Abstract
The focus of this study was to determine which chemokine receptors are present on oral fibroblasts and whether these receptors influence proliferation, migration, and/or the release of wound healing mediators. This information may provide insight into the superior wound healing characteristics of the oral mucosa. The gingiva fibroblasts expressed 12 different chemokine receptors (CCR3, CCR4, CCR6, CCR9, CCR10, CXCR1, CXCR2, CXCR4, CXCR5, CXCR7, CX3CR1, and XCR1), as analyzed by flow cytometry. Fourteen corresponding chemokines (CCL5, CCL15, CCL20, CCL22, CCL25, CCL27, CCL28, CXCL1, CXCL8, CXCL11, CXCL12, CXCL13, CX3CL1, and XCL1) were used to study the activation of these receptors on gingiva fibroblasts. Twelve of these fourteen chemokines stimulated gingiva fibroblast migration (all except for CXCL8 and CXCL12). Five of the chemokines stimulated proliferation (CCL5/CCR3, CCL15/CCR3, CCL22/CCR4, CCL28/CCR3/CCR10, and XCL1/XCR1). Furthermore, CCL28/CCR3/CCR10 and CCL22/CCR4 stimulation increased IL‐6 secretion and CCL28/CCR3/CCR10 together with CCL27/CCR10 upregulated HGF secretion. Moreover, TIMP‐1 secretion was reduced by CCL15/CCR3. In conclusion, this in‐vitro study identifies chemokine receptor‐ligand pairs which may be used in future targeted wound healing strategies. In particular, we identified the chemokine receptors CCR3 and CCR4, and the mucosa specific chemokine CCL28, as having an predominant role in oral wound healing by increasing human gingiva fibroblast proliferation, migration, and the secretion of IL‐6 and HGF and reducing the secretion of TIMP‐1.
Keywords: chemokine, cytokine, gingiva, migration, proliferation
Abbreviations
- ACKR
atypical chemokine receptor
- ECM
extracellular matrix
- ELISA
enzyme‐linked immunosorbent assay
- HGF
hepatic growth factor
- hTERT
human telomerase reverse transcriptase
- IL
interleukin
- MEC
mucosae‐associated epithelial chemokine
- TIMP‐1
tissue‐inhibitor‐of‐metalloproteinase‐1
1. INTRODUCTION
Fibroblasts are the principal cell type present within connective tissues (Sriram, Bigliardi, & Bigliardi‐Qi, 2015). Since their primary functions are to maintain the extracellular matrix (ECM) and to promote an inflammatory response (Häkkinen, Larjava, & Fournier, 2014; Kendall & Feghali‐Bostwick, 2014), fibroblasts can be considered to be key players in the wound healing process. Interestingly, the oral mucosa shows faster wound healing and significantly reduced scar formation compared to skin (Shannon, McKeown, Lundy, & Irwin, 2006). The superior wound healing characteristics of the oral mucosa may in part be attributed to the oral fibroblasts. It has already been shown in vitro that oral fibroblasts proliferate and migrate more than skin fibroblasts (Boink et al., 2016; Häkkinen et al., 2014). Research into the mechanisms which regulate oral fibroblast proliferation and migration and their inflammatory response to trauma is therefore of interest for future wound healing and regenerative therapies.
Chemokines are key players which regulate the process of wound healing (Balaji et al., 2015; Ding & Tredget, 2014; Rees, Greaves, Baguneid, & Bayat, 2015). These small chemotactic cytokines have been shown to be responsible for directional migration of cells, increasing proliferation, and modulating inflammatory cytokine release (Rees et al., 2015; van den Broek et al., 2014). Chemokines are classified according to their protein structure (CC, CXC, XC, or CX3C), followed by L for a ligand or R for a receptor, and then by a number (Bachelerie et al., 2014). Some ligands are able to interact with only one specific receptor, while others can bind to multiple receptors (e.g., CXCL8 can bind to CXCR1 and CXCR2). A single receptor may also be able to interact with multiple ligands. In humans there are 18 known chemokine receptors and at least four atypical chemokine receptors (ACKR) (Bachelerie et al., 2014). The atypical chemokine receptors act as chemokine scavengers and do not induce migration as the typical chemokine receptors do. Of note CXCR7, also known as ACKR3, is a scavenger of CXCL11 and CXCL12, but has also been shown to be involved in cell migration (Bachelerie et al., 2014). A chemokine of particular interest is CCL28 (receptor CCR3 and CCR10), also named mucosae‐associated epithelial chemokine (MEC), because of its association with mucosal inflammation (Hieshima et al., 2003; Kosten, Buskermolen, Spiekstra, de Gruijl, & Gibbs, 2015; Xiong, Fu, Hu, Xia, & Yang, 2012). Its counterpart CCL27, which is the only other ligand able to bind to CCR10, is in contrast predominantly expressed in skin (Homey et al., 2002; Xiong et al., 2012). It is of interest to determine how chemokine receptor‐ligand pairs stimulate oral fibroblasts during wound healing as this will provide valuable information on the superior wound healing characteristics of oral mucosa.
The aim of this study was to identify which chemokine receptors promote oral fibroblast proliferation, migration, and secretion of wound healing mediators in order to identify targets for wound healing strategies. Expression of all the 18 known human chemokine receptors and the ACKR CXCR7 was determined by flow cytometry in both primary and hTERT‐immortalized human gingiva fibroblasts. Since the receptor expression of hTERT‐immortalized gingiva fibroblasts was identical to their primary counterpart, further extended experiments were only performed with the immortalized fibroblasts. Proliferation over 3 days was investigated using a DNA quantification assay and migration was investigated using a wound healing scratch assay. Secretion of wound healing mediators which are known to influence inflammation during wound healing (IL‐6 and CXCL8), scar formation (Hepatic Growth Factor [HGF], and tissue remodeling (tissue‐inhibitor‐of‐metalloproteinase‐1 [TIMP‐1]) were investigated by ELISA (Jackson, Nesti, & Tuan, 2012; Liechty, Adzick, & Crombleholme, 2000; Morandini et al., 2012; Stephens et al., 2001).
