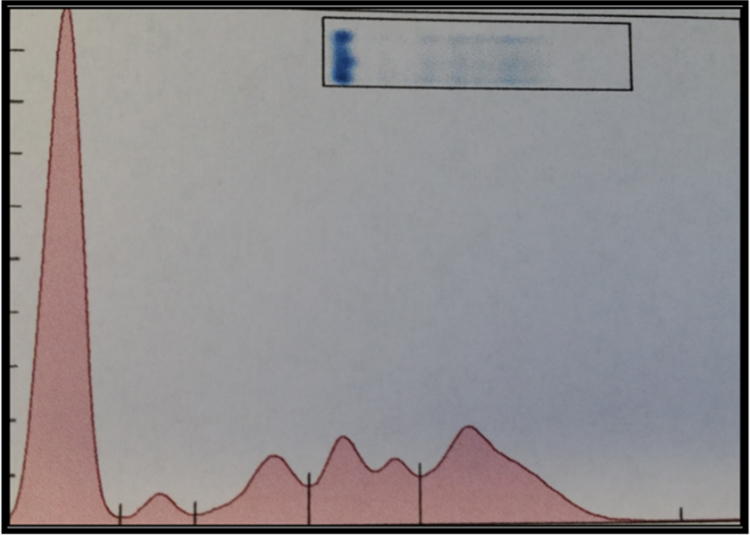
A complete blood count was performed in January 2016 and was found to be mostly unremarkable, except a hemoglobin value of 10.8 g/dL and platelet count of 154. Iron studies showed an elevated ferritin, and otherwise normal iron and total iron binding capacity. An initial SPEP/SIFE showed a monoclonal IgG kappa band at a concentration of about 0.68 mg/dL, in the anodal gamma region (Fig. 1).
Fig. 1.
SPEP and SIFE performed in January 2016 reveal a monoclonal IgG kappa band located in the anodal gamma region.
A second SPEP/SIFE performed in April 2016 showed a M-spike, at a concentration of 0.47 g/dL, but in the cathodal gamma region, in contrast to the anodal region spike reported in January suggesting an error in specimen labeling(Fig. 2). SIFE showed the peak to be IgG lambda, further supporting the contention that the two samples were from two different patients. A repeat SPEP/SIFE was recommended to rule out pre-analytical errors, and showed a IgG Lambda peak similar to that shown in Fig. 2. Bone marrow examination performed after the second SPEP showed 30% plasma cells with lambda restriction. UIFE revealed a prominent lambda monoclonal light chain spike. Serial serum protein and immunoglobulin quantitation levels are listed in Table 1.
Fig. 2.
SPEP and SIFE performed, allegedly, on the same patient in April 2016 showed a monoclonal IgG lambda spike, and, it was now located in the cathodal gamma region. A repeat test on a new specimen confirmed that the results shown in Fig. 2 were correct and were reproduced. The SIFE was done on the specimen in Fig. 2 because the peak was noted to be located in a different part of the gamma region as compared to that shown in Fig. 1.
Table 1.
Serial serum quantitation levels.
| Test | January 2016 | April 2016 | May 2016 |
|---|---|---|---|
| Albumin (g/dL) | 3.92 | 4.37 | 4.64 |
| Alpha 1 Globulin (g/dL) | 0.27 | 0.24 | 0.06 |
| Alpha 2 Globulin (g/dL) | 0.77 | 0.78 | 0.74 |
| Beta Globulin (g/dL) | 1.19 | 1.07 | 0.95 |
| IgA (mg/dL) | <15 | <15 | 21 |
| IgG (mg/dL) | 864 | 792 | 797 |
| IgM (mg/dL) | 18 | 16 | 15 |
The results of the various tests were not meaningfully different, leading us to conclude that only SPEP was done on the wrong specimen in January and other tests were conducted on the correct specimen.
4. Discussion
Investigation of monoclonal gammopathy includes electrophoretic analysis of serum and urine. Once the identity of the monoclonal spike is established, repeat SIFE is not recommended if the spike is in the same location as noted previously [5]. If a peak is not detectable on SPEP, and SIFE is recommended due the greater sensitivity of the latter.
