Abstract
We have developed a modified RNA interference (RNAi) method for generating gene knock-outs in Drosophila melanogaster. We used the sequence of the yellow (y) locus to construct an inverted repeat that will form a double-stranded hairpin structure (y-IR) that is under the control of the upstream activating sequence (UAS) of the yeast transcriptional activator GAL4. Hairpins are extremely difficult to manipulate in Escherichia coli, so our method makes use of a heterologous 330 bp spacer encoding sequences from green fluorescent protein to facilitate the cloning steps. When the UAS–y–IR hairpin is expressed under the control of different promoter–GAL4 fusions, a high frequency of y pigment phenocopies is obtained in adults. Consequently this method for producing gene knock-outs has several advantages over previous methods in that it is applicable to any gene within the fly genome, greatly facilitates cloning of the hairpin, can be used if required with GAL4 drivers to avoid lethality or to induce RNAi in a specific developmental stage and/or tissue, is useful for generating knock-outs of adult phenotypes as reported here and, finally, the system can be manipulated to investigate the trans-acting factors that are involved in the RNAi mechanism.
INTRODUCTION
Drosophila melanogaster is undoubtedly one of the most versatile and useful organisms in molecular genetics research and presents some advantages over other models such as mouse, yeast or Arabidopsis. Yet until very recently, Drosophila suffered from an inadequacy not shared with some of its fellows: it was not suitable for reverse genetics because it is difficult to ‘knock-out’ a gene identified on the basis of its sequence or position. Although large collections of P-element mutagenised flies are readily available at various stock centres, insertions within the desired coding sequences are often absent. This drawback has become more of an issue given that the recently completed Drosophila genome project has made available a wealth of new sequence information (1).
RNA interference (RNAi) is a powerful method for silencing genes at a post-transcriptional stage. Introduction of double-stranded RNA (dsRNA) triggers degradation of the mRNA bearing the same sequence in a variety of organisms (2–4). The mechanism by which this is achieved is as yet poorly understood but one of the earliest steps seems to consist of the degradation of the dsRNA into short fragments (5,6). Injection of specific dsRNA into the early Drosophila embryo results in interference persisting throughout the embryonic development (4,7,8) while interference of gene function at the adult stage seems to be greatly reduced (7). Apart from its transient nature, interference mediated by dsRNA injection is not heritable in a Mendelian fashion, so that each individual analysed in an experiment is necessarily the result of direct manipulation, making the collection of large data sets extremely labour intensive.
In order to overcome these hurdles we have developed an alternative method for administering dsRNA to Drosophila, which relies on transcription from an integrated construct consisting of two inverted repeats (IR), separated by a unrelated DNA sequence that acts as a spacer, to give a hairpin–loop shaped RNA. A similar method has been recently used to trigger RNAi in the nematode Caenorhabditis elegans (9) and in the fly (10,11). However, unlike these previous reports, we exploited the GAL4–upstream activating sequence (UAS) binary system (12) in order to drive expression of the transgene to produce knock-out of an adult phenotype. We describe here the generation of yellow (y) phenocopies in which the expression of the construct UAS–y–IR is driven by GAL4 under the control of the Actin5c, daughterless, timeless or heat-shock promoters.
MATERIALS AND METHODS
Plasmid construction
A 1120 bp fragment from the coding sequence of the y gene was amplified using the pair of primers 5′-CTTTGACTTGACCACGGATAC-3′ and 5′-ATGATGCCACCACCCAGATTG-3′ and cloned in the T vector pDK101 (13) to yield pDK–y. A 950 bp subfragment was then excised using the endogenous EcoRI and PstI sites and cloned at the EcoRI–PstI sites of the pBC KS+ vector (Stratagene). In parallel, the primer pair 5′-ACGGCCTGCAGTGCTTCAGC-3′ and 5′-GAGCTGCAGGCTGCCGTCCT-3′ was used to amplify a fragment of the green fluorescent protein (GFP) coding sequence, resulting in a 330 bp DNA sequence with PstI sites at both ends. pDK–y was then linearised by digesting with PstI (from the yellow fragment) and SalI (from the vector polylinker) and re-circularised together with the 330 bp PstI–PstI GFP fragment and the 950 bp y fragment excised from pBC using SalI and PstI. Finally, the 2.2 kb EcoRI–EcoRI fragment containing the two y IR separated by the GFP spacer was cloned into the Drosophila transformation vector pUAST (12) to give UAS–y–IR.
