Abstract
The action of phytohormones in plants requires the spatiotemporal regulation of their accumulation and responses at various levels. Recent studies reveal an emerging relationship between the function of phytohormones and epigenetic modifications. In particular, evidence suggests that auxin biosynthesis, transport, and signal transduction is modulated by microRNAs and epigenetic factors such as histone modification, chromatin remodeling, and DNA methylation. Furthermore, some phytohormones have been shown to affect epigenetic modifications. These findings are shedding light on the mode of action of phytohormones and opening up a new avenue of research on phytohormones as well as revealing mechanisms regulating the epigenetic modifications.
Introduction
Epigenetic modifications regulate mitotically or meiotically heritable gene expression without altering any changes in the genomic DNA sequences, and therefore contribute to flexible and reversible gene expression regulation. Epigenetic modifications involve histone modification, chromatin remodeling, non-coding RNAs, and DNA methylation. Each of these modifications alone, or in combination with one another, and the interplay between different epigenetic modifications, control gene expression patterns. Numerous studies show that genetic programming can be overridden by altering epigenetic modifications in response to environmental conditions, thus contributing to flexible survival strategies of sessile plants (Kim et al., 2008; Dowen et al., 2012). Intriguing underexplored aspects in this field of research include the biochemical signals that alter the epigenome, and the transduction of these signals to control the downstream epigenetic pathways. An increasing number of studies suggest a tight link between epigenetic regulation and plant hormone signaling (Zhu, 2010). The plant hormone auxin is perceived by the nuclear auxin receptors TRANSPORT INHIBITOR RESPONSE1 (TIR)/AUXIN SIGNALING F BOX PROTEINS (AFBs), leading to the activation of AUXIN RESPONSE FACTORS (ARFs), the transcriptional factors that activate auxin induced gene expression (Salehin et al., 2015). Emerging evidence indicates that the ARF-dependent induction of auxin responsive genes is modulated by microRNAs as well as by multiple epigenetic factors, such as histone modifications and the chromatin remodeling factor PICKLE (PKL) (Jones-Rhoades and Bartel 2004; Long et al., 2006; Navarro et al., 2006; Chen et al., 2010; Rhoades et al., 2002; Mallory et al., 2005; Wu et al., 2006; Zhu, 2010; Weiste and Droge-Laser, 2014). Interestingly, auxin has also been implicated in the regulation of changes in the epigenome, suggesting an auxin-linked epigenetic regulation loop. In this review, we discuss recent literature on the interconnection between epigenetic control and phytohormone signaling with a focus on auxin signaling.
Histone modification machinery and plant hormone signaling
(i) Histone acetylation and plant hormones
Eukaryotic chromatin is a highly organized complex of DNA and proteins, and is composed of the basic repeat element, the nucleosome. Each nucleosome contains two copies of the histone protein H2A, H2B, H3 and H4, and is typically enfolded by 147 bp of DNA. Modifications of histone tails such as acetylation, methylation, phosphorylation and ubiquitination play an important role in epigenetic regulation. One major histone modification, which occurs at the ε-amino group of conserved lysine residue, is acetylation mediated by the reversible activity of histone acetyl transferases (HAT) and histone deacetylases (HDAC). Both histone acetylation and deacetylation play an important role in gene regulation and have been implicated in hormone signaling (Sieberer et al., 2003; Zhou et al., 2005; Long et al., 2006; Chen et al., 2010; Chen and Wu 2010; Zhu, 2010). Acetylation neutralizes the positive charges of lysine residues on the histone N-terminal tail, thereby decreasing the interaction between histone protein and negative charged DNA, leading to a more open and loose chromatin conformation (Shahbazian and Grunstein, 2007). There are four histone acetylase (HAT) families. GCN5 (general control nonderepressible 5) belongs to the Gcn5 N-acetyltransferase (GNAT) subfamily and is the best characterized HAT in yeast, mammals and plants (Baker and Grant, 2007; Chen and Tian, 2007; Lee and Workman, 2007). Arabidopsis GCN5 acetyltransferase and the transcription factor (TF) adaptor proteins ADA2a and ADA2b (also known as PROPORZ1) interact with each other, and are the subunits of the transcriptional adaptor complex SAGA (Spt-Ada-Gcn5-Acethyltransferase) (Servet et al., 2010). GCN5’s HAT activity is modulated by ADA2b in Arabidopsis (Mao et al., 2006). Genome-wide analysis showed that the expression of ~5% of all genes is changed in gcn5 and ada2b/prz1 mutants (Benhamed et al., 2008). However, some reports indicate that specific genetic pathways are controlled by GCN5 or ADA2.
gcn5/hag1 mutants have a short root phenotype with defects in the columella differentiation layer and in QC marker gene expression (Vlachonasios et al., 2003; Kornet and Scheres., 2009), implicating the GCN5 complex in the maintenance of root stem cell niche in Arabidopsis. PLT1 and PLT2 genes encode AP2 domain TFs induced by auxin in an ARF-dependent manner, and play a major role in the specification of root stem cells (Aida et al., 2004; Galinha et al., 2007). Interestingly, GCN5 acts in the same genetic pathway as the PLT genes, and the short root phenotype of gcn5/hag1 mutant results from severely reduced expression of PLT genes, suggesting a chromatin modification-based mechanism that underlies the PLT-dependent stem cell specification. However, whether the GCN5 acetylase complex is recruited to the promoter of PLT genes directly to activate PLT gene expression remains obscure (Kornet and Scheres., 2009).