2. MATERIALS AND METHODS
2.1. Cell culture of primary and hTERT immortalized human gingiva fibroblasts
Healthy human gingiva tissue was used in an anonymous fashion in accordance with the “Human Tissue and Medical Research: Code of conduct for responsible use” as formulated by the Federation of Dutch Medical Scientific Societies (www.federa.org). Primary human gingiva fibroblasts were isolated as previously described (Buskermolen et al., 2016). In short, after enzymatic separation of the epithelium and lamina propria, the lamina propria was digested in collagenase type II (Gibco, Grand Island, NY) for 3 hr. After passing the tissue suspension through a 40 μm cell strainer (Corning, Oneonta, NY) and washing with PBS, the cells were cultured in fibroblast medium consisting of DMEM (Gibco), supplemented with 5% Fetal Clone III (GE, Logan, UT) and 1% penicillin‐streptomycin (Gibco). Fibroblasts were used between passage 2 and 4. The hTERT‐immortalized human gingiva fibroblasts (T0026, purchased from ABM, Richmond, BC, Canada) were also cultured in fibroblast medium and used until passage 30.
2.2. Flow cytometry
Gingiva fibroblasts and hTERT‐immortalized gingiva fibroblasts were examined for cell‐surface expression of chemokine receptors. In short, after trypsinization, the fibroblasts were washed in PBS containing 0.1% bovine serum albumin and 0.1% sodium azide, incubated with chemokine receptor antibodies, or isotype controls (BD Biosciences, San Jose, CA or R&D Systems, Minneapolis, MN) (Table 1) for 1 hr and re‐suspended in the same solution for flow cytometric analysis (FACScalibur flow cytometer, Beckton Dickinson, San Jose, CA). The data was analyzed using CellQuestPro software. Three different fibroblast donors and three different immortalized fibroblast passages (p = 5, 16, and 18) were analyzed.
Table 1.
Antibodies used for flow cytometry
| Receptor | Product number, supplier | Isotype |
|---|---|---|
| CCR1 | FAB 145, R&D Systems | Mouse, IgG2b |
| CCR2 | FAB 151, R&D Systems | Mouse, IgG2b |
| CCR3 | 558165, BD Biosciences | Mouse, IgG2b |
| CCR4 | 551120, BD Biosciences | Mouse, IgG1 |
| CCR5 | 555993, BD Biosciences | Mouse, IgG2a |
| CCR6 | 559562, BD Biosciences | Mouse, IgG1 |
| CCR7 | 552176, BD Biosciences | Rat, IgG2a |
| CCR8 | FAB 142, R&D Systems | Rat, IgG2b |
| CCR9 | 561607, BD Biosciences | Mouse, IgG2a |
| CCR10 | FAB 3478, R&D Systems | Rat, IgG2a |
| CXCR1 | 555940, BD Biosciences | Mouse, IgG2b |
| CXCR2 | 555933, BD Biosciences | Mouse, IgG1 |
| CXCR3 | 557185, BD Biosciences | Mouse, IgG1 |
| CXCR4 | FAB 170, R&D Systems | Mouse, IgG2a |
| CXCR5 | 558112, BD Biosciences | Rat, IgG2b |
| CXCR6 | FAB 699, R&D Systems | Mouse, IgG2b |
| CXCR7 | FAB 42271, R&D Systems | Mouse, IgG2a |
| CX3CR1 | FAB 5204, R&D Systems | Mouse, IgG1 |
| XCR1 | FAB 857P, R&D Systems | Goat, IgG |
2.3. Chemokine mediated proliferation
For determining the influence of different chemokines on proliferation, hTERT‐immortalized gingiva fibroblasts were seeded into a 48 well‐plate at 5 × 103 cells per well. The cells were cultured in DMEM supplemented with 0.5% Fetal Clone III and 1% penicillin‐streptomycin and exposed to serial dilutions (0, 31, 63, 125, and 250 ng/ml) of 14 different chemokines (Table 2) for 3 days. All chemokines were purchased from PeproTech, Rocky Hill, NJ. After 3 days of culture, cells were harvested and DNA content per well was determined using a DNA quantification kit (CyQUANT, Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions. For quantification of cell number, a serial dilution of a known amount of fibroblasts was used. Four individual experiments were performed, each with intra‐experiment duplicates. Cell numbers were calculated relative to the unexposed controls for each experiment.
Table 2.
Overview of chemokine receptor‐ligand interactions on gingiva fibroblasts
| Receptor | Alias | Expressed | Ligand | Alias | Proliferation | Migration | IL‐6 | HGF | TIMP‐1 |
|---|---|---|---|---|---|---|---|---|---|
| CCR1 | CD191 | No | |||||||
| CCR2 | CD192 | No | |||||||
| CCR3 | CD193 | Yes | CCL5 CCL15 | RANTES | + | + | − | − | − |
| HCC‐2 | + | + | − | − | ‐‐ | ||||
| CCL28 | MEC | + | + | + | + | − | |||
| CCR4 | CD194 | Yes | CCL22 | MDC | + | + | + | − | − |
| CCR5 | CD195 | No | |||||||
| CCR6 | CD196 | Yes | CCL20 | MIP‐3α | − | + | − | − | − |
| CCR7 | CD197 | No | |||||||
| CCR8 | CDw198 | No | |||||||
| CCR9 | CDw199 | Yes | CCL25 | TECK | − | + | − | − | − |
| CCR10 | Yes | CCL27 CCL28 | CTACK | − | + | − | + | − | |
| MEC | + | + | + | + | − | ||||
| CXCR1 | CD181 | Yes | CXCL8 | IL‐8 | − | − | − | − | − |
| CXCR2 | CD182 | Yes | CXCL1 | GROα | − | + | − | − | − |
| CXCL8 | IL‐8 | − | − | − | − | − | |||
| CXCR3 | CD183 | No | |||||||
| CXCR4 | CD184 | Yes | CXCL12 | SDF‐1α | − | − | − | − | − |
| CXCR5 | CD185 | Yes | CXCL13 | BLC | − | + | − | − | − |
| CXCR6 | CD186 | No | |||||||
| CXCR7 | ACKR3 | Yes | CXCL11 | I‐TAC | − | + | − | − | − |
| CXCL12 | SDF‐1α | − | − | − | − | − | |||
| CX3CR1 | Yes | CX3CL1 | Fractalkine | − | + | + | − | − | |
| XCR1 | No | XCL1 | Lymphotactin α | + | + | − | − | − |
Chemokine receptors that were expressed on gingiva fibroblasts were stimulated with corresponding chemokine ligands to study proliferation, migration, and secretion of wound healing mediators. +indicates an increase compared to control, −indicates no difference from control, and ‐‐indicates a decrease compared to control.