In patients with multiple myeloma, treatment, especially stem cell transplantation may lead to the emergence of peaks in different locations and an oligoclonal pattern is not uncommon in post-stem cell transplantation state. Similarly, treatment with monoclonal antibodies may reveal a band incongruous with the original monoclonal immunoglobulin [9].
In this case, a disparity in the location of the peak was noted between the SPEP in January and the second one in April. The patient had not received any treatment for the monoclonal gammopathy in the interim. The difference in the monoclonal peak in two specimens was confirmed by the SIFE results that showed kappa chain in the January specimen and lambda chain in the April specimen. A third specimen was obtained and the identity of the peak was confirmed to be IgG lambda. IgG lambda nature of the lesion was also validated by bone marrow study and urine examination in May 2016.
The practice of documenting the location of the peak would have caused a delta check flag in this patient even if SIFE had shown IgG kappa in the second specimen, being that IgG kappa is the commonest monoclonal immunoglobulin. A retrospective look at all of the SPEP/SIFEs done on and around day of the first SPEP in January did not reveal another specimen with the SPEP/SIFE findings seen in the second and third specimens. The immunoglobulin quantification results of the three specimens were similar. Due to the time lapse between January and April SPEPs on the patient, we were not able to retest the specimens from January. It appeared that there was not a switch between two patients, but an incorrect sample was labeled as being from this patient for SPEP only, as the immunoglobulin quantitation results were similar in all three test episodes. It also appeared that an SPEP had not been requested on the specimen erroneously analyzed in January.
It is recommended that the location of the peak of a monoclonal immunoglobulin be recorded to facilitate detection of specimen mislabeling. The practice would also allow judicious use of SIFE [5]. There would be no additional cost to this maneuver but it would result is increased patient safety. It is possible that IT solutions may allow the densitometric scanners to compare the location of peaks with earlier peaks, as is the case with routine delta checks. The computerized comparison of location of the peaks promises to make the process more objective than the current subjective reading of the location.
Funding sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- 1.College of American Pathologists. When a rose is not a rose. http://www.cap.org/apps/portlets/contentViewer/show.do?printFriendly=true&contentReference=practice_management/directips/mislabeled_specimens.html Accessed May 29, 2016.
- 2.Killeen J.P., Chan T.C., Jones K. Impact of bar coding technology and computerized physician order entry on reducing laboratory specimen mis-identification errors in the emergency department. Acad. Emerg. Med. 2005;12(suppl 1):49. [Google Scholar]
- 3.American Association of Clinical Chemistry. Fixing the Problem of Mislabeled Specimens in Clinical Labs. (2014, April 1). Accessed May 29, 2016.
- 4.M. Astion. Right Patient, Wrong Sample. Patient Safety Net. https://psnet.ahrq.gov/webmm/case/142 Accessed May 29, 2016.
- 5.Heaton C., Vyas S.G., Singh G. Audit of use and overuse of serum protein immunofixation electrophoresis and serum free light chain assay in tertiary health care: a case for algorithmic testing to optimize laboratory utilization. Am. J. Clin. Pathol. 2016;145(4):531–537. doi: 10.1093/ajcp/aqw026. [DOI] [PubMed] [Google Scholar]
- 6.Kyle R.A. Sequence of testing for monoclonal gammopathies: serum and urine assays. Arch. Pathol. Lab Med. 1999;123:114–118. doi: 10.5858/1999-123-0114-SOTFMG. [DOI] [PubMed] [Google Scholar]
- 7.Dimopoulos M., Kyle R., Fermand J.-P. consensus recommendations for standard investigative workup: report of the International Myeloma workshop consensus panel 3. Blood. 2011;117:4701–4705. doi: 10.1182/blood-2010-10-299529. (Epub 2011 Feb 3) [DOI] [PubMed] [Google Scholar]
- 8.Zia H.M., Singh G. Optimization of utilization of serum protein analysis: role of the electronic medical record in promoting consultation by pathology. Am. J. Clin. Pathol. 2013;139(6):793–797. doi: 10.1309/AJCP1ZRZ7KLYSLTG. [DOI] [PubMed] [Google Scholar]
- 9.van de Donk Niels W.C.J., Moreau Philippe. Clinical efficacy and management of monoclonal antibodies targeting CD38 and SLAMF7 in multiple myeloma. Blood. 2016;127:681–695. doi: 10.1182/blood-2015-10-646810. [DOI] [PubMed] [Google Scholar]