The prz1 (proporz1) mutant was isolated based on the phenotype of ectopic callus tissue formation in root under auxin treatment (Sieberer et al., 2003). The PRZ1 gene encodes for ADA2b, and the observed phenotype in prz1mutant is at least partially caused by misexpression of KIP RELATED PROTEIN (KRP) family genes (Sieberer et al., 2003). Auxin treatment did appear to have an impact on histone acetylation at the whole chromatin level. However, ChIP experiments showed that ADA2b/PRZ1 is associated with the KRP7 locus, and auxin treatment decreased histone H3Kac9 and H3Kac14 levels in the KRP7 locus, which correlated with the reduction in expression of the KRP7 gene. Interestingly, the auxin-mediated reduction in KRP7 expression was more obvious in the prz1 mutant. Furthermore, constitutively reduced histone H3Kac9 and H3Kac14 levels were observed in the KRP7 locus in prz1 mutant. Collectively, these studies support the hypothesis that auxin reduces histone acetylation level, whereas ADA2b/PRZ1 oppose the auxin mediated suppression signal to control appropriate KRP7 expression (Anzola et al., 2010). Future areas of research will involve auxin regulation of histone acetylation at a specific locus.
Elongator was first identified as an RNA polymerase II-associated protein complex in yeast (Otero et al., 1999). This elongator protein complex consists of six subunits (ELP1–ELP6), with ELP3 containing a HAT domain (Wittschieben et al., 1999). Some publications reported that mutations in elongator subunits cause pleiotropic phenotypes including ABA, auxin, ethylene and jasmonic acid (JA) related phenotypes (Nelissen et al., 2005; Chen et al., 2006; Ding and Mou, 2015). ChIP experiment indicated that the SHORT HYPOCOTYL2 (SHY2)/IAA3 and auxin influx carrier LIKE AUXIN RESISTANT 2 (LAX2) genes were direct targets of elongator HAT activity. Interestingly, SHY2/IAA3 is also a target of the GCN5 HAT (Benhamed et al., 2006), thus indicating a complex regulatory mechanism where two different HATs modulate SHY2/IAA3 gene expression.
(ii) HDACs and plant hormone responses
Histone deacetylation has also been implicated in the regulation of hormone responses in plants. Histone deacetylation is mediated by the HDAC complex, which is composed of HDAC and other components. The Arabidopsis genome encodes 18 HDACs, and the largest and most characterized HDAC family is RPD3/HDA1 that can be divided into three classes (class I–III) based on sequence similarity (Hollender and Liu, 2008; Alinsug et al., 2009). HDA6, 7, 9 and 19 belong to the class I family of RPD3/HDA1. Class II has three members, HDA5, HDA15, and HDA18. Class III is comprises the plant-specific HD2A, HD2B and HD2C (Pandey et al., 2002; Hollender and Liu, 2008). In contrast to HATs, HDACs repress transcription activity. Similarly to HATs, the recruitment of HDACs to DNA seems to occur both globally and at specific gene loci. For example, hda19 knockout and knockdown mutants show pleiotropic phenotypes, implicating HDA19 in the regulation of various developmental processes such as seed dormancy, and embryo, leaf, and flower development (Tian and Chen 2001; Tian et al., 2003; Tian et al., 2005; Long et al., 2006). The observed pleiotropic effects suggest HDA19’s global role in gene regulation. However, HDA19 is also implicated in the specific regulation of auxin signaling (more details of this point are discussed later).
Several studies suggest an important role for HDA6 and HDA19 in the regulation of plant hormone responses. The expression of HDA6 and 19 is induced by plant hormones ethylene and JA (Zhou et al., 2005), and knocking out HDA6 and 19 causes ABA hypersensitivity (Chen et al., 2010; Chen and Wu 2010). The transcriptional repressors (JASMONATE ZIM-DOMAIN) JAZ proteins and the TFs ETHYLENE INSENSTIVE 3 (EIN3) and its homolog EIN3-LIKE 1 (EIL1) act as master regulators for JA and ethylene signaling, respectively (Alonso et al., 2003; Chini et al., 2007; Yan et al., 2007; Thines et al., 2007; An et al., 2010; Zhong et al., 2009). JAZ inhibits the EIN3/EIL1 function, thus JAZ2 and EIN3 act at the crosstalk point of JA inducible ethylene regulated gene expression. HDA6 interacts with both EIN3 and JAZ proteins, and act as a repressor for EIN3-mediated transcription and JA signaling through histone acetyltransferase activity (Zhu et al., 2011). This evidence highlights a mechanism that HDAC can be recruited to specific target locus through physical interaction with TFs and its associated proteins. In addition, the SIN3-LIKE1 (SNL) component of HDACs has been shown to regulate plant responses to these hormones. Arabidopsis SNL1 and SNL2 belong to SWI-Independent 3 (SIN3)-like (SNL) protein family (Bowen et al., 2010). In yeast and mammals, the SIN3 protein acts as a scaffold recruiting histone binding protein and HDACs (Ahringer 2000; Grzenda et al., 2009). Arabidopsis SNL1 interacts with HDA19, thus SNL1 is presumed to be a component of the HDAC repressor complex, analogous to yeast and animal systems (Wang et al., 2013). Notably, SNL1 and SNL2 enhance ABA responses and suppress ethylene signaling, through the deacetylation of H3K9 and H3K18 associated with ABA- and ethylene-related genes, respectively. Moreover, the snl1 and snl2 knockouts show reduced dormancy and enhanced triple response phenotype to ACC, respectively (Wang et al., 2013), which is consistent with the fact that ABA plays an important role in seed maturation and dormancy establishment, whereas ethylene promotes seed germination and represses seed dormancy (Gutierrez et al., 2007; Finkelstein et al., 2008; Linkies and Leubner-Metzger., 2012). It is yet to be determined how ABA and ethylene modulate the recruitment of the HDA19 to regulate specific sets of genes; it is possible that the TPL-EAR repressome described below could provide a unifying mechanism for the specific regulation of epigenetic regulation by various hormones.
