Abstract
We have developed a new method for quantification of promoter activity in cell lines transfected with recombinant plasmids containing the reporter gene encoding chloramphenicol acetyl transferase (CAT) by real-time PCR. As the efficiency of transfection has a direct influence on the total mRNA produced, we have used the neomycin-resistance gene present within the same vector DNA to normalize the measurement of mRNA levels. Three promoters from genes encoding toxins (pre-synaptic neurotoxin phospholipase A2, post-synaptic α neurotoxin and cardiotoxin), believed to have evolved from the same ancestor but exhibiting different promoter activities, have been employed in this study to demonstrate the feasibility and accuracy of the method in CAT gene reporter analysis.
INTRODUCTION
Methods available for investigating promoter activity usually involve linking the promoter from a gene of interest to the coding sequence of an unrelated reporter gene. The reporter gene usually codes for a specific enzyme, which is absent in the host and can easily be detected. To date, a number of reporter gene systems have been established for measuring the gene expression. These include the genes encoding chloramphenicol acetyltransferase (CAT; 1), firefly luciferase (2) and β-galactosidase (3). In the CAT assay, the CAT enzyme activity is measured from the expressed proteins, using 14C-labeled chloramphenicol as the substrate. In the luciferase assay, a bioluminescent reaction catalyzed by luciferase is used and in the β-galactosidase assays, the β-galactosidase activity is measured colorimetrically. Although the method of measuring the CAT enzyme activity can be correlated to gene expression, it does suffer from disadvantages such as the use of radioisotopes, reduced sensitivity of the assay and extensive procedures such as thin layer chromatography, autoradiography, etc. These will undoubtedly affect the accuracy of measurements of the reporter construct. Furthermore, when dealing with an inducible system, the duration of protein turnover must also be taken into account (4). Thus, measurement of mRNA levels will be the most direct method for investigating the gene activity. Techniques such as northern hybridization, S1 nuclease and RNase I protection assays can be used for such purposes. However, all these procedures are again considerably laborious and are not always useful for simultaneous analysis of many different gene constructs. Reverse transcription–polymerase chain reaction (RT–PCR) has been widely used for the detection of the mRNA molecules and offers a possible alternative method to quantifying gene expression level. The quantification of PCR products mainly relies on the phase of the reaction. Various laboratories have developed competitive PCR assays. However, competitive PCR requires accurate quantification of target and competitor amplicons at the end of the reaction and this usually entails laborious post-PCR processing steps. Recently, the ABI PRISM™ 7700 Sequence Detection System (SDS) coupled with TaqMan® chemistry has been shown to provide a rapid, sensitive method for quantifying nucleic acids (5–7). In this assay, reactions are characterized on a real-time basis at the onset of PCR, using gene-specific fluorogenic probes which are subjected to 5′ nucleotidase activity of the DNA polymerase. The higher the copy number of the mRNAs or cDNAs, the sooner a significant increase in fluorescence is observed which is detected as a CT (threshold cycle) value. The system can support multiplex PCR and, hence, more than one species of mRNA/DNA per sample can be measured simultaneously. In this communication, we report the use of the real-time PCR in the quantification of promoter activity of three related toxin genes, using CAT mRNAs as the target.
MATERIALS AND METHODS
Cell culture and DNA transfection
The Chinese hamster ovary (CHO) cell line was maintained in α-MEM (8) medium supplemented with 10% fetal bovine serum, 50 U penicillin and 50 µg/ml streptomycin. The day before transfection, CHO cells were subcultured, and trypsinized cells at a density of 3.0 × 105 cells per well were inoculated into a six-well plate. Transfections were carried out with 6 µg of reporter plasmids by calcium phosphate method (9). After 24 h, transfection medium was replaced with fresh α-MEM and incubated at 37°C for a further 24 h before harvesting cells for CAT assays or RNA isolation. To normalize the transfection efficiencies, 6 µg of pSV-β-Gal, a plasmid expressing β-galactosidase (Promega, WI, USA) was co-transfected with the test plasmids in each experiment.
For CAT assays, cytoplasmic extracts of cells were prepared by the freeze–thaw method and the enzyme activity was determined by using a Beckman liquid scintillation counter as described by Seed and Sheen (10). All results were normalized by using pSV-β-gal as an internal control and are the means of determinations ± SD from six individual experiments.