2.4. Chemokine mediated migration
Migration of hTERT‐immortalized gingiva fibroblasts under influence of chemokines was studied using a wound healing scratch assay (Monsuur et al., 2016). In short: the fibroblasts were cultured in fibroblast‐medium (section 2.1) in 48 well‐plates until 100% confluency was reached. Subsequently, a scratch was made across the middle of the well with a 1 ml pipet tip. After washing with PBS, DMEM was added supplemented with 0,1% BSA and 1% penicillin‐streptomycin and serial dilutions (0, 31, 63, 125, and 250 ng/ml) of 14 different chemokines (PeproTech) (Table 2). The experiments were performed in triplicate with an intra‐experiment duplicate of each condition. Unexposed controls and positive controls (1 ng/ml EGF) were performed in quadruplicate within an experiment. Phase contrast micrographs were made at day 0 and 4 days after exposure. At the end of the experiment the culture supernatant were collected for ELISA (section 2.5). Surface area of the scratch before and after cell migration was measured using an image processing algorithm that has been described in detail in previous work (Monsuur et al., 2016; Topman, Sharabani‐Yosef, & Gefen, 2012). Reduction of the scratch surface area was calculated relative to the unexposed controls for each experiment.
2.5. Chemokine induced secretion of wound healing mediators
The culture supernatant collected after exposure of fibroblasts to 125 ng/ml of chemokines at the end of the migration experiments was used to analyze the secretion of IL‐6, CXCL8, HGF, and TIMP‐1. This concentration was chosen because of the general positive effect this chemokine concentration had on proliferation and migration. ELISA's were performed in accordance with the manufacturer's specifications (CXCL8: Sanquin, Amsterdam, The Netherlands; IL‐6, HGF, and TIMP‐1: R&D) as previously described (Spiekstra et al., 2005).
2.6. Statistics
Statistical analysis was performed with the aid of GraphPad Prism, version 6 (GraphPad Software, Inc.). Flow cytometric data of primary and TERT‐immortalized gingiva fibroblasts were compared with multiple t‐test corrected with Holm–Sidak for multiple comparisons. All other data were analyzed with the Kruskal–Wallis test followed by Dunn's multiple comparison test. Differences were considered significant when p < 0.05. Data are represented as mean ± standard error of mean (SEM); *p < 0.05; **p < 0.01; ***p < 0.001.
3. RESULTS
3.1. Chemokine receptors expressed on gingiva fibroblasts
Of the 19 chemokine receptors studied, 12 were clearly expressed on the surface of gingiva fibroblasts. CCR3, CCR4, CCR6, CCR9, CCR10, CXCR1, CXCR4, CXCR5, and CXCR7 were expressed on over 90% of the gingiva fibroblasts and CXCR2, CX3CR1, and XCR1 were expressed on over 50% of the gingiva fibroblasts (Figure 1). In contrast, CCR1, CCR2, CCR5, CCR7, CCR8, and CXCR3 were on average found on less than 5% of the gingiva fibroblasts and CXCR6 on less than 15% of the gingiva fibroblasts. Since the isotype controls for these low expressed receptors overlapped with the specific antibodies we can conclude that these receptors are most likely not expressed on gingiva fibroblasts. On the histograms a single peak was observed, indicating a homogeneous receptor expression within the entire fibroblast population. There were no differences between the primary and hTERT‐immortalized gingiva fibroblasts in number of positive cells or mean fluorescent intensity. Therefore, all following experiments were performed with the hTERT‐immortalized cells. Only the receptors that were found to be expressed on the surface of gingiva fibroblasts were further investigated.
Figure 1.
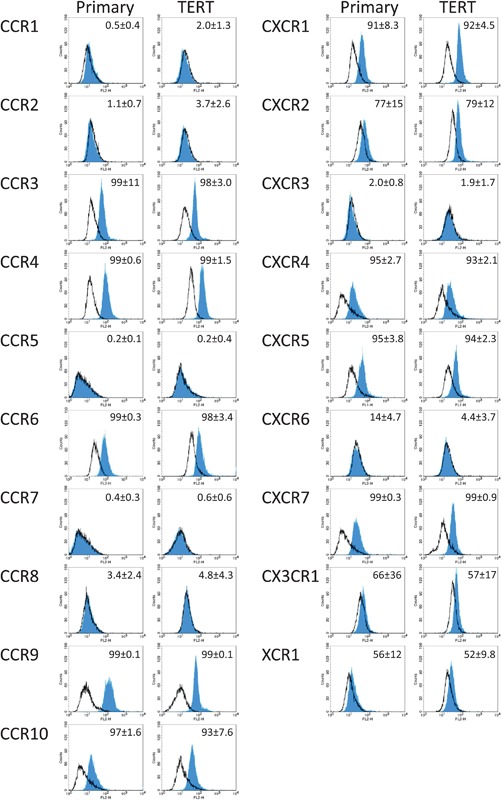
Chemokine receptor expression on gingiva fibroblasts. The expression of chemokine receptors on primary (left column) and hTERT‐immortalized (right column) human gingiva fibroblasts was analyzed by flow cytometry. Open lines represent isotype controls and closed lines the chemokine receptor specific antibody. Mean of percent positive cells and standard deviation of three individual experiments are shown in the upper right corner of each histogram. A total of 12 of the 19 chemokine receptors were expressed
3.2. Chemokine receptor‐ligand pairs regulating proliferation and migration of gingiva fibroblasts
Ligands were chosen to investigate the influence of chemokine receptor activation on gingiva fibroblast migration and proliferation. Where possible a ligand was chosen that only bound to one of the expressed receptors (Table 2). However, all ligands that interact with CXCR1 also interact with CXCR2. In contrast, CXCR2 does have monospecific ligands. Therefore, CXCL8 was chosen to investigate the co‐activation of CXCR1 and CXCR2, and CXCL1 was used to activate CXCR2 alone. Furthermore, CXCL12 is the only ligand that interacts with CXCR4, but it also interacts with CXCR7. Therefore, CXCL12 was used to investigate the co‐activation of CXCR4 and CXCR7, while CXCL11 was used to investigate the activation of CXCR7 alone. Finally, although CCL28 interacts with both CCR3 and CCR10, this ligand was specifically investigated because of its association with mucosa.
