Abstract
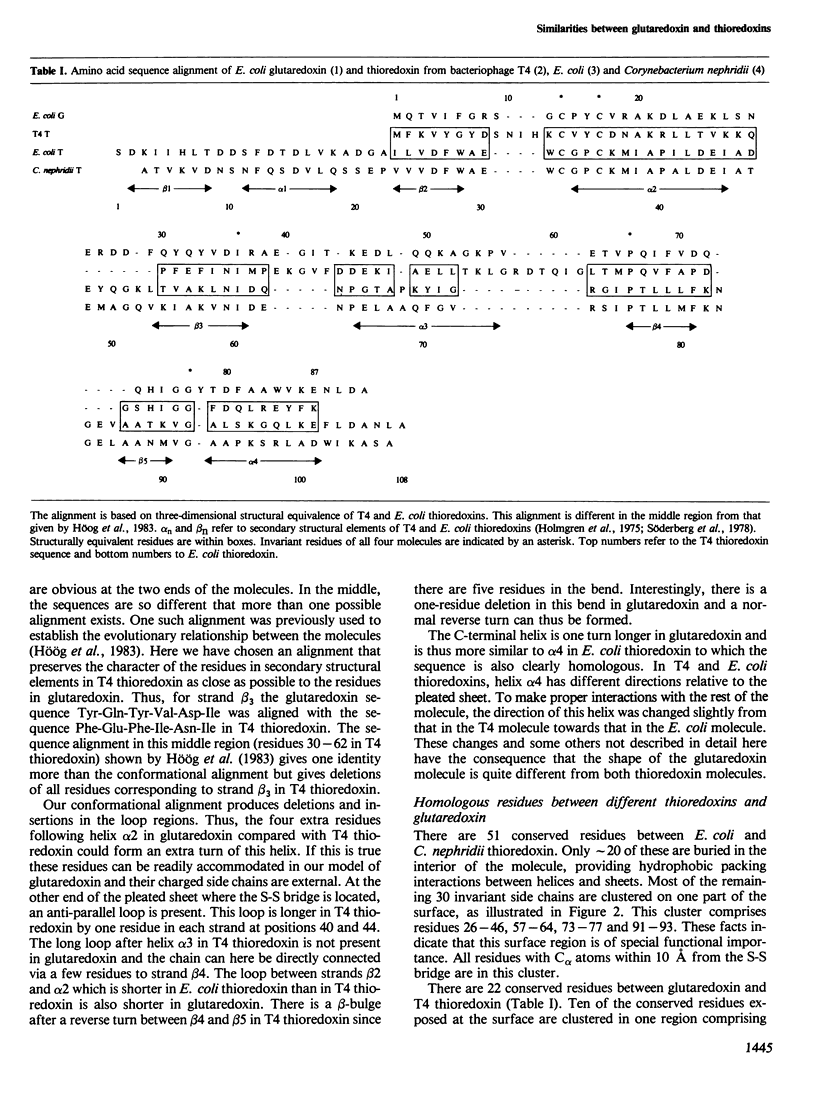
The tertiary structures of thioredoxin from Escherichia coli and bacteriophage T4 have been compared and aligned giving a common fold of 68 C alpha atoms with a root mean square difference of 2.6 A. The amino acid sequence of glutaredoxin has been aligned to those of the thioredoxins assuming that glutaredoxin has the same common fold. A model of the glutaredoxin molecule was built on a vector display using this alignment and the T4 thioredoxin tertiary structure. By comparison of the model with those of the thioredoxins, we have identified a molecular surface area on one side of the redox-active S-S bridge which we suggest is the binding area of these molecules for redox interactions with other proteins. This area comprises residues 33-34, 75-76 and 91-93 in E. coli thioredoxin; 15-16, 65-66 and 76-78 in T4 thioredoxin and 12-13, 59-60 and 69-71 in glutaredoxin. In all three molecules, this part of the surface is flat and hydrophobic. Charged groups are completely absent. In contrast, there is a cluster of charged groups on the other side of the S-S bridge which we suggest participates in the mechanisms of the redox reactions. In particular, a lysine residue close to an aromatic ring is conserved in all molecules.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berglund O., Holmgren A. Thioredoxin reductase-mediated hydrogen transfer from Escherichia coli thioredoxin-(SH)2 to phage T4 thioredoxin-S2. J Biol Chem. 1975 Apr 25;250(8):2778–2782. [PubMed] [Google Scholar]
- Berglund O. Identification of a thioredoxin induced by bacteriophage T4. J Biol Chem. 1969 Nov 25;244(22):6306–6308. [PubMed] [Google Scholar]
- Hall D. E., Baldesten A., Holmgren A., Reichard P. Yeast thioredoxin. Amino-acid sequence around the active-center disulfide of thioredoxin I and II. Eur J Biochem. 1971 Nov 11;23(2):328–335. doi: 10.1111/j.1432-1033.1971.tb01625.x. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Glutathione-dependent enzyme reactions of the phage T4 ribonucleotide reductase system. J Biol Chem. 1978 Oct 25;253(20):7424–7430. [PubMed] [Google Scholar]
- Holmgren A. Glutathione-dependent synthesis of deoxyribonucleotides. Purification and characterization of glutaredoxin from Escherichia coli. J Biol Chem. 1979 May 10;254(9):3664–3671. [PubMed] [Google Scholar]
- Holmgren A. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2275–2279. doi: 10.1073/pnas.73.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A., Kallis G. B., Nordström B. A mutant thioredoxin from Escherichia coli tsnC 7007 that is nonfunctional as subunit of phage T7 DNA polymerase. J Biol Chem. 1981 Mar 25;256(6):3118–3124. [PubMed] [Google Scholar]
- Holmgren A., Ohlsson I., Grankvist M. L. Thiroedoxin from Escherichia coli. Radioimmunological and enzymatic determinations in wild type cells and mutants defective in phage T7 DNA replication. J Biol Chem. 1978 Jan 25;253(2):430–436. [PubMed] [Google Scholar]
- Holmgren A. Reduction of disulfides by thioredoxin. Exceptional reactivity of insulin and suggested functions of thioredoxin in mechanism of hormone action. J Biol Chem. 1979 Sep 25;254(18):9113–9119. [PubMed] [Google Scholar]
- Holmgren A., Roberts G. Nuclear magnetic resonance studies of redox-induced conformational changes in thioredoxin from Escherichia coli. FEBS Lett. 1976 Dec 1;71(2):261–265. doi: 10.1016/0014-5793(76)80946-5. [DOI] [PubMed] [Google Scholar]