Plasmid construction
Plasmid pMAMneoCAT (Clontech, CA, USA) was digested with NheI and NdeI to remove the mouse mammary tumor virus (MMTV) promoter. Toxin gene promoters with different promotional strength available in the laboratory were first amplified by PCR, and then subcloned into promoterless CAT reporter gene vector pMAMneoCAT plasmid in normal and reverse direction. The sequences of the constructs have also been verified by DNA sequencing (Automated DNA sequencer, Applied Biosystems, CA, USA).
In vitro transcription and translation
In vitro transcription was carried out by using RiboMAX Kit (Promega). The reactions were carried out according to instructions from the manufacturer.
RNA isolation and real-time PCR
Total RNA was isolated by a single step method using TRIZOL reagent (Life Technologies, CA, USA) from CHO cells 48 h after transfection. Before real-time PCR, the RNA samples were further treated by RNase-free DNase I at 37°C for 30 min.
TaqMan probes and primers
SDS primers and TaqMan probes for the CAT gene and the Neomycin gene have been designed using PrimerExpress 1.0 software (Perkin Elmer, MA, USA). The SDS forward and reverse primers for the Neo gene were 5′-CTCCTGCCGAGAAAGTATCCA-3′ and 5′-GCCGGATCAAGCGTATGC-3′, respectively, while the SDS forward and reverse primers for the CAT gene were 5′-GCCGCTGGCGATTCAG-3′ and 5′-TTCATTAAGCATTCTGCCGACAT-3′. The TaqMan probe for CAT gene, 5′-TCATCATGCCGTCTGTGATGGCTTC-3′ and the probe for Neo gene, 5′-CGCCGCATTGCATCAGCCAT-3′, have been labeled with a reporter fluorescent dye, 6-carboxyfluorescein (FAM), and VIC dye, respectively, at their 5′ ends. The 3′ ends of the probes have been labeled with a fluorescent dye, 6-carboxy-tetramethyl-rhodamine (TAMRA), as the quencher. The specificity of SDS primers was examined under the standard PCR conditions prior to quantitation by real-time PCR. The primers and probes have been synthesized by Applied Biosystems.
Real-time quantitative PCR
Real-time PCR was carried out using the TaqMan® EZ RT–PCR reagents. Both RT and DNA polymerization were carried out in the same tube without subsequent addition of enzymes or buffer.
The RT–PCR was performed in a MicroAmp Optical 96-well reaction plate using rTth DNA polymerase. This enzyme functions as both a thermostable reverse transcriptase and a thermostable DNA polymerase. Each RT–PCR reaction contained the following: 1× TaqMan buffer, 3 mM manganese acetate, 300 µM each of dATP, dCTP and dGTP, 600 µM of dUTP, 200 nM primers, 100 nM TaqMan probe, 2.0 U rTth DNA polymerase, 0.5 U AmpErase UNG and 50 ng total RNA, in 50 µl of reaction volume. RT was carried out for 30 min at 60°C following an initial 2 min incubation at 50°C for UNG activity to prevent carryover reactions. The reaction was terminated by heating at 95°C for 5 min. The PCR amplification was then performed for 40 cycles with each cycle at 94°C for 20 s and 60°C for 1 min. All reactions were carried out in triplicate using the ABI Prism 7700 SDS. The threshold cycle, CT, values were averaged from the values obtained from each reaction, and the promoter activity was calculated as a relative level of expression (CAT gene using FAM-labeled probe) to the calibrator (Neomycin gene using VIC-labeled probe). The FAM and VIC spectral data were collected from reactions carried out in separate tubes using the same stock of RNA and other reagents to avoid the spectral overlap among VIC/FAM/TAMRA and to avoid limitation of reagents and dNTPs.