First, the influence of chemokine receptor activation on proliferation of gingiva fibroblasts was investigated (Figure 2 and Table 2). Of the 14 chemokines tested in a dose response only five receptor‐ligand pairs increased gingiva fibroblast proliferation (CCL5/CCR3, CCL15/CCR3, CCL22/CCR4, CCL28/CCR3/CCR10, and XCL1/XCR1). A concentration of 125 ng/ml of CCL5, CCL22, and CCL28 was most potent in increasing proliferation. For CCL15 a trend was seen at 125 ng/ml and a significant increase in proliferation at 250 ng/ml. For XCL1 an overall increase in proliferation was observed. The remaining chemokines did not induce a proliferative response in the gingiva fibroblasts (CCL20/CCR6, CCL25/CCR9, CCL27/CCR10, CXCL1/CXCR2, CXCL8/CXCR1/CXCR2, CXCL11/CXCR7, CXCL12/CXCR4/CXCR7, CXCL13/CXCR5, and CXC3CL1/CX3CR1).
Figure 2.
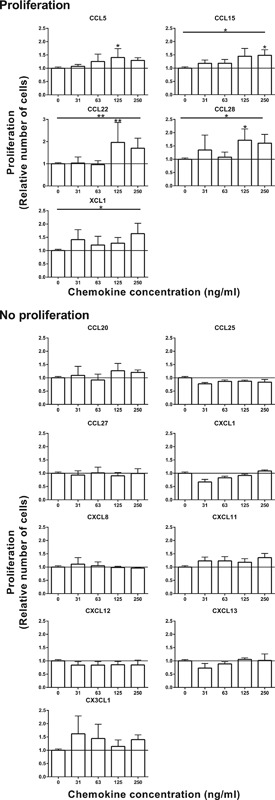
Proliferation of gingiva fibroblasts stimulated by chemokines. Proliferation of gingiva fibroblasts over 3 days of stimulation with chemokines was assessed by DNA quantification. Four individual proliferation experiments in duplicate are expressed as mean ± SEM relative to unstimulated controls. A total of 5 of the 14 studied chemokines increased proliferation of gingiva fibroblast. *p < 0.05; **p < 0.01; Kruskal–Wallis test followed by Dunn's multiple comparison test
Next the migration of gingiva fibroblasts was investigated in a wound healing scratch assay (Figure 3 and Table 2). A total of 12 of the 14 chemokines significantly increased gingiva fibroblast migration. CCL20 caused a significantly increase of migration from a concentration of 31 ng/ml. CCL5, CCL20, CCL22, CCL25, CCL27, CCL28, CXCL1, CXCL13, and CX3CL1 significantly increased gingiva fibroblast migration from a concentration of 125 ng/ml. A strong trend was also visible at this concentration for CXCL11 and CCL15. CXCL11 and CX3CL1 significantly increased migration at 63 ng/ml whereas a concentration of 250 ng/ml was required for CCL15. The differences found relating to chemokine concentration and maximum effect are most likely due to the in vitro assay set up (proliferation and migration assay) and the biological activity of the different recombinant proteins. The two chemokine‐ligand pairs that did not significantly affect migration were CXCL8/CXCR1/CXCR2 and CXCL12/CXCR4.
Figure 3.
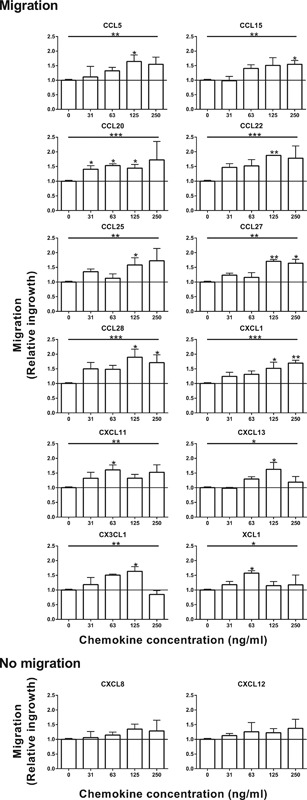
Migration of gingiva fibroblasts stimulated by chemokines. Migration of gingiva fibroblasts over 4 days of stimulation with chemokines was studied using a wound healing scratch assay. Fibroblast migration into the scratch of three individual experiments in duplicate are expressed as mean ± SEM relative to unstimulated controls. A total of 12 of the 14 studied chemokines increased migration of gingiva fibroblasts. *p < 0.05; **p < 0.01; ***p < 0.001; Kruskal–Wallis test followed by Dunn's multiple comparison test
3.3. Chemokine receptor‐ligand pairs stimulating wound healing mediator release
Next it was determined whether the chemokines could stimulate wound healing mediator release from gingiva fibroblasts. Fibroblasts were exposed to a chemokine concentration of 125 ng/ml, because our proliferation and migration experiments had already shown that this was a general biologically active chemokine concentration. The secretion of the wound healing mediators IL‐6, CXCL8, HGF, and TIMP‐1 by the gingiva fibroblasts was influenced by different chemokine receptor‐ligand pairs (Figure 4). More IL‐6 was secreted in the culture supernatant of gingiva fibroblasts exposed to CCL22 (CCR4), CCL28 (CCR3 and CCR10), or CX3CL1 (CX3CR1) than in unexposed gingiva fibroblasts. CXCL8 was not significantly upregulated in any of the conditions (data not shown). HGF secretion was increased by CCL27 and CCL28. Both these chemokines are able to bind to CCR10. CCL28 is also able to bind CCR3. TIMP‐1 secretion by gingiva fibroblasts was reduced by the chemokine CCL15, which binds CCR3.