(iii) Histone acetylation and deacetylation regulate TPL-EAR repressome-mediated expression of auxin-responsive genes
How are the HAT and HDAC complexes recruited to target gene loci in response to hormonal or environmental signal, and how is the cumulative action of histone acetylation and deacetylation controlled or integrated into the plant hormone signaling pathway? The studies of TOPLESS (TPL) co-suppressor for auxin signaling have begun to unravel the answers to these interesting questions (Long et al., 2006). Auxin response factors (ARFs) are TFs that bind auxin-responsive elements (AuxRE) to regulate auxin-responsive genes (Tiwari et al., 2003, Ulmasov et al., 1997). AUX/IAAs act as repressors of ARFs (Figure 1). The AUX/IAA proteins contain four conserved sequence motifs, domains I–IV, and each domain is supposed to have distinct interacting partners and functions. For example, the auxin receptor, TIR1, interacts with AUX/IAA through domain II, and this binding recruits AUX/IAA to the SCFTIR E3 complex for ubiquitination and subsequent degradation by the 26S proteosome (Dharmasiri et al., 2005; Kepinski and Leyser., 2005). Point mutations in conserved residues of domain II cause the inability of this domain to bind TIR1, leading to the dominant negative effect, such as the bdl mutation on IAA12 that results in the insensitivity to auxin induction of gene expression (Hamam et al., 2002). Domains III and IV are the regions that enable AUX/IAA-ARFs interactions to repress auxin signaling (Guilfoyle and Hagen., 2012; Figure 1).
Figure 1. The model for the regulation of TIR-mediated auxin signaling pathway trough the HDAC and GCN5 complex.
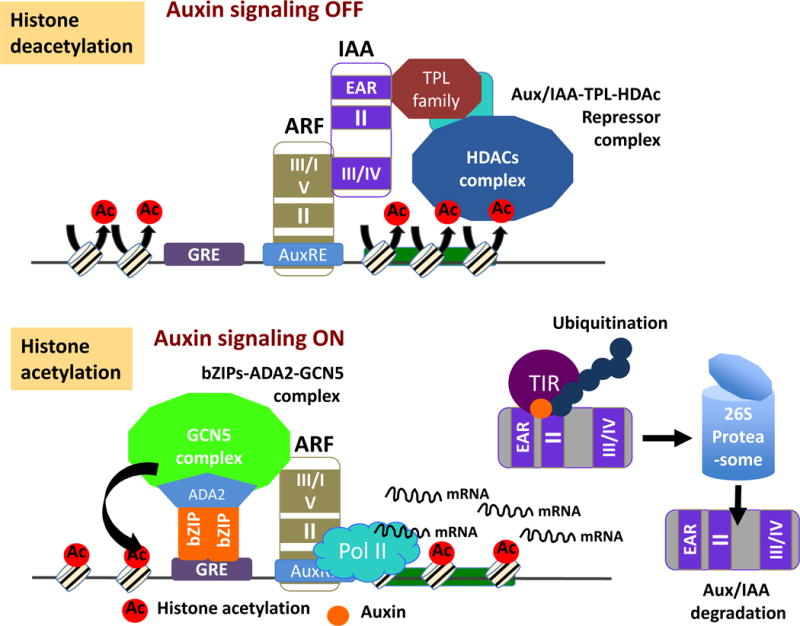
AuxRE and GRE are the sites for recruiting these suppressor and activator complexes.
tpl-1 is a very interesting temperature sensitive gain-of-function mutant that exhibits ectopic root formation at the apical pole of embryo in restrictive temperature (29°C) conditions. In lower temperature conditions, tpl-1 mutants fail to form a shoot apical meristem and have a varying degree of fused cotyledon phenotype (Long et al., 2002; Long et al., 2006). The TPL protein contains LisH (lissencephaly homology) and C-terminal WD40 repeats domains, and belongs to the evolutionally conserved Gro/Tup1 transcriptional co-repressor family (Long et al., 2006). This LisH domain, found in a large number of eukaryotic proteins, is involved in protein-protein interactions (Emes et al., 2001). Importantly, IAA12/BDL has been identified as a TPL interacting protein, suggesting a link between TIR1-dependent auxin signaling and TPL. Moreover, double mutant analysis showed that the tpl-1 mutation suppressed the bdl severe phenotype, such as basal patterning defects and reduction in cotyledon vasculature development. This supports the biological significance of TPL as a co-repressor of the IAA complex (Szemenyei et al., 2008). The EAR motif, defined by the consensus sequence of LXLXL, in the N-terminus of IAA12/BDL, is necessary and sufficient for the interaction with TPL. The EAR motif-mediated repressome is postulated to recruit the HDAC complex to the promoter of target genes (Song et al., 2005). In this scenario, HDAC is supposed to be recruited to the AUX/IAA repressor complex through EAR mediated IAA-TPL interaction. Consistently, suppressor screening of the tpl-1 mutant identified the GCN5/HAG1 histone acetylase gene (Long et al., 2006). Moreover, T-DNA insertion allele of hda19 mutant has several tpl-1 like phenotypes at the restrictive temperature (29°C), suggesting that TPL and HD19 act on the same target. Collectively, these studies support the model that the co-repressor TPL recruits HDA19 to the AUX/IAA repressor in an EAR motif-dependent manner, and GCN5/HAG1 histone acetylase has an opposing role to the IAA12/BDL-TPL-HDA19 repressor complex in the ARF-dependent expression of auxin responsive genes (Figure 1) (Long et al., 2006; Szemenyei et al., 2008).