RESULTS AND DISCUSSION
CAT assays on cardiotoxin-2, neurotoxin-2 and phospholipase A2 promoters
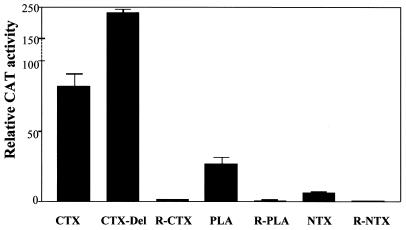
The promoters of cardiotoxin-2 (CTX-2; 11), neurotoxin-2 (NTX-2; 12) and phospholipase A2 (PLA2; 13) genes were sub-cloned upstream of CAT gene into pMAMneoCAT promoterless reporter vector in both correct and reverse orientations and transfected separately into CHO cells. We have shown previously (11) that the epithelial-derived CHO cells yielded comparatively similar levels of expression of toxin genes and hence can be used as a cell line for the investigation of the toxin genes’ promoter activity. A deletion construct of CTX promoter containing the highly active TATA box (Del 4; 11) was also used. Both the CAT assay and real-time PCR analysis were carried out for all the constructs. Figure 1 shows that the promoter activity of the CTX-2 gene, is ∼5-fold higher than that of the PLA2 gene. However, lower promoter activity was observed for NTX promoter (4-fold less than that of PLA2). The CTX-deletion construct showed the highest promoter activity (∼3-fold higher than the intact CTX-2 promoter). The promoters cloned in reverse orientation showed no CAT activity as expected. The ratio of CAT activity for promoters CTX:PLA2:NTX was found to be 12:4:1 (Fig. 1).
Figure 1.
CAT assays on different toxin promoters. PLA, CTX and NTX represent PLA2, CTX-2 and NTX-2 promoters; R-PLA, R-CTX and R-NTX represent the respective promoters in reverse orientation. CTX-Del denotes a selected deletion construct (Del 4; 11) of CTX-2 promoter.
Real-time PCR quantitation
The TaqMan assay utilizes the 5′→3′ exonuclease activity of Taq DNA polymerase and a fluorogenic probe for automated quantification of DNA in a real-time manner. The CT value refers to the threshold cycle at which a statistically significant increase in fluorescence is first detected by the sequence detection system. The increase in fluorescence is directly proportional to the exponential increase in PCR products and the measurement of signal is carried out in a real-time manner. Therefore, samples containing low concentration of target molecules would require more PCR cycles (hence higher CT) to amplify enough copies to produce a significant fluorescent signal.
We have used the expression of the Neomycin gene present in the recombinant plasmid as an internal standard in these experiments. The optimal concentration of the total RNA for real-time detection was found to be 50 ng. A no-template control, in which sterile buffer was replaced for template RNA, was used in each experiment. This control was used to subtract any fluorescence that is not related to PCR amplification. The amount of target, normalized (ΔCTS) to an endogenous reference (Neomycin gene) was calculated by subtracting the CT value of the reference (CTR) from the CT value of the sample (CTS; ΔCTS = CTS – CTR). The relative expression (2–ΔΔCT) to an arbitary calibrator (e.g. rCTX) is determined by subtracting the ΔCTcalibrator from the ΔCTS value (ΔΔCT = ΔCTS – ΔCTcalibrator). The ratio obtained from the results for the CAT mRNA produced by CTX:PLA2:NTX was 2.01:1.5:1 (Table 1, column 6). These results corresponded to our previous study on the mRNA levels in the venom gland of Naja naja sputatrix determined by in situ hybridization, where CTX:PLA2:NTX ratio was 2:1.3:1, respectively, (14). The venom protein profiles obtained by LC-MS (14) showed that the ratio of CTX:PLA:NTX in the venom was 13:3:1. Similarly, in a separate report, Tan (15) documented that CTX in N.sputatrix comprise almost 60% of venom dry weight, whereas PLA2 and NTX comprise 14 and 4.5%, respectively, approximating to a similar ratio of 13:3:1. These differences between the mRNA and the protein content may be due to slow turnover rate of toxin transcripts and high translational efficiency in a manner similar to the observation made by other laboratories (16–19). These observations also imply that regulatory elements determining the mRNA stability and translational efficiency may be located within the toxin promoters. However, these speculations require further investigation.