Figure 4.
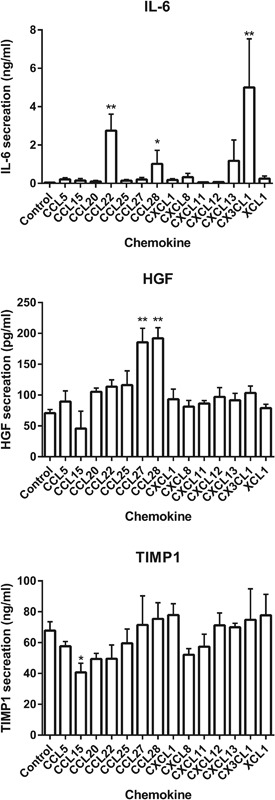
Wound healing mediator release of gingiva fibroblasts stimulated by chemokines. Cumulative secretion of IL‐6, HGF, and TIMP‐1 by gingiva fibroblasts over 4 days of stimulation with 125 ng/ml of 14 different chemokines in a scratch assay. Data represent mean ± SEM of three individual experiments in duplicate. IL‐6 secretion was increased by CCL22, CCL28, and CX3CL1. HGF secretion was increased by CCL27 and CCL28. TIMP‐1 secretion was decreased by CCL15. *p < 0.05; **p < 0.01; ***p < 0.001; Dunn's multiple comparison test against unstimulated controls
4. DISCUSSION
Despite the clear association of chemokines with wound healing, scar formation and their potential as therapeutic targets, surprisingly few studies describe chemokine receptor expression and function on oral mucosa fibroblasts (Rees et al., 2015). Our study provides an extensive overview of chemokine receptor expression on gingiva fibroblasts and describes which ligands stimulate in vitro wound healing (see summary Table 2 and Figure 5). The specific chemokine concentrations which affected gingiva fibroblast proliferation and migration in this study reflect the in vitro assay set up and the biological activity of the different recombinant proteins and are not indicative of physiologically relevant endogenous chemokine concentrations. In line with our findings, CCR6 and CXCR4 expression on gingiva fibroblasts has been reported (Hosokawa, Hosokawa, Ozaki, Nakae, & Matsuo, 2005, Hosokawa, Hosokawa, Ozaki, Nakae, Murakami, et al., 2005; Sun, Nemoto, Hong, & Sasaki, 2016). Also in line with us CXCR6 expression was reported as absent on gingiva fibroblasts and in gingiva tissue derived from clinically healthy patients (Hosokawa, Hosokawa, Ozaki, Nakae, & Matsuo, 2007). However, in a later study the same authors did detect CXCR6 expression and found that its ligand CXCL16 could induce gingiva fibroblast proliferation (Hosokawa, Hosokawa, Ozaki, Nakae, & Matsuo, 2009). These contradictory results may possibly be explained by different culture conditions and the inflammatory state of the gingiva tissue in the different studies. We found no other results in the literature describing gingiva fibroblast chemokine receptor expression. Here, we show that the receptors CCR3, CCR4, CCR6, CCR10, CXCR1, CXCR2, and CXCR4, which we previously found to be expressed on skin fibroblasts were are also expressed on gingiva fibroblasts (Kroeze et al., 2009). This indicates that these receptors have a general role in mediating a fibroblast wound healing response that is not restricted to oral fibroblasts. The receptors CCR9, CXCR5, CXCR6, CXCR7, CX3CR1, and XCR1 were not analyzed in our previous skin fibroblast study, nor could we find any other studies that analyzed the expression of these receptors on human skin fibroblasts.
Figure 5.
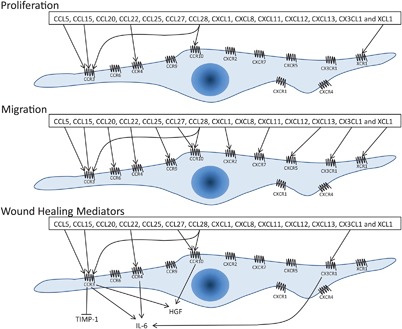
Overview of chemokine receptors and ligand function on gingiva fibroblasts. Expressed chemokine receptors on gingiva fibroblasts are depicted on the cell membrane. Arrows represent an positive effect and the blunt arrow represents a negative effect of chemokines on the proliferation (top), migration (middle), or secretion of wound healing mediators (bottom) by gingiva fibroblasts
We found that the chemokine receptors CCR3 and CCR4 are potent stimulators of in vitro gingiva fibroblast wound healing. In addition to stimulating proliferation and migration, CCR3 activation of gingiva fibroblasts with CCL15 reduced TIMP‐1 secretion. The reduction of TIMP‐1 increases ECM turnover in wound healing, possibly increasing fibroblast movement into a wound site and reducing scar formation (Stephens et al., 2001). IL‐6 secretion was increased by stimulation with CCL28, which binds to both CCR3 and CCR10. Therefore, it is not possible to say which of the two receptors was responsible for the increased IL‐6 secretion and thereby potentially stimulating the inflammatory response, as IL‐6 has been described to stimulate the inflammatory response in wound healing (Liechty et al., 2000). Together these results show that CCR3 clearly has a major role in the oral wound healing process. CCR4 activation of gingiva fibroblasts also stimulated proliferation and migration and increased secretion of the pro‐inflammatory cytokine IL‐6. It has been found that CCL17, a monospecific ligand to CCR4, is upregulated in gingiva inflammation (Hosokawa, Hosokawa, Ozaki, Nakae, & Matsuo, 2008). In mice it has been found that CCL17 accelerated the early stage of wound closure (Kato et al., 2011). These findings together indicate the potent role of CCR4 in the oral wound healing. Among the cytokines that gingiva fibroblasts upregulate when they are in a pro‐inflammatory environment are the chemokines CCL2 and CCL5 (Buskermolen et al., 2016; Kosten et al., 2015; Verardi, Page, Ammons, & Bordin, 2007). These chemokines bind to CCR4 and CCR3, respectively. Here, we found that this in turn will cause gingiva fibroblasts to proliferate, migrate, and secrete inflammatory cytokines, thus potentially initiating an autologous feedback loop that stimulates the wound healing environment. CCR3 and CCR4 activation has also been found to stimulate the migration of skin fibroblasts (Gaspar et al., 2013; Kroeze et al., 2009). Taken together these results show that CCR3 and CCR4 may be potent targets for wound healing therapeutics.