The EAR repressome is plant-specific and appears to be widely used to regulate plant hormone signaling. For example, the EAR motif was identified in several proteins, including NOVEL INTERACTOR OF JAZ (NINJA), ABI-FIVE BINDING PROTEINS (AFPs), BRASSINAZOLE RESISTANT1 (BZR1) and BRI1-EMS-SUPPRESSOR 1 (BES1), and are involved in ethylene, JA, ABA, gibberellic acid (GA) and brassinosteroid (BR) signaling (Pauwels et al., 2010; Oh et al., 2014; Fukazawa et al., 2014; Ryu et al., 2014). In particular, the EAR repressome and the signaling pathways leading to the degradation of the EAR repressors are essentially paralleled in auxin and JA signaling (Pérez and Goossens., 2013). TPL and its family proteins TOPLESS-RELATED (TPRs) are implicated in auxin, JA, GA and BR signaling (Pauwels et al., 2010; Oh et al., 2014; Fukazawa et al., 2014; Ryu et al., 2014). The same mechanisms involving HDACs via TPL may be employed by other plant hormone regulatory machineries. However, a direct interaction between TPL and HDA19 has not been observed (Gonzalez et al., 2007; Causier et al., 2012), suggesting an additional bridge protein is required for HDAC recruitment. To control the specificity for each hormonal signaling pathway, plants must have evolved specific partner proteins of TPL. Further studies will be necessary to reveal the mechanism for the functional specificity of TPL family proteins for EAR repressome-mediated plant hormone signaling.
ARF transcription factors bind AuxRE elements in the promoter of auxin responsive genes to activate or inactivate their expression (Tiwari et al., 2003; Ulmasov et al., 1997; Ulmasov et al., 1999; Guilfoyle and Hagen., 2007). Binding of AUX/IAA to ARF allows the TPL-DHAC complex to come into proximity with the AuxRE element of chromatin, inducing its repression status. However, how the GCN5/HAG1 histone acetylase complex is recruited to the auxin responsive promoter to activate gene expression has yet to be elucidated. A possible mechanism involves an auxin inducible bZIP11 TF that binds ADA2b, and the bZIP11-ADA2b complex is then targeted to the G-box-related elements (GREs) motif, the binding site for bZIP TFs (Jakoby et al., 2002). bZIP11-ADA2b is able to act as a key adaptor protein complex in the recruitment of GCN5/HAG1 acetylase to the GH3.3 promoter, inducing the activation of GH3.3 expression (Figure 1).
ADA2a showed a weaker ability to interact with bZIP proteins compared with ADA2b (Weiste and Droge-Laser., 2014). This might explain the observation that only the prz1/ada2b knockout displays auxin and morphological phenotypes (Sieberer et al., 2003; Vlachonasion et al., 2003; Weiste and Droge-Laser., 2014). However, the interaction of bZIP11with ADA2b cannot explain all the auxin related phenotypes found in the ada2b single knockout. Most probably, the ADA2b-GCN5 complex also controls a number of auxin responsive genes. AuxRE on the promoter of auxin responsive gene is a well-studied motif, which is the site for recruiting ARF-IAA-TPL protein complexes. Both AuxRE and GREs motifs are supposed to be necessary for sensitive and quantitative regulation of auxin signal in response to varying developmental and environmental changes (Weiste and Droge-Laser., 2014). To date, there is only evidence for bZIPs/ADA2b recruitment to the GREs motif in the GH3.3 gene promoter. It will be important to determine whether this attractive model can be applied to other auxin responsive genes as a general mechanism for auxin responses.
(iv) Histone methylation and plant hormones
Unlike acetylation, methylation does not change the charges of the histone tail. However, it increases the affinity of the histone for negatively charged DNA. The H3K4me3 or H3K27me3 modifications of histone protein are catalyzed by regulatory proteins of the Trithorax-group (Trx-G) and Polycomb-group (Pc-G) (Zhang et al., 2007). Histone methylation has been implicated in the regulation of several hormones. In particular, H3K4me3, a well known epigenetic mark of active transcription, has been shown to mark several genes that affect hormone functions in plants. ARABIDOPSIS HOMOLOG OF TRITHORAX1 (ATX1) directly targets the 9-cis-epoxycarotenoid dioxygenase 3 (NCED3), which plays a key role in the ABA biosynthesis pathway, and regulates its transcriptional activity. Consequently, atx1 knockout shows various ABA-related phenotypes during dehydration stress (Ding et al., 2011).
