Abstract
One single glucoamylase gene could be identified in the chromosomal DNA of Aspergillus niger by Southern blot analysis. This glucoamylase gene was isolated from a genomic library of A. niger DNA. The glucoamylase gene is situated on a 2.5-kb EcoRI-EcoRV fragment and contains five intervening sequences in the coding region. One 169-bp intron is involved in differential mRNA processing leading to the two different glucoamylase enzymes G1 and G2; the other four introns are all very small ranging from 55 to 75 bp in length. One intron has a significant homology to the coding region which immediately follows, and it contains the internal conserved sequence TACTAAC, which is also found in yeast chromosomal gene introns, and is thought to participate in mRNA splicing. Two transcription initiation sites and a typical eukaryotic promoter region with TATAAT and CAAT boxes are located upstream from the gene.
Full text
PDF

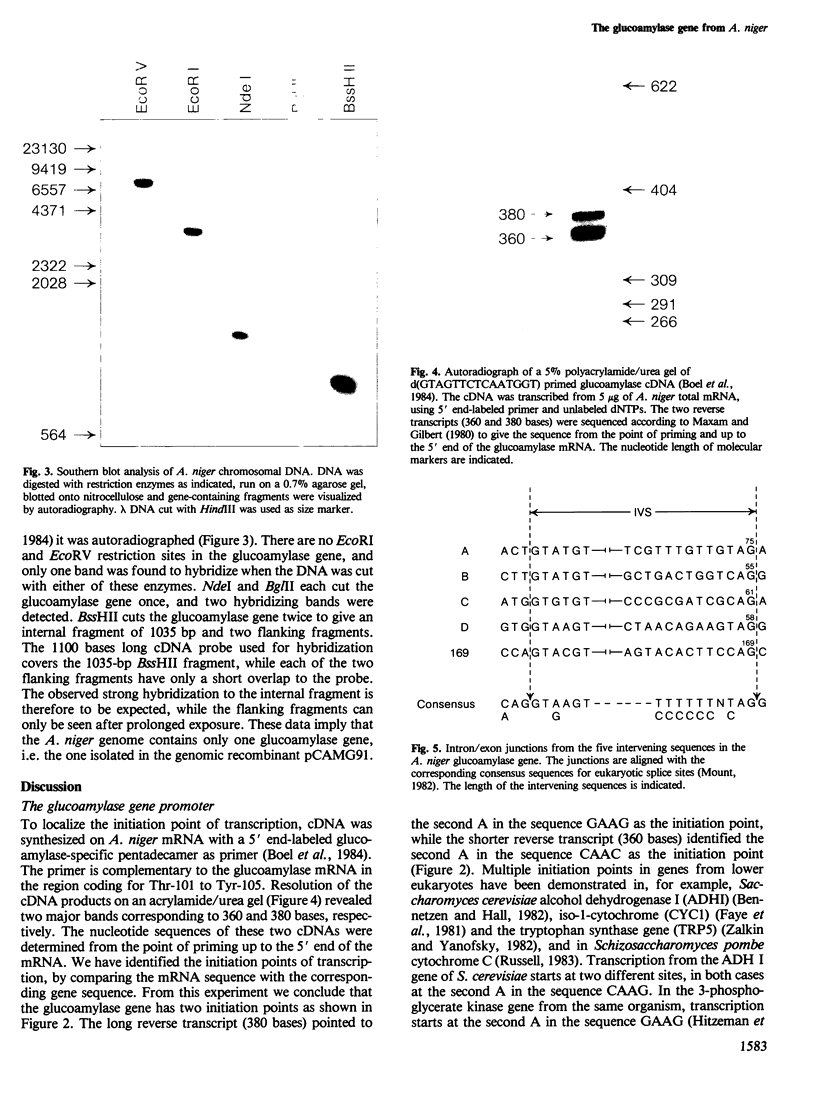


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Meselson M. Plasmid screening at high colony density. Methods Enzymol. 1983;100:333–342. doi: 10.1016/0076-6879(83)00066-x. [DOI] [PubMed] [Google Scholar]
- Hitzeman R. A., Hagie F. E., Hayflick J. S., Chen C. Y., Seeburg P. H., Derynck R. The primary structure of the Saccharomyces cerevisiae gene for 3-phosphoglycerate kinase. Nucleic Acids Res. 1982 Dec 11;10(23):7791–7808. doi: 10.1093/nar/10.23.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAZUR J. H., ANDO T. The action of an amyloglucosidase of Aspergillus niger on starch and malto-oligosaccharides. J Biol Chem. 1959 Aug;234(8):1966–1970. [PubMed] [Google Scholar]
- Pazur J. H., Knull H. R., Cepure A. Glycoenzymes: structure and properties of the two forms of glucoamylase from Aspergillus niger. Carbohydr Res. 1971 Nov;20(1):83–96. doi: 10.1016/s0008-6215(00)84951-4. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Teem J. L., Rosbash M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: implications for U1 RNA and metazoan mRNA splicing. Cell. 1983 Sep;34(2):395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Rogers J., Wall R. A mechanism for RNA splicing. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1877–1879. doi: 10.1073/pnas.77.4.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P. R. Evolutionary divergence of the mRNA transcription initiation mechanism in yeast. Nature. 1983 Jan 13;301(5896):167–169. doi: 10.1038/301167a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tollervey D., Wise J. A., Guthrie C. A U4-like small nuclear RNA is dispensable in yeast. Cell. 1983 Dec;35(3 Pt 2):753–762. doi: 10.1016/0092-8674(83)90108-3. [DOI] [PubMed] [Google Scholar]
- Wise J. A., Tollervey D., Maloney D., Swerdlow H., Dunn E. J., Guthrie C. Yeast contains small nuclear RNAs encoded by single copy genes. Cell. 1983 Dec;35(3 Pt 2):743–751. doi: 10.1016/0092-8674(83)90107-1. [DOI] [PubMed] [Google Scholar]
- Zalkin H., Yanofsky C. Yeast gene TRP5: structure, function, regulation. J Biol Chem. 1982 Feb 10;257(3):1491–1500. [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]