We found that CX3CL1/CX3CR1 stimulated migration and IL‐6 secretion of gingiva fibroblasts. This chemokine receptor‐ligand pair has been shown to play an important role in skin inflammation (Sugaya, 2015). Furthermore, in CX3CR1 knock‐out mice skin wound closure was slower and the presence of myofibroblasts was reduced (Ishida, Gao, & Murphy, 2008). These results indicate that CX3CL1/CX3CR1 plays a role in oral as well as in skin wound healing.
For CCL28 it cannot be concluded whether it stimulates proliferation and migration via CCR3 or CCR10. However, since the other chemokines that bind CCR3 did increase proliferation while CCL27 (which specifically binds CCR10) did not, it is likely that the proliferative effect of CCL28 was via the CCR3 receptor. In contrast, CCL27 and CCL28 were the only chemokines found to stimulate HGF secretion, indicating an effect via CCR10. HGF has been described to have anti‐fibrotic properties and to promote regeneration, thus reducing scarring after injury (Jackson et al., 2012; Ono et al., 2004). HGF is secreted in higher amounts by oral fibroblasts than by skin fibroblasts which, together with its anti‐fibrotic properties, suggest that it may be involved in the superior wound healing observed in oral mucosa compared to skin (Okazaki, Yoshimura, Uchida, & Harii, 2002; Shannon et al., 2006). Since we found that CCR10 stimulation increased HGF production and it has been shown that CCR10 stimulation on keratinocytes increased wound healing in vitro and in animal studies, this receptor could be a target for wound healing therapies (Inokuma et al., 2006; van den Broek et al., 2014). CCL28 was the only chemokine acting on CCR3 and/or CCR10 that increased the secretion of the pro‐inflammatory cytokine IL‐6. Taken together, the effect of the mucosa associated chemokine CCL28 on gingiva fibroblast proliferation, migration, and cytokine secretion suggests an important role in oral wound healing.
Although the appropriate receptors for CXCL8 and CXCL12 are expressed on gingiva fibroblasts, these chemokines did not affect proliferation, migration, or the secretion of wound healing mediators. Both CXCL8 and CXCL12 have been shown to influence skin fibroblast migration and CXCL12 is even thought to be a major factor in stem cell homing to sites of injury (Akazawa et al., 2015; Stuermer et al., 2015). For CXCL8, our results may possibly be explained by the fact that gingiva fibroblasts secrete large quantities of endogenous CXCL8 which would mask any effect of the recombinant CXCL8 used in this study (Buskermolen et al., 2016; Kosten et al., 2015). This is in line with reports describing CXCL8 being highly expressed in oral inflammation and attracting infiltrating immune cells into the connective tissue (Ertugrul, Sahin, Dikilitas, Alpaslan, & Bozoglan, 2013). The unresponsiveness of gingiva fibroblasts to CXCL12 is in line with previous work, where we have found that in contrast to skin, gingiva only secretes very low amounts of CXCL12 and whereas the CXCL12/CXCR4 axis is pivotal in skin Langerhans Cell migration, this chemokine receptor pair does not appear to play such a role in the gingiva (Kroeze, Jurgens, Doulabi, van Milligen, Scheper, & Gibbs 2015; Kosten, Spiekstra, de Gruijl, & Gibbs, 2016). Others have also shown that, in contrast to Il‐6 and CXCL8, the chemokine CXCL12 in not upregulated after TLR2 stimulation of gingiva fibroblasts (Morandini et al., 2012). It is however possible that CXCR7 scavenges CXCL12 thus down regulating the CXCR4 mediated wound healing response. However, this needs further investigation.
This study shows that the TERT‐immortalized human gingiva fibroblasts expressed the same receptors as the primary gingiva fibroblasts. Previously we (Buskermolen et al., 2016) incorporated these TERT‐immortalized fibroblasts into a gingiva equivalent model together with TERT‐immortalized gingiva keratinocytes. When full thickness wounds were introduced into the gingiva equivalent, the fibroblasts migrated into the wound region. The fibroblast migration into the wounded region of the gingiva equivalent together with the receptor expression correlation in this study, further validates this cell line and underlines its value for replacing primary cells in in vitro models.
In conclusion, this study identifies chemokine receptor‐ligand pairs which may be used in future targeted wound healing strategies. In particular, we identify the chemokine receptors CCR3 and CCR4, and the mucosa specific chemokine CCL28 (also named MEC), as having a predominant role in oral wound healing by increasing human gingiva fibroblast proliferation, migration, and the secretion of IL‐6 and HGF secretion, and reducing the secretion of TIMP‐1 in our in vitro models.
ACKNOWLEDGMENTS
The authors would like to thank the dentistry practices Uijlenbroek & Partners Tandartsen and Tandheelkunde & Implantologie Aemstelwijck for their help in obtaining patient material. The authors would also like to thank Albert Feilzer, Astrid Bakker, and Cees Kleverlaan for their guidance during the research.
CONFLICTS OF INTEREST
Susan Gibbs is co‐founder of the VU university spin off company (SME) A‐Skin BV.