P-element mediated transformation
Transformation of Drosophila embryos was carried out according to Spradling (14). Several transformant lines were obtained with autosomal insertions, two of which (5F and 7A) were analysed for their ability to trigger dsRNAi upon activation of transgene transcription by crossing to daughterless–GAL4 (da–GAL4), Actin5c–GAL4 (Act5c–GAL4), timeless–GAL4 (tim–GAL4) and heat-shock–GAL4 (hsp70–GAL4) driver strains. In the latter case, expression of the driver was triggered by placing vials, containing Drosophila at different developmental stages, in a 37°C water bath for 1 h.
The chromosome harbouring the UAS–y–IR insert(s) was determined with standard genetic crosses and the insert was then balanced with dominantly marked, multiple inverted chromosomes and later made homozygous. In situ hybridisation on third instar salivary gland chromosomes was used to pinpoint the map position and detect the number of inserts in each line using the y–IR probe (the two y IR separated by the GFP spacer), by means of the DIG-DNA Labeling and Detection Kit (Boehringer Mannheim) (15).
RESULTS
In situ hybrydisation on polytene chromosomes revealed that transgenic line 5F was homozygous for an insert mapping at position 98–99 (chromosome 3R), while line 7A carried two homozygous inserts at map positions 69F (3L) and 97–98 (3R). Additionally, both lines showed the presence of another copy of the UAS–y–IR insert, mapping at 101A-B (chromosome 4) in line 5F and at 59 (2R) in line 7A. These additional copies must have hitch-hiked during the various crosses performed in order to make the two transformant lines homozygous for chromosome 3 and therefore must be present in the transgenic populations at a somewhat low frequency. In case of the insert mapping at position 101A-B, there is a high probability that this transgene falls in a heterochromatic portion of the genome and is therefore not expressed.
Transgenic flies carrying the UAS–y–IR construct were crossed to the GAL4 drivers in order to activate transcription from the hairpin-encoding transgene (Fig. 1) and the progeny were inspected for the colour of cuticle, wings and bristles. In order to estimate the efficiency of the system in promoting gene silencing, we set up individual crosses for each line using single homozygous UAS–y–IR flies, to homozygous (da–GAL4, tim–GAL4, hsp70–GAL4) or balanced (Act5c–GAL4/TM6c,Tb) driver lines. We estimated the silencing efficiency by examining the percentage of progeny that gave a y-like phenotype. Crosses UAS–y–IR × Act5c–GAL4/TM6c, Tb resulted in a 100% of progeny [after removal of the Tubby (Tb) pupae, devoid of the Act5c–GAL4 driver] mimicking y1-type mutants, displaying yellow-like cuticle, wings and bristles (Fig. 2), irrespective of the UAS–y–IR line used for the cross. We retrieved 109 y-like males and 127 y-like females from crossing Act5c–GAL4 with line 5F, and 90 and 74 yellow males and females, respectively, with UAS–y–IR 7A (Table 1). As expected, all Tb flies displayed a wild-type pigmentation.
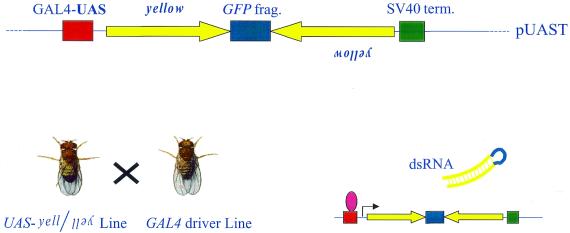
Figure 1.
Scheme for the generation of transgenic, hairpin-mediated RNAi. A coding fragment of the yellow gene was cloned as an inverted repeat into the Drosophila transformation vector pUAST, using a GFP fragment to separate the repeats and therefore facilitate the cloning. The resulting transformants were then crossed to various GAL4 strains to drive expression of the hairpin-encoding transgene.
Figure 2.
Effect of UAS–y–IR-encoded dsRNA transcription on the pigmentation of male and female adult D.melanogaster. y+ flies (w1118; da-GAL4/+) and y1-type mutants are shown for comparison. Expression of UAS–y–IR-encoded dsRNA mediated by a daughterless–GAL4 driver blocks melanin pigment deposition in cuticle and wing structures but not in bristles, mimicking the colour pattern of y2-type mutants. Act5c–GAL4 driven dsRNA transcription triggers effective RNAi also in bristles, producing y1-like phenocopies.