Table 1. The threshold cycle (CT) values of the promoter constructs.
| |
CTS Fam (CAT) |
CTR Vic (NEO) |
ΔCTS |
ΔΔCT rel
to R-CTX |
2–ΔΔCT rel
to R-CTX |
ΔΔCT rel
to pMAM |
2–ΔΔCT rel
to pMAM |
ΔΔCT rel
to CHO |
2–ΔΔCT rel
to CHO |
| NTC | 36.5 ± 0.07 | 35.01 ± 0.10 | – | – | – | – | – | – | – |
| CHO | 40.0 ± 0 | 32.6 ± 0.06 | 7.4 | 0.36 | 1.28 | –4.27 | 0.05 | 0 | 1.00 |
| pMAM | 33.32 ± 0.13 | 30.20 ± 0.15 | 3.13 | –3.91 | 15.03 | 0 | 1.00 | –4.27 | 19.3 |
| R-CTX | 28.15 ± 0.10 | 21.11 ± 0.07 | 7.04 | 0 | 1.00 | 3.91 | 0.067 | –0.36 | 1.28 |
| CTX | 22.05 ± 0.12 | 20.63 ± 0.13 | 1.42 | –5.62 | 49.18 | –1.71 | 3.27 | –5.98 | 63.12 |
| NTX | 24.05 ± 0.08 | 21.71 ± 0.10 | 2.43 | –4.61 | 24.42 | –0.79 | 1.72 | –4.97 | 31.34 |
| PLA | 20.23 ± 0.06 | 18.35 ± 0.13 | 1.88 | –5.16 | 35.75 | –1.25 | 2.37 | –5.52 | 45.89 |
| CTX-Del | 19.73 ± 0.09 | 20.15 ± 0.10 | –0.42 | –7.46 | 176.07 | –3.55 | 11.71 | –7.82 | 225.97 |
The calculated ΔCTS values in column 4 are based on the CT values of the CAT and NEO gene. 2–ΔΔCT relative to R-CTX (column 6) has been calculated with references to the control, CTX promoter cloned in reverse orientation. Columns 8 and 10 represent the activity of the toxin gene promoter relative to MMTV promoter (pMAM) and rRNA in CHO cells (CHO), respectively. CTX-Del indicates the deletion construct of CTX promoter and NTC forms the no-template control.
We obtain similar real-time results when the mRNA produced in each case has been normalized to either the pMAMneo promoter activity or to the rRNA in CHO cells (Table 1, columns 8 and 10, respectively). Hence, the results obtained by real-time PCR represent the actual amount of mRNA produced in vivo. From our CAT assays (based on the enzyme activity), it is clear that the values form an approximate estimation of the promotional activity of the promoters. We validated this by examining the expression of one of the toxin genes, CTX. Stable CHO clones containing the CTX promoter proximal to the CAT gene (in the pMAMneo-CAT plasmid) were prepared (11) and used in the measurement of CAT mRNA and CAT enzyme content. A ratio of CAT mRNA to CAT enzyme, 1:9, somewhat similar to the above results, was obtained.
In this study, we have also carried out an in vitro transcription of the CAT gene utilizing the CTX-2 promoter. Nuclear extracts from both the venom gland and CHO cells have been used. The results were found to be similar in both cases (Table 2) indicating that CHO cells form an alternative cell line for the analysis of toxin gene promoter activities. However, only a 2-fold increase in activity for the CTX-deletion construct (in contrast to the expected 4-fold increase) has been observed. We speculate that this may be due to the non-physiological environment present in in vitro assays. In our previous study (11), we demonstrated that the CTX promoter can initiate transcripts from the correct sites (similar to snake venom gland cells) when expressed in CHO cells. In vitro footprinting assays, using nuclear extracts from snake venom glands and CHO cells gave identical DNase I protection patterns, to demonstrate that CHO cells could be used as alternative cell lines to snake venom gland cells to investigate the expression toxin genes (11). Besides, Marchot et al. (20) have reported that CHO cells can be used to express and process toxin gene products.
Table 2. In vitro transcription on CTX-2 promoter.
|
In vitro transcription |
CT FAM (CAT) |
ΔCT rel
to R-CTX |
ΔΔCT rel
to R-CTX |
2–ΔΔCT |
| Vm R-CTX |
19.3 ± 0.04 |
0 |
0 |
1.0 |
| Vm-CTX |
14.8 ± 0.09 |
–4.5 |
–4.5 |
22.63 |
| Vm-CTX-Del |
13.7 ± 0.10 |
–5.6 |
–5.6 |
48.5 |
| CHO-R-CTX |
19.81 ± 0.06 |
0 |
0 |
1.0 |
| CHO-CTX |
15.40 ± 0.04 |
–4.41 |
–4.41 |
21.26 |
| CHO-CTX-Del | 14.21 ± 0.03 | –5.60 | –5.60 | 48.5 |
Vm and CHO refer to nuclear extracts from venom gland and CHO cells, respectively.