Buskermolen JK, Roffel S, Gibbs S. Stimulation of oral fibroblast chemokine receptors identifies CCR3 and CCR4 as potential wound healing targets. J Cell Physiol. 2017;232: 2996–3005. https://doi.org/10.1002/jcp.25946
REFERENCES
- Akazawa, Y. , Hasegawa, T. , Yoshimura, Y. , Chosa, N. , Asakawa, T. , Ueda, K. , … Iwamoto, T. (2015). Recruitment of mesenchymal stem cells by stromal cell‐derived factor 1alpha in pulp cells from deciduous teeth. International Journal of Molecular Medicine, 36, 442–448. [DOI] [PubMed] [Google Scholar]
- Bachelerie, F. , Ben‐Baruch, A. , Burkhardt, A. M. , Combadiere, C. , Farber, J. M. , Graham, G. J. , … Zlotnik, A. (2014). International union of basic and clinical pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacological Reviews, 66, 1–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji, S. , Watson, C. L. , Ranjan, R. , King, A. , Bollyky, P. L. , & Keswani, S. G. (2015). Chemokine involvement in fetal and adult wound healing. Advances in Wound Care, 4, 660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boink, M. A. , Van Den Broek, L. J. , Roffel, S. , Nazmi, K. , Bolscher, J. G. M. , Gefen, A. , … Gibbs, S. (2016). Different wound healing properties of dermis, adipose, and gingiva mesenchymal stromal cells. Wound Repair and Regeneration, 24, 100–109. [DOI] [PubMed] [Google Scholar]
- Buskermolen, J. K. , Reijnders, C. M. A. , Spiekstra, S. W. , Steinberg, T. , Kleverlaan, C. J. , Feilzer, A. J. , … Gibbs, S. (2016). Development of a full thickness human gingiva equivalent constructed from immortalized keratinocytes and fibroblasts. Tissue Engineering Part C, 22, 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, J. , & Tredget, E. E. (2014). The role of chemokines in fibrotic wound healing. Advances in Wound Care 0, 48, 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertugrul, A. S. , Sahin, H. , Dikilitas, A. , Alpaslan, N. , & Bozoglan, A. (2013). Comparison of CCL28, interleukin‐8, interleukin‐1β and tumor necrosis factor‐alpha in subjects with gingivitis, chronic periodontitis and generalized aggressive periodontitis. Journal of Periodontal Research, 4, 673–686. [DOI] [PubMed] [Google Scholar]
- Gaspar, K. , Kukova, G. , Bunemann, E. , Buhren, B. A. , Sonkoly, E. , Szollosi, A. G. , … Homey, B. (2013). The chemokine receptor CCR3 participates in tissue remodeling during atopic skin inflammation. Journal of Dermatological Science, 71, 12–21. [DOI] [PubMed] [Google Scholar]
- Häkkinen, L. , Larjava, H. , & Fournier, B. P. J. (2014). Distinct phenotype and therapeutic potential of gingival fibroblasts. Cytotherapy, 16, 1171–1186. [DOI] [PubMed] [Google Scholar]
- Hieshima, K. , Ohtani, H. , Shibano, M. , Izawa, D. , Nakayama, T. , Kawasaki, Y. , … Yoshie, O. (2003). CCL28 has dual roles in mucosal immunity as a chemokine with broad‐spectrum antimicrobial activity. The Journal of Immunology, 170, 1452–1461. [DOI] [PubMed] [Google Scholar]
- Homey, B. , Alenius, H. , Müller, A. , Soto, H. , Bowman, E. P. , Yuan, W. , … Zlotnik, A. (2002). CCL27‐CCR10 interactions regulate T cell‐mediated skin inflammation. Nature Medicine, 8, 157–165. [DOI] [PubMed] [Google Scholar]
- Hosokawa, Y. , Hosokawa, I. , Ozaki, K. , Nakae, H. , & Matsuo, T. (2005). Increase of CCL20 expression by human gingival fibroblasts upon stimulation with cytokines and bacterial endotoxin. Clinical and Experimental Immunology, 142, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, Y. , Hosokawa, I. , Ozaki, K. , Nakae, H. , Murakami, K. , Miyake, Y. , & Matsuo, T. (2005). CXCL12 and CXCR4 expression by human gingival fibroblasts in periodontal disease. Clinical and Experimental Immunology, 141, 467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, Y. , Hosokawa, I. , Ozaki, K. , Nakae, H. , & Matsuo, T. (2007). CXC chemokine ligand 16 in periodontal diseases: Expression in diseased tissues and production by cytokine‐stimulated human gingival fibroblasts. Clinical and Experimental Immunology, 149, 146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, Y. , Hosokawa, I. , Ozaki, K. , Nakae, H. , & Matsuo, T. (2008). CC Chemokine ligand 17 in periodontal diseases: Expression in diseased tissues and production by human gingival fibroblasts. Journal of Periodontal Research, 43, 471–477. [DOI] [PubMed] [Google Scholar]
- Hosokawa, Y. , Hosokawa, I. , Ozaki, K. , Nakae, H. , & Matsuo, T. (2009). Human gingival fibroblasts express functional chemokine receptor CXCR6. Clinical and Experimental Immunology, 156, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuma, D. , Abe, R. , Fujita, Y. , Sasaki, M. , Shibaki, A. , Nakamura, H. , … Shimizu, H. (2006). CTACK/CCL27 accelerates skin regeneration via accumulation of bone marrow‐derived keratinocytes. Stem Cells, 24, 2810–2816. [DOI] [PubMed] [Google Scholar]
- Ishida, Y. , Gao, J. L. , & Murphy, P. M. (2008). Chemokine receptor CX3CR1 mediates skin wound healing by promoting macrophage and fibroblast accumulation and function. The Journal of Immunology 180, 569–579. [DOI] [PubMed] [Google Scholar]
- Jackson, W. M. , Nesti, L. J. , & Tuan, R. S. (2012). Mesenchymal stem cell therapy for attenuation of scar formation during wound healing. Stem Cell Research & Therapy, 3, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, T. , Saeki, H. , Tsunemi, Y. , Shibata, S. , Tamaki, K. , & Sato, S. (2011). Thymus and activation‐regulated chemokine (TARC)/CC chemokine ligand (CCL) 17 accelerates wound healing by enhancing fibroblast migration. Experimental Dermatology, 20, 669–674. [DOI] [PubMed] [Google Scholar]