Table 1. Summary of data relative to the crosses between UAS–y–IR transgenic lines and GAL4 driver strains.
| Parental genotype |
Wild-type male |
Malea |
y male |
% y males |
Wild-type female |
Femalea |
y female |
% y females |
% y total |
|
UAS–y–IR 5F × da–GAL4 |
33 |
138 |
119 |
41 |
33 |
122 |
95 |
38 |
40 |
|
UAS–y–IR 7A × da–GAL4 |
38 |
192 |
138 |
37 |
51 |
166 |
141 |
39 |
38 |
|
UAS–y–IR 5F × Act5c–GAL4 |
– |
– |
109 |
100 |
– |
– |
127 |
100 |
100 |
|
UAS–y–IR 7A × Act5c–GAL4 |
– |
– |
90 |
100 |
– |
– |
74 |
100 |
100 |
|
UAS–y–IR 5F × hsp70–GAL4 |
– |
44 |
32 |
42 |
– |
71 |
21 |
23 |
32 |
| UAS–y–IR 7A × hsp70–GAL4 | – | – | 171 | 100 | – | – | 154 | 100 | 100 |
The number of wild type progeny are indicated, together with that of yellow phenocopies. y2-type for crosses with da–GAL4 and y1-type when the UAS–y–IR is activated by the Act5c–GAL4 driver. Percentage of y phenocopies is computed taking into account only the number of y-like flies and considering the incomplete yellow phenocopies as wild-type. Figures indicated for the UAS–y–IR × hsp70–GAL4 crosses refer to progeny obtained after reiterated heat treatments.
aThe incomplete yellow phenotype observed with the da–GAL4 driver and those resulting from crosses with hsp70–GAL4.
When da–GAL4 was used to drive UAS–y–IR expression, the resulting phenotype resembled y2-type mutants (16), with yellow-like cuticle and wings but wild-type bristles (Fig. 2) and overall interference efficiency was significantly reduced (down to ∼37%, with similar values in both UAS–y–IR lines, Table 1) compared to Act5c–GAL4. A high proportion of the progeny also displayed a yellowish cuticle, especially visible on the pale areas of abdomen and thorax, what we refer to as an ‘incomplete y phenotype’. The degree of variation of this yellow hue is rather broad, ranging from almost y-like to almost wild-type. For this reason we classified those ‘incomplete y’ flies as wild-type when computing the knock-out frequency, thereby generating an extremely conservative estimate of efficiency.
Crosses with the tim–GAL4 driver did not generate y individuals, consistent with the reported expression of tim within internal organs that have circadian function (17).
UAS–y–IR expression was also driven with an hsp70–GAL4 transgene. A once-in-a-lifetime, 1 h heat pulse could trigger RNAi with a low efficiency when administered at the late third instar/early pupa stage, resulting in ∼5–10% of the flies displaying a y1-like phenotype while for a higher proportion (∼30%) y silencing was only restricted to the bristles (these flies had yellow-brown bristles but wild-type wings and body). No y silencing was observed when the heat treatment was given to embryos, first or second instar larvae, older pupae or adults. The frequency of interference was dramatically enhanced in the double transformants hsp70–GAL4; UAS–y–IR line 7A (originated in the cross hsp70–GAL4 UAS–y–IR 7A; 100%, Table 1) when Drosophila were treated with a 1 h heat pulse per day from the late third instar stage until hatching. The same treatment on hsp70–GAL4; UAS–y–IR 5F resulted in a high frequency of flies with a body colour reminiscent of the incomplete y obtained in the da–GAL4 crosses (yellowish body and dark pigmented areas) readily discernible from their untreated siblings and y bristles (∼60%), while the remaining flies also had a mosaic pigment pattern of the tergites.
In summary, the overall results indicate that the presence of a heterologous 330 bp fragment separating the two inverted repeats does not obstruct the ability of the construct to trigger RNAi.
DISCUSSION
We have obtained a knock-out of the y gene by expressing a transcript in Drosophila, consisting of two IRs which presumably fold to give a dsRNA. These flies, although genotypically y+, mimic amorphic mutations of the y locus, resulting in a yellow-brown cuticle colour (16). The difference in the phenotypes obtained with Act5c, da, tim and hsp70 promoters will presumably reflect not only their intrinsic expression patterns but also their promoter strength and insert positions. The hsp70 promoter may drive a high but transient expression of GAL4 (and therefore of the UAS–y–IR hairpin), that results in a weak silencing effect. On the other hand, reiterated heat treatments may maintain the expression level of the y–IR transcript above a certain threshold necessary to evoke RNAi in line 7A. In line 5F the average y–IR transcript level may be lower since this line carries a single copy of the UAS–y–IR construct (albeit some individuals may carry an extra copy on chromosome 4), although a position effect cannot be ruled out. It is noteworthy that a dose/position effect can be observed with the hsp70–GAL4 driver but not when UAS–y–IR transcription is activated by da–GAL4. Such dose/position effects in modulating the outcome of RNAi can have practical advantages when trying to silence lethal genes, as they could allow survivors to display unexpected phenotypes.