Real-time PCR has been widely used in the field of developmental studies (21), clinical diagnostics/prognosis (22–24) and the detection/screening of residual or very low copy numbers of gene or gene products (25,26). In this paper, we have demonstrated that real-time PCR could be used for quantifying the gene expression level. Compared to the traditional CAT assay, real-time PCR has provided a very rapid, sensitive and reproducible approach for quantitation of gene activity. The level of sensitivity (pg level) obtained by real-time PCR is rather difficult for assays such as northern hybridization and RNase protection. Real-time PCR also allows a large number of different RNA samples to be analysed and quantitated simultaneously. This method can also be used over a range of concentration of total RNA. Besides, the variations in results can be normalized using an internal control as, in this case, the expression of the neomycin gene. Hence, co-transfection experiments to determine the efficiency of transfections are not required. Thus, real-time PCR analysis forms the most direct and accurate method that is available to date for gene expression studies.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a Research Grant (R-183-000-031-112) from the National University of Singapore. D.M. received a University Scholarship.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +65 874 3248; Fax: +65 779 1453; Email: AF097000, AF241224, AF101235
References
- 1.Gorman C.M., Moffat,L.F. and Howard,B.H. (1982) Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell Biol., 2, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen V.T., Morange,M. and Bensaude,O. (1988) Firefly luciferase luminescence assays using scintillation counters for quantitation in transfected mammalian cells. Anal. Biochem., 171, 404–408. [DOI] [PubMed] [Google Scholar]
- 3.Alam J. and Cook,J.L. (1990) Reporter genes: application to the study of mammalian gene transcription. Anal. Biochem., 188, 245–254. [DOI] [PubMed] [Google Scholar]
- 4.Kain S.R., Adams,M., Kondepudi,A., Yang,T.T., Ward,W.W. and Kitts,P. (1995) Green fluorescent protein as a reporter of gene expression and protein localization. Biotechniques, 19, 650–655. [PubMed] [Google Scholar]
- 5.Gibson U.E., Heid,C.A. and Williams,P.M. (1996) A novel method for real time quantitative RT–PCR. Genome Res., 6, 995–1001. [DOI] [PubMed] [Google Scholar]
- 6.Desjardin L.E., Chen,Y., Perkins,M.D., Teixeira,L., Cave,M.D. and Eisenach,K.D. (1998) Comparison of the ABI 7700 system (TaqMan) and competitive PCR for quantification of IS6110 DNA in sputum during treatment of tuberculosis. J. Clin. Microbiol., 36, 1964–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishida K., Zhu,B. and Maeda,H. (2000) Novel approach to quantitative reverse transcription PCR assay of mRNA component in autopsy material using the TaqMan fluorogenic detection system: dynamics of pulmonary surfactant apoprotein A. Forensic Sci. Int., 113, 127–131. [DOI] [PubMed] [Google Scholar]
- 8.Stanners C.P., Eliceiri,G.L. and Green,H. (1971) Two types of ribosome in mouse–hamster hybrid cells. Nat. New Biol., 230, 52–54. [DOI] [PubMed] [Google Scholar]
- 9.Jordan M., Schallhorn,A. and Wurm,F.M. (1996) Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res., 24, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seed B. and Sheen,J.Y. (1988) A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene, 67, 271–277. [DOI] [PubMed] [Google Scholar]
- 11.Ma D.H., Armugam,A. and Jeyaseelan,K. (2001) Expression of cardiotoxin-2 gene: cloning, characterization and deletion analysis of the promoter. Eur. J. Biochem., 268, 1844–1850. [DOI] [PubMed] [Google Scholar]