Pc-G was shown to methylate H3K27 via its histone methyltransferase subunit to maintain the silent state of gene expression in Drosophila (Cao et al., 2002; Czermin et al., 2002). In Arabidopsis, a large number of genes appear to be regulated by H3K27me3, which is catalyzed by PRC2 proteins CURLY LEAF (CLF) and WINGER (SWN) (Lafos et al., 2011). Whole-genome tiling array analysis identified genes involved in auxin biosynthesis, transport and signaling as targets of H3K27me3 (Lafos et al., 2011). For example, H3K27me3 targets 14 AUX/IAA genes directly. Furthermore, all previously reported miRNAs and ta-si RNAs that regulate ARF genes are H3K27me3 targets, suggesting H3K27me3 controls ARF gene expression trough miRNA loci indirectly (Lafos et al., 2011). The gene loci encoding the auxin transporter genes PIN-FORMED 1 (PIN1), PIN4, PIN7 and PIN8 are differentially methylated at H3K27 in leaves and meristems (Lafos et al., 2011). Differential H3K27me3 levels are correlated with tissue-specific PIN1, PIN4, PIN7 and PIN8 expression patterns in the leaf and meristem, suggesting that H3K27me3 is one of the major determinants of tissue-specific PIN gene expression among the PIN gene family (Lafos et al., 2011).
HETEROCHROMATIN PROTEIN 1 (HP1) was originally named because of its protein enrichment in heterochromatic region and is known to be involved in the formation and maintenance of heterochromatin (Allshire et al., 1995). However a role for HP proteins in euchromatic gene regulation has also been demonstrated in mammals and Drosophila (Piacentini et al., 2003; Cryderman et al., 2005; Vakoc et al., 2005). THRMINAL FLOWER2 (TFL2), also known as LHP1, is the only homolog of HP1 in Arabidopsis that regulate specific genes in euchromatin but not in heterochromatin (Nakahigashi et al., 2005). TFL2/LHP1 specifically associates with target genes marked by H3K27me3 to repress the expression of many genes targeted by Polycomb repressive complex 2 (PRC2) (Turck et al., 2007; Exner et al., 2009). Interestingly, the tfl2 KO mutant showed weak expression of auxin biosynthesis genes, such as YUCCA, and had elevated endogenous auxin contents (Rizzardi et al., 2011). Moreover, TFL2/LHP1 directly target YUCCA genes in an auxin dependent manner suggesting the potential function of TFL2/LHP1 in auxin regulated positive transcriptional control of YUCCA genes (Rizzardi et al., 2011). However the association of TFL2/LHP1 enrichment in YUCCA loci and its positive regulation of expression were not observed in all YUCCA genes (Rizzardi et al., 2011). The function of HP1 protein seemed to be controlled by complex mechanisms, many of which still remain unknown in Arabidopsis. It will be necessary to uncover the precise mechanism of auxin regulation of TFL2/LHP1 enrichment in YUCCA loci.
The CHD3 chromatin remodeling factor PKL regulates plant hormone signaling
Chromatin remodeling is another major machinery that determines chromatin structure and gene expression in yeast, mammal and plants. Chromatin remodeling factors can directly change nucleosome structures in an ATP-dependent manner by altering histone-DNA interactions that affect the accessibility of nucleosomal DNA (Clapier and Cairns., 2009). Thus, it is anticipated that chromatin remodeling factors may regulate responses to various hormones that regulate gene expression. Genetic analysis suggested that BRAHAMA (BRM), which encodes a homolog of the conserved SWI/SNF chromatin remodeling ATPase, participates in the modulation of GA and CK (Cytokinins) responses (Archacki et al., 2013; Efroni et al., 2013), and in auxin distribution by controlling the expression of several PIN genes. ChIP assays showed that PIN 2, 3, 4, and 7 are direct targets of BRM (Yang et al., 2015). Furthermore, SWI3B interacts with several proteins involved in ABA responses, such as HYPERSENSITIVE TO ABA1 (HAB1), ABA INSENSITIVE 1 (ABI1), and ABI2, and modulates ABA signaling (Saez et al., 2008). Moreover, SWI3C interacts with DELLA proteins and knockout mutant shows altered responses to ethylene, ABA, BR and GA (Sarnowska et al., 2013). These findings strongly suggest important roles of chromatin remodeling in plant hormone signaling.
Arabidopsis PICKLE (PKL), a homolog of CHD3/Mi-2 ATP-dependent chromatin remodeling factors in animals, prevents inadvertent activation of the embryonic programs in post germination processes (Eshed et al., 1999; Ogas et al., 1997, 1999) and affects responses to several hormones in Arabidopsis. First, PKL is required for normal GA responsiveness and biosynthesis (Henderson et al., 2004). Second, ABI3 and ABI5 gene expression is up-regulated in PKL knockout, leading to ABA hypersensitive phenotype (Perruc et al., 2007). Third, loss of function mutations in PKL cause cytokinin (CK) hypersensitive phenotype and enhance growth and greening of callus in detached hypocotyls independent of CK (Furuta et al., 2011). In addition, the PKL function in auxin responses has also been suggested by suppressor mutations of the solitary root (slr) mutant. slr doesn’t produce any lateral root (LR), due to a dominant mutation in the SLR/IAA14 gene, which causes the stabilization of the SLR/IAA14 protein and blocks auxin-inducible LR formation (Fukaki et al., 2002). Interestingly, mutations in PKL abolished the auxin insensitive phenotype for LR formation in slr mutant (Fukaki et al., 2006). In animals, HDACs are found in the Mi-2/NuRD complex, and Mi-2/NuRD is thought to act as transcriptional repressors (Ahringer 2000). In agreement with this, treatment with the histone deacetylase inbibitor TSA induces LR formation in slr mutant (Fukaki et al., 2006). These effects of pkl mutations and TSA treatment for LR induction in the slr mutant require ARF7 and ARF19 functions. Therefore, it is likely that PKL controls auxin/ARF-dependent cell fate specification and cell cycle progression through changes in histone modifications during the LR formation process (Fukaki et al., 2006). Key aspects to elucidate include whether the PKL-mediated chromatin organization that suppresses auxin-induced gene expression is directly regulated by auxin signaling, and whether PKL and TPL-AUX/IAA mediated repressor complexes for auxin signaling act independently or synergistically (Figure 2A).