Since previous reports indicate that a very limited number of dsRNA molecules are sufficient to induce interference (4,18), we speculate that the hsp70–GAL4 result could mean that the hairpin RNA is rapidly degraded in Drosophila and possibly does not undergo an amplification step as reported in C.elegans (19), Arabidopsis (20) and Neurospora (21). Absence of an RNA amplification step is in agreement with findings that RNAi in Drosophila is independent of RNA synthesis (22).
In fact it is conceivable that dsRNA degradation may explain why the y2-like bristle pattern is obtained with the da–GAL4 construct, even though da is expressed in the cuticle and peripheral sense organs (23,24). After differentiation of the proneural cluster, da transcription may be switched off, leading to a rapid degradation of the y–IR transcript before the late third instar stage, when its expression would be required to silence cuticular y mRNA.
In order to facilitate the cloning of the yellow inverted repeat construct, we used a spacer between the two repeats, consisting of an unrelated DNA sequence, a 330 bp GFP fragment. As reported in the literature (25) and also in our experience, inverted repeats are cloned in Escherichia coli at an extremely inefficient rate and the introduction of this spacer greatly increased the efficiency. The spacer’s length appears to be a critical factor since the use of a shorter spacer (150 bp) did not result in any improvement of the cloning efficiency. We attempted to remove the GFP fragment from the pDK–y–IR (the complete construct cloned in pDK101 rather than in the P-element vector pUAST), an operation requiring a simple PstI digest followed by electrophoretic separation of the resulting two fragments and re-circularisation of the 5 kb band. However, even from this sterically advantaged ligation reaction, we were unable to recover any transformant colony.
We can confirm that the difficulty in cloning the two IRs was not a peculiarity of the y sequences we used for the work described here, since we met the same problems when preparing two analogous constructs for performing RNAi of the tim2 and cry genes of Drosophila (A.Piccin and E.Rosato, unpublished observations). Also in those cases, the use of a ∼300 bp spacer dramatically increased the cloning efficiency. Given the high levels of knock-out recovered after induction of the hairpin-encoding transgene, we can infer that the presence of a relatively long spacer (approximately one-third of the repeat length) does not significantly obstruct the ability of the dsRNA in mediating interference, while greatly facilitating the IR cloning steps.
This method of producing virtual knock-out of a given gene appears to be more practical than the homologous recombination recently obtained at the y locus by Rong and Golic (26). A difficulty with the latter technique is that homologous recombination may occur via a double-strand breakage-induced replication mechanism, therefore limiting the efficacy of the method to genes located near the tip of the chromosome (27). A similar strategy adopted by a different group indeed failed to produce any recombinants at the less distal white locus (28). In any case, our method is more flexible since the appropriate choice of the GAL4 driver can limit gene silencing to the tissue and/or timing chosen by the investigator, thus circumventing problems of lethality. Moreover, the relatively small constructs needed for transgenic RNAi integrate within the fly genome with high efficiency, therefore making this technique a rather suitable approach to functional genomic studies.
A possible downfall of transgenic RNAi as a mean of generating knock-outs is that dsRNA fragments may mediate post-transcriptional silencing of an undesired gene which happens to fortuitously share sequence homology with the chosen target. Future experiments should assess the correlation between degree of homology of dsRNA and mRNA and effectiveness of interference. Meanwhile, careful designing of the IR construct should reduce this problem.
Finally, UAS–y–IR transgenic flies can be used as a valuable model system in the identification of trans-acting factors involved in establishing dsRNA-mediated genetic interference, by subjecting it to a mutagenesis screening and selecting for individuals resistant to RNAi.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Dr Marina de Bernard (Dipartimento di Scienze Biomediche, Padova, Italy) for technical help with a Bio-Rad MRC 1024ES confocal microscope and Dr Giacomo Cavalli (Institut de Genetique Humaine, Montpellier, France) for kindly supplying the da–GAL4 driver strain. The above research was supported by grants from the European Community (ERBIO4CT960096), Ministero dell’Università e Ricerca Scientifica e Tecnologica (MURST) PRIN and CRUI–MURST–British Council.