- Holmgren A., Söderberg B. O., Eklund H., Brändén C. I. Three-dimensional structure of Escherichia coli thioredoxin-S2 to 2.8 A resolution. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2305–2309. doi: 10.1073/pnas.72.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979 Oct 10;254(19):9627–9632. [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin. 6. The amino acid sequence of the protein from escherichia coli B. Eur J Biochem. 1968 Dec 5;6(4):475–484. doi: 10.1111/j.1432-1033.1968.tb00470.x. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Tryptophan fluorescence study of conformational transitions of the oxidized and reduced form of thioredoxin. J Biol Chem. 1972 Apr 10;247(7):1992–1998. [PubMed] [Google Scholar]
- Hög J. O., Jörnvall H., Holmgren A., Carlquist M., Persson M. The primary structure of Escherichia coli glutaredoxin. Distant homology with thioredoxins in a superfamily of small proteins with a redox-active cystine disulfide/cysteine dithiol. Eur J Biochem. 1983 Oct 17;136(1):223–232. doi: 10.1111/j.1432-1033.1983.tb07730.x. [DOI] [PubMed] [Google Scholar]
- Ladenstein R., Epp O., Bartels K., Jones A., Huber R., Wendel A. Structure analysis and molecular model of the selenoenzyme glutathione peroxidase at 2.8 A resolution. J Mol Biol. 1979 Oct 25;134(2):199–218. doi: 10.1016/0022-2836(79)90032-9. [DOI] [PubMed] [Google Scholar]
- Mark D. F., Richardson C. C. Escherichia coli thioredoxin: a subunit of bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1976 Mar;73(3):780–784. doi: 10.1073/pnas.73.3.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng M., Hogenkamp H. P. Purification, characterization, and amino acid sequence of thioredoxin from Corynebacterium nephridii. J Biol Chem. 1981 Sep 10;256(17):9174–9182. [PubMed] [Google Scholar]
- Ohlsson I., Nordström B., Brändén C. I. Structural and functional similarities within the coenzyme binding domains of dehydrogenases. J Mol Biol. 1974 Oct 25;89(2):339–354. doi: 10.1016/0022-2836(74)90523-3. [DOI] [PubMed] [Google Scholar]
- Sjöberg B. M., Holmgren A. Studies on the structure of T4 thioredoxin. II. Amino acid sequence of the protein and comparison with thioredoxin from Escherichia coli. J Biol Chem. 1972 Dec 25;247(24):8063–8068. [PubMed] [Google Scholar]
- Slabý I., Holmgren A. Structure and enzymatic functions of thioredoxin refolded by complementation of two tryptic peptide fragments. Biochemistry. 1979 Dec 11;18(25):5584–5591. doi: 10.1021/bi00592a010. [DOI] [PubMed] [Google Scholar]
- Stryer L., Holmgren A., Reichard P. Thioredoxin. A localized conformational change accompanying reduction of the protein to the sulfhydryl form. Biochemistry. 1967 Apr;6(4):1016–1020. doi: 10.1021/bi00856a009. [DOI] [PubMed] [Google Scholar]
- Söderberg B. O., Sjöberg B. M., Sonnerstam U., Brändén C. I. Three-dimensional structure of thioredoxin induced by bacteriophage T4. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5827–5830. doi: 10.1073/pnas.75.12.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander L. Reaction mechanism of ribonucleoside diphosphate reductase from Escherichia coli. Oxidation-reduction-active disulfides in the B1 subunit. J Biol Chem. 1974 Aug 10;249(15):4858–4862. [PubMed] [Google Scholar]
- Thelander L., Reichard P. Reduction of ribonucleotides. Annu Rev Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- Tsugita A., Maeda K., Schürmann P. Spinach chloroplast thioredoxins in evolutionary drift. Biochem Biophys Res Commun. 1983 Aug 30;115(1):1–7. doi: 10.1016/0006-291x(83)90960-9. [DOI] [PubMed] [Google Scholar]