- 12.Afifiyan F., Armugam,A., Tan,C.H., Gopalakrishnakone,P. and Jeyaseelan,K. (1999) Postsynaptic α-neurotoxin gene of the spitting cobra, Naja naja sputatrix: structure, organization, and phylogenetic analysis. Genome Res., 9, 259–266. [PMC free article] [PubMed] [Google Scholar]
- 13.Jeyaseelan K., Armugam,A., Donghui,M. and Tan,N.H. (2000) Structure and phylogeny of the venom group I phospholipase A(2) gene. Mol. Biol. Evol., 17, 1010–1021. [DOI] [PubMed] [Google Scholar]
- 14.Lachumanan R., Armugam,A., Durairaj,P., Gopalakrishnakone,P., Tan,C.H. and Jeyaseelan,K. (1999) In situ hybridization and immunohistochemical analysis of the expression of cardiotoxin and neurotoxin genes in Naja naja sputatrix. J. Histochem. Cytochem., 47, 551–560. [DOI] [PubMed] [Google Scholar]
- 15.Tan N.H. (1991) The biochemistry of venoms of some venomous snakes of Malaysia—a review. Trop. Biomed., 8, 91–103. [Google Scholar]
- 16.Sato S., Yoshida,H., Abe,H. and Tamiya,N. (1969) Properties and biosynthesis of a neurotoxic protein of the venoms of sea snakes Laticauda laticaudata and Laticauda colubrina. Biochem. J., 115, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kochva E., Tonsing,L., Louw,A.I., Liebenberg,N.V. and Visser,L. (1982) Biosynthesis, secretion and in vivo isotopic labelling of venom of the Egyptian cobra, Naja haje annulifera. Toxicon, 20, 615–635. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa T., Onoue,H., Nakagawa,K., Nomura,S., Sueishi,K., Hattori,S., Kihara,H. and Ohno,M. (1995) Localization and expression of phospholipases A2 in Trimeresurus flavoviridis (habu snake) venom gland. Toxicon, 33, 1645–1652. [DOI] [PubMed] [Google Scholar]
- 19.Takeda M., Yoshida,H. and Tamiya,N. (1974) Biosynthesis of erabutoxins in the sea snake, Laticauda semifasciata. Toxicon, 12, 633–641. [DOI] [PubMed] [Google Scholar]
- 20.Marchot P., Prowse,C.N., Kanter,J., Camp,S., Ackermann,E.J., Radic,Z., Bougis,P.E. and Taylor,P. (1997) Expression and activity of mutants of fasciculin, a peptidic acetylcholinesterase inhibitor from Mamba venom. J. Biol. Chem., 272, 3502–3510. [DOI] [PubMed] [Google Scholar]
- 21.Steuerwald N., Cohen,J., Herrera,R.J. and Brenner,C.A. (2000) Quantification of mRNA in single oocytes and embryos by real-time rapid cycle fluorescence monitored RT–PCR. Mol. Hum. Reprod., 6, 448–453. [DOI] [PubMed] [Google Scholar]
- 22.Fujimaki S., Funato,T., Harigae,H., Imaizumi,M., Suzuki,H., Kaneko,Y., Miura,Y. and Sasaki,T. (2000) A quantitative reverse transcriptase polymerase chain reaction method for the detection of leukaemic cells with t(8;21) in peripheral blood. Eur. J Haematol., 64, 252–258. [DOI] [PubMed] [Google Scholar]
- 23.Kreuzer K.A., Lass,U., Nagel,S., Ellerbrok,H., Pauli,G., Pawlaczyk-Peter,B., Siegert,W., Huhn,D. and Schmidt,C.A. (2000) Applicability of an absolute quantitative procedure to monitor intra-individual bcr/abl transcript kinetics in clinical samples from chronic myelogenous leukemia patients. Int. J. Cancer, 86, 741–746. [DOI] [PubMed] [Google Scholar]
- 24.Pfitzner T., Engert,A., Wittor,H., Schinkothe,T., Oberhauser,F., Schulz,H., Diehl,V. and Barth,S. (2000) A real-time PCR assay for the quantification of residual malignant cells in B cell chronic lymphatic leukemia. Leukemia, 14, 754–766. [DOI] [PubMed] [Google Scholar]
- 25.Zerr D.M., Huang,M.L., Corey,L., Erickson,M., Parker,H.L. and Frenkel,L.M. (2000) Sensitive method for detection of human herpesviruses 6 and 7 in saliva collected in field studies. J. Clin. Microbiol., 38, 1981–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miley W.J., Suryanarayana,K., Manns,A., Kubota,R., Jacobson,S., Lifson,J.D. and Waters,D. (2000) Real-time polymerase chain reaction assay for cell-associated HTLV type I DNA viral load. AIDS Res. Hum. Retroviruses, 16, 665–675. [DOI] [PubMed] [Google Scholar]