Figure 2. (A) TPL mediated EAR repressome and PKL chromatin remodeling factor control plant hormone signaling.
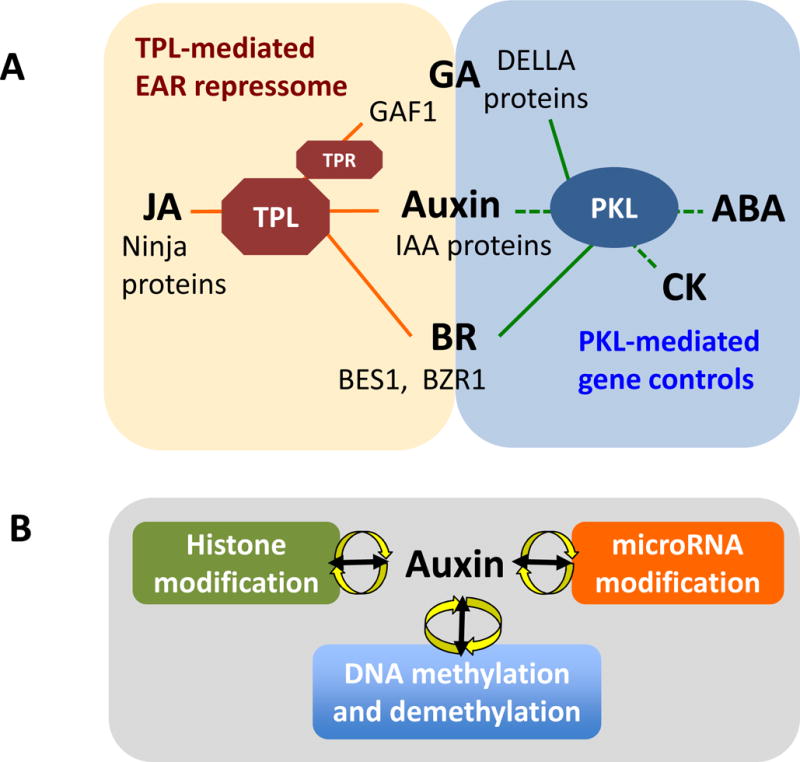
Some overlapping direct or indirect functional association are in the middle. Solid lines indicate direct interaction and dotted lines indicate functional association or regulation. Ninja protein (Pauwels et al., 2010), GAF1 (Fukazawa et al., 2014), IAA proteins (Fukaki et al., 2006; Long et al., 2006), BES1 (Ryu et al., 2014), BZR1 (Oh et al., 2014) and DELLA proteins (Zhang et al., 2104). (B) A hypothetical auxin-linked epigenetic regulation loop.
It is clear that PKL has important roles in most plant hormone responses, but it is unclear how it regulates a specific hormone signaling pathway. In animals, interactions with more specialized proteins allow Mi2/NuRD complexes to have a large array of biological functions. MTA3 is a cell type specific subunit of Mi2/NuRD and interacts with master regulator for cell fate determination (Fujita et al., 2004). PKL may regulate a common molecular machinery for each plant hormonal signaling depending on the plant hormone specific factors they associate with (Figure 2A).
A recent publication has demonstrated a role for PKL in the regulation of gene expression mediated by GA and BRs (Zhang et al., 2014). The authors showed that PKL functions not only in repression, but also in the activation of gene expression. The basic helix-loop-helix transcription factor PHYTOCHROME INTERACTING FACTOR (PIF) family proteins accumulate in the nucleus under dark conditions to promote gene expression that is required for cell elongation (Leivar et al., 2008; Shin et al., 2009). Both BR and GA promote the cell elongation related gene expression. After BRs are perceived by BRASSINOSTEROID INSENSITIVE 1 (BRI1), the downstream signaling cascade dephosphorylates and translocates BZR1 from the cytosol to the nucleus, allowing BZR1 to promote the expression of cell elongation related genes, such as INDOLE-3-ACETIC ACID INDUCIBLE 19 (IAA19) and PACLOBUTRAZOL RESISTANCE1 (PRE1) (Nakamura et al., 2003; Zhang et al., 2009; Kim and Wang, 2010; Wang et al., 2012). Arabidopsis have 5 DELLA family proteins including GIBBERELLIC ACID INSENSITIVE (GAI), REPRESSOR OF ga1-3 (RGA), RGA LIKE 1(RGL1), RGL2 and RGL3, that negatively regulate cell elongation. GA promotes the degradation of DELLA proteins through proteasome activity, which result in cell elongation (Sun and Gubler., 2004; Silverstone et al., 2001; Sun., 2011). PKL negatively regulates photomorphogenesis (Jing and Lin., 2013). PKL physically interacts with PIF3 and BZR1 and represses H3K27me3 at the promoter of cell elongation related gene, thus promotes gene expression. Conversely, DELLA proteins suppress the expression of cell elongation related genes by inhibiting the binding between PKL and PIF3, decreasing the accumulation of the PKL protein in the promoter of these genes. Moreover, GA and BR treatments promote PKL accumulation at the promoter of cell elongation genes and reduce H3K27me3 in the promoter (Zhang et al., 2014). Taken together, these findings suggest that the interaction between PKL and different partner proteins enables the integration of different signaling pathways to control appropriate histone modification and gene expression levels (repression or activation) in response to external and internal signals. Future studies should identify specific partner proteins of PKL for each plant hormone signaling pathway in order to understand how PKL functions to coordinate multiple hormone signaling pathways (Figure 2A).