References
- 1.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe D.C. (1999) Gene silencing: RNA makes RNA makes no protein. Curr. Biol., 9, R599–R601. [DOI] [PubMed] [Google Scholar]
- 3.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 4.Kennerdell J.R. and Carthew,R.W. (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the Wingless pathway. Cell, 95, 1017–1026. [DOI] [PubMed] [Google Scholar]
- 5.Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 6.Yang D., Lu,H. and Erickson,J.W. (2000) Evidence that processed small dsRNA may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr. Biol., 10, 1191–1200. [DOI] [PubMed] [Google Scholar]
- 7.Misquitta L. and Paterson,B.M. (1999) Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation. Proc. Natl Acad. Sci. USA, 96, 1451–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsuda M., Kamimura,K., Nakato,H., Archer,M., Staatz,W., Fox,B., Humphrey,M., Olson,S., Futch,T., Kaluza,V. et al. (1999) The cell-surface proteoglycan Dally regulates Wingless signalling in Drosophila. Nature, 400, 276–280. [DOI] [PubMed] [Google Scholar]
- 9.Tavernarakis N., Wang,S.L., Dorovkov,M., Ryazanov,A. and Driscoll,M. (2000) Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet., 24, 180–183. [DOI] [PubMed] [Google Scholar]
- 10.Kennerdell J.R. and Carthew,R.W. (2000) Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol., 18, 896–898. [DOI] [PubMed] [Google Scholar]
- 11.Lam G. and Thummel,C.S. (2000) Inducible expression of double-stranded RNA directs specific genetic interference in Drosophila. Curr. Biol., 10, 957–963. [DOI] [PubMed] [Google Scholar]
- 12.Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 13.Kovalic D., Kwak,J.H. and Weisblum,B. (1991) General method for direct cloning of DNA fragments generated by the polymerase chain reaction. Nucleic Acids Res., 19, 4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spradling A.C. (1986) P element-mediated transformation. In Roberts,D.B. (ed), Drosophila: A Practical Approach. IRL Press, Oxford, pp. 175–197.
- 15.Schmidt E.R., Keyl,H.L. and Hankeln,T. (1988) In situ localization of two haemoglobin gene clusters in the chromosomes of 3 species of Chironomus. Chromosoma, 96, 353–359. [Google Scholar]
- 16.Wilson R., Burnet,B., Eastwood,L. and Connolly,K. (1976) Behavioural pleiotropy of the yellow gene in Drosophila melanogaster. Genet. Res., 28, 75–88. [DOI] [PubMed] [Google Scholar]
- 17.Plautz J.D., Kaneko,M., Hall,J.C. and Kay,S.A. (1997) Independent photoreceptive circadian clocks throughout Drosophila. Science, 278, 1632–1635. [DOI] [PubMed] [Google Scholar]
- 18.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 19.Smardon A., Spoerke,J.M., Stacey,S.C., Klein,M.E., Mackin,N. and Maine,E.M. (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol., 10, 169–178. [DOI] [PubMed] [Google Scholar]
- 20.Dalmay T., Hamilton,A., Rudd,S., Angell,S. and Baulcombe,D.C. (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- 21.Cogoni C. and Macino,G. (1999) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature, 399, 166–169. [DOI] [PubMed] [Google Scholar]
- 22.Yang D., Lu,H. and Erickson,J.W. (2000) Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr. Biol., 10, 1191–1200. [DOI] [PubMed] [Google Scholar]
- 23.Cronmiller C., Schedl,P. and and Cline,T.W. (1988) Molecular characterization of daughterless, a Drosophila sex determination gene with multiple roles in development. Genes Dev., 2, 1666–1676. [DOI] [PubMed] [Google Scholar]
- 24.Cronmiller C. and Cummings,C.A. (1993) The daughterless gene product in Drosophila is a nuclear protein that is broadly expressed throughout the organism during development. Mech. Dev., 42, 159–169. [DOI] [PubMed] [Google Scholar]
- 25.Hagan C.E. and Warren,G.J. (1983) Viability of palindromic DNA is restored by deletions occurring at low but variable frequency in plasmids of Escherichia coli. Gene, 24, 317–326. [DOI] [PubMed] [Google Scholar]
- 26.Rong Y.S. and Golic,K.G. (2000) Gene targeting by homologous recombination in Drosophila. Science, 288, 2013–2018. [DOI] [PubMed] [Google Scholar]
- 27.Engels W.R. (2000) Reversal of fortune for Drosophila geneticists? Science, 288, 1973–1975. [DOI] [PubMed] [Google Scholar]
- 28.Bellaiche Y., Mogila,V. and Perrimon,N. (1999) I-SceI endonuclease, a new tool for studying DNA double-strand break repair mechanisms in Drosophila. Genetics, 152, 1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]