- Kendall, R. T. , & Feghali‐Bostwick, C. A. (2014). Fibroblasts in fibrosis: Novel roles and mediators. Frontiers in Pharmacology, 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosten, I. J. , Buskermolen, J. K. , Spiekstra, S. W. , de Gruijl, T. D. , & Gibbs, S. (2015). Gingiva equivalents secrete negligible amounts of key chemokines involved in langerhans cell migration compared to skin equivalents. Journal of Immunology Research, 2015, 627125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosten, I. J. , Spiekstra, S. W. , de Gruijl, T. D. , & Gibbs, S. (2016). MUTZ‐3 Langerhans cell maturation and CXCL12 independent migration in reconstructed human gingiva. Alternativen zu Tierexperimenten, 33, 423–434. [DOI] [PubMed] [Google Scholar]
- Kroeze, K. L. , Jurgens, W. J. , Doulabi, B. Z. , van Milligen, F. J. , Scheper, R. J. , & Gibbs, S. (2009). Chemokine‐mediated migration of skin‐derived stem cells: Predominant role for CCL5/RANTES. Journal of Investigative Dermatology, 129, 1569–1581. [DOI] [PubMed] [Google Scholar]
- Liechty, K. W. , Adzick, N. S. , & Crombleholme, T. M. (2000). Diminished interleukin 6 (IL‐6) production during scarless human fetal wound repair. Cytokine, 12, 671–676. [DOI] [PubMed] [Google Scholar]
- Monsuur, H. N. , Boink, M. A. , Weijers, E. M. , Roffel, S. , Breetveld, M. , Gefen, A. , … Gibbs, S. (2016). Methods to study differences in cell mobility during skin wound healing in vitro. Journal of Biomechanics, 49, 1381–1387. [DOI] [PubMed] [Google Scholar]
- Morandini, A. C. F. , Chaves Souza, P. P. , Ramos‐Junior, E. S. , Brozoski, D. T. , Sipert, C. R. , Souza Costa, C. A. , & Santos, C. F. (2012). Toll‐Like receptor 2 knockdown modulates interleukin (IL)‐6 and IL‐8 but not stromal derived factor‐1 (SDF‐1/CXCL12) in human periodontal ligament and gingival fibroblasts. Journal of Periodontology, 1, 1–11. [DOI] [PubMed] [Google Scholar]
- Okazaki, M. , Yoshimura, K. , Uchida, G. , & Harii, K. (2002). Elevated expression of hepatocyte and keratinocyte growth factor in cultured buccal‐mucosa‐derived fibroblasts compared with normal‐skin‐derived fibroblasts. Journal of Dermatological Science, 30, 108–115. [DOI] [PubMed] [Google Scholar]
- Ono, I. , Yamashita, T. , Hida, T. , Jin, H. Y. , Ito, Y. , Hamada, H. , … Jimbow, K. (2004). Local administration of hepatocyte growth factor gene enhances the regeneration of dermis in acute incisional wounds. Journal of Surgical Research, 120, 47–55. [DOI] [PubMed] [Google Scholar]
- Rees, P. A. , Greaves, N. S. , Baguneid, M. , & Bayat, A. (2015). Chemokines in wound healing and as potential therapeutic targets for reducing cutaneous scarring. Advances in Wound Care, 4, 687–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, D. B. , McKeown, S. T. W. , Lundy, F. T. , & Irwin, C. R. (2006). Phenotypic differences between oral and skin fibroblasts in wound contraction and growth factor expression. Wound Repair and Regeneration, 14, 172–178. [DOI] [PubMed] [Google Scholar]
- Spiekstra, S. W. , Toebak, M. J. , Sampat‐Sardjoepersad, S. , van Beek, P. J. , Boorsma, D. M. , Stoof, T. J. , … Gibbs, S. (2005). Induction of cytokine (interleukin‐1alpha and tumor necrosis factor‐alpha) and chemokine (CCL20, CCL27, and CXCL8) alarm signals after allergen and irritant exposure. Experimental Dermatology, 14, 109–116. [DOI] [PubMed] [Google Scholar]
- Sriram, G. , Bigliardi, P. L. , & Bigliardi‐Qi, M. (2015). Fibroblast heterogeneity and its implications for engineering organotypic skin models in vitro. European Journal of Cell Biology, 94, 483–512. [DOI] [PubMed] [Google Scholar]
- Stephens, P. , Davies, K. J. , Occleston, N. , Pleass, R. D. , Kon, C. , Daniels, J. , … Thomas, D. W. (2001). Skin and oral fibroblasts exhibit phenotypic differences in extracellular matrix reorganization and matrix metalloproteinase activity. British Journal of Dermatology, 144, 229–237. [DOI] [PubMed] [Google Scholar]
- Stuermer, E. K. , Lipenksy, A. , Thamm, O. , Neugebauer, E. , Schaefer, N. , Fuchs, P. , … Koenen, P. (2015). The role of SDF‐1 in homing of human adipose‐derived stem cells. Wound Repair and Regeneration, 23, 82–89. [DOI] [PubMed] [Google Scholar]
- Sugaya, M. (2015). Chemokines and skin diseases. Archivum Immunologiae et Therapiae Experimentalis, 63, 109–115. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Nemoto, E. , Hong, G. , & Sasaki, K. (2016). Modulation of stromal cell‐derived factor 1 alpha (SDF‐1α) and its receptor CXCR4 in Porphyromonas gingivalis‐induced periodontal inflammation. BMC Oral Health, 17, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topman, G. , Sharabani‐Yosef, O. , & Gefen, A. (2012). A standardized objective method for continuously measuring the kinematics of cultures covering a mechanically damaged site. Medical Engineering and Physics, 34, 225–232. [DOI] [PubMed] [Google Scholar]
- van den Broek, L. J. , Kroeze, K. L. , Waaijman, T. , Breetveld, M. , Sampat‐Sardjoepersad, S. C. , Niessen, F. B. , … Gibbs, S. (2014). Differential response of human adipose tissue‐derived mesenchymal stem cells, dermal fibroblasts, and keratinocytes to burn wound exudates: Potential role of skin‐specific chemokine CCL27. Tissue Engineering Part A, 20A, 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verardi, S. , Page, R. C. , Ammons, W. F. , & Bordin, S. (2007). Differential chemokine response of fibroblast subtypes to complement C1q. Journal of Periodontal Research, 42, 62–68. [DOI] [PubMed] [Google Scholar]
- Xiong, N. , Fu, Y. , Hu, S. , Xia, M. , & Yang, J. (2012). CCR10 and its ligands in regulation of epithelial immunity and diseases. Protein Cell, 3, 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]