MicroRNAs modulate multiple regulatory layers of plant hormone signaling
Small RNAs are commonly involved in the epigenetic regulation of gene expression. Plant microRNAs (miRNAs) are small endogenous non-coding RNA that are generated from the processing of local hairpin precursor structures. Mature miRNAs can target mRNAs for cleavage, leading to the destabilization of target mRNAs and thereby suppressing specific gene expression (Bartel et al., 2004; He and Hannon., 2004; Bologna and Voinnet., 2014). MICRORNA 393 (miR393) targets TIR1, AFB1, AFB2 and AFB3, and thus modulates auxin sensitivity in plants (Jones-Rhoades and Bartel 2004; Navarro et al., 2006; Chen et al., 2011). miR393 negatively regulates TIR1, AFB1, AFB2 and AFB3 in response to pathogen attack. Overexpression of miR393 leads to a decrease in TIR1 transcript levels and enhances bacterial resistance (Navarro et al., 2006). miR393 targets all auxin receptor F-box genes, TIR1/AFBs, suggesting that this miRNA has evolved as a conserved mechanism to regulate auxin responses. However, miR393 might have also evolved as a mechanism to regulate specific TIR1/AFBs under certain conditions. It was shown that miR393 expression is induced by nitrate treatment, but only changes in the AFB3 transcript level are negatively correlated with miR393 expression level after nitrate treatment. It is likely that additional factor(s) are needed to specifically destabilize the AFB3 transcripts by miR393 in response to nitrate treatments (Vidal et al., 2010), though the mechanism underlying the specificity remains unknown. Multiple components of the auxin signaling pathway are under the control of miRNAs. In addition to the TIR/AFB family genes, ARF6 and ARF8 are targets of miR167, while RF10, ARF16, and ARF17 are targeted by miR160 (Rhoades et al., 2002; Mallory et al., 2005; Wu et al., 2006). miR160 and its target gene, ARF10, play a role for shoot regeneration from somatic culture cells that is mediated by a balance between auxin and CK. Although ARF10 overexpression does not have an impact on shoot regeneration, miR160 resistant mARF10 transgenic plants have greater shoot regeneration ability than wild type. Thus, the abundance of ARF10 transcripts seems to be mainly controlled by miR160, indicating a precise regulatory mechanism during regeneration processes (Qiao et al., 2012; Qiao and Xiang, 2013).
AuxRE motifs were found in the promoters of several miRNAs that might target auxin-related genes in rice, suggesting a potential role for miRNAs in the auxin signaling feedback loop (Meng et al., 2010). Feedback regulations between miRNA and TF have been demonstrated in animals (Johnston et al., 2005; Tsang et al., 2007). In Arabidopsis, positive and negative feedback regulation mechanisms were observed in miR390 and ARFs involved in trans-acting short-interfering RNA (ta-siRNA) mediated LR growth (Marin et al., 2010). ta-siRNAs are plant specific small RNAs, whose biogenesis requires the siRNA pathway. However, their production requires the cleavage of TRANSACTING SIRNA (TAS) transcripts by action of specific microRNA-ARGONAUTE 7 (AGO7) complex, and unlike miRNA, the activity of RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) and SUPPRESSOR OF GENE SILENCING 3 (SGS3) are necessary for the subsequent production of the 21 nucleotide ta-siRNA mediated by DCL4, DICER-LIKE 4 (DCL4) (Allen et al., 2005; Gasciolli et al., 2005; Peragine et al., 2004; Vazquez et al., 2004; Xie et al., 2005; Yoshikawa et al., 2005). TAS3 precursor requires miR390 to produce ta-siRNA (ta-siARF) and target ARF2, ARF3 and ARF4 for transcript cleavage. ARF2, 3 and 4 inhibit LR growth. Auxin induces miR390 production and consequently promotes LR growth by the reduction of ARF2, 3, and 4 expression through the ta-siRNA function (Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Hunter et al., 2006; Marin et al., 2010). Interestingly, the level of auxin inducible miR390 expression was significantly higher in both arf4 knockout mutant and transgenic line that carried ta-si ARF resistant form of ARF3 gene compared with that of control, indicating that the expression levels of these ARF genes and miR390 accumulation are connected by both negative- and positive-feedback loops. The authors further showed exciting data that the negative feedback loops between miR390 and ARF4 play a pivotal role for spatial regulation of miR390 expression during LR growth (Marin et al., 2010). miRNA mediated feedback regulation was also observed between CK and its activator HD-ZIPII TF PABULOSA (PHB). The destabilization of PHB by miR165 was negatively regulated by CK in roots (Dello Ioio et al., 2012). It is conceivable that feedback loop mechanisms between miRNA and TF may be conserved in plants.
One notable roles of ta-siARF has been demonstrated in leaf development. ARF3 is required for abaxial fate in leaf development (Allen et al., 2005; Pekker et al., 2005). The expression of pARF3:ARF3-GUS is present mainly on the abaxial side, and ta-siARF resistant pARF3:ARF3m-GUS is expressed ubiquitously in young leaf primordia. ARF3 mRNA expression in the adaxial side is supposed to be destabilized by ta-siARF. Moreover, the gradient of ta-siARF expression from the adaxial to abaxial sides has been observed in older leaves. The expression of essential components for ta-siARF biogenesis, AGO7 and TAS, is limited to the central region of the adaxial-most cell layers of leaves, suggesting that ta-siARF moves from the adaxial to the abaxial side to create a gradient of ARF3 expression (Chitwood et al., 2009). The moving ta-siARF provides a new regulatory layer for the auxin control of leaf polarity, which is different from well-established roles of polar auxin transport and distribution. Intercellular movement of small RNAs to regulate hormone action appears to be a common mechanism. miR165 and miR166, which target CK-regulated genes PHABULOSA (PHB) (Dello Ioio et al., 2012; Carlsbecker et al., 2010), also move from the endodermis to the stele, creating the gradient of target class III homeodomain-leucin zipper (HD-ZIPIII), PHABULOSA (PHB), REVOLUTA, ATHB8 and ATHB15 expression, although the movement is limited in several cell layers in this case (Prigge et al., 2005; Carlsbecker et al., 2010). The study of moving small RNAs may provide a new paradigm of miRNA-mediated gradients of plant hormone signaling.
As discussed above, the studies of miRNA shed light on additional regulatory layers of hormone signaling/action. A recent report proposes a molecular basis for a new concept, “microRNA timer”, involved in regenerative capacity (Zhang et al., 2015). Plant cells from a piece of differentiated tissue have the competence of regeneration. In general, cells of young plants have higher competence for regeneration compared with cells from old plant tissues. miR156 targets SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) TFs, and thus regulates the juvenile-to-adult phase transition (Chuck et al., 2007; Wu et al., 2009; Wang et al., 2011; Bergonzi et al., 2013; Zhou et al., 2013). These authors found that miR156 expression is abundant in early leaves and targets SPL9 transcript. SPL9 protein can interact with B-type ARR protein, ARR2, and affect its transcriptional activity. Type-B ARRs are positive regulators of cytokinin signaling that control cytokinin-regulated gene expression (Hwang and Sheen, 2001; Sakai et al., 2001; Tajima et al., 2004; Mason et al., 2005; Taniguchi et al., 2007; Yokoyama et al., 2007). Interestingly, B-type arr knockout mutants show decreased regeneration capacity. These results suggest that SPL9 protein negatively regulates the expression of CK responsible genes trough its interaction with B-type ARABIDOPSIS RESPONSE REGULATORs (ARR). Moreover, SPL9 expression becomes higher with age due to the reduction in miR156 abundance, and therefore early leaves exhibit a higher regeneration rate than late leaves (Zhang et al., 2015). An interesting question is what regulates the decrease in miR156 expression during the plant life cycle: accumulated cellular damage, deregulation of the epigenome, or specific developmental mechanism?
Does hormone signaling regulate DNA methylation or demethylation?
DNA methylation is a relatively stable but reversible epigenetic mark regulating gene expression and suppressing transposon activities in plants and animals. DNA methylation occurs at the three different cytosine sequence contexts (CG, CHG and CHH, where H is C, A or T) in plants (Henderson and Jacobsen., 2007). DNA cytosine methylation in the CG and CHG contexts is maintained by METHYLTRANSFERASE1 (MET1) that is the ortholog of mammalian DNA methyltransferase Dnmt1, and the plant specific DNA methyltransferase CHROMOMETHYLASE 3 (CMT3), respectively (Finnegan and Dennis., 1933; Jackson et al., 2002; Kankel et al., 2003; Saze et al., 2003). The maintenance of CHH sequence contexts are controlled by DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2) and CMT2 (Law and Jacobsen., 2010; Zemech et al., 2013; Stroud et al., 2014). De novo DNA cytosine methylation requires 24-nt small interfering RNAs (siRNA) that direct DRM2 to methylate all three sequence contexts via the RNA-directed DNA methylation (RdDM) pathway (Matzke et al., 2009, Zhang et al., 2012). Active DNA demethylation depends on the function of the REPRESSOR OF SILENCING1 (ROS1) family genes that encode 5-methylcytosine DNA glycosylase/lyases. ROS1 and its paralogues, DEMETER-like 2 (DML2) and DEMETER-like 3 (DML3) are required for the prevention of hypermethylation at thousands of genomic regions and play a critical role in the regulation of transgenes, transposable elements and some endogenous gene expression (Gong et al., 2002; Penterman et al., 2007; Zhu et al., 2007; Qian et al., 2012; Yamamuro et al., 2014). DME is expressed in the central cell, and its function is required for active DNA demethylation of maternal allele and gene imprinting in endosperm development (Gehring et al., 2006).
Some studies suggest a potential link between the maintenance of DNA methylation and plant hormone signaling. met1 null allele embryos exhibit a wide range of developmental phenotypes, including delayed transition from vegetative phase to reproductive phase (Soppe et al., 2000; Kankel et al., 2003) and embryo abnormalities, which are reminiscent of mutants that have defects in auxin distribution (Xiao et al., 2006). Indeed the DR5:GFP transgene, which has been used to report auxin-induced gene transcription (Ottenschläger et al., 2003; Friml et al., 2002a; Friml et al., 2002b), and PIN1, which encodes an auxin efflux carrier and is required for auxin distribution in the early embryo (Friml 2003, Weijers et al., 2005), are expressed abnormally in met1 null allele abnormal embryos. However, DNA methylation at the PIN1 gene locus was not observed in neither wild type nor met1 null allele, indicating that although an auxin gradient formation in early embryos requires proper MET1-meintained DNA methylation level, MET1 influences PIN1 expression indirectly (Xiao et al., 2006).
WUSCHEL (WUS) encodes a homeodomain containing TF that plays an important role as a master gene for both stem cell fate determinations in the shoot apical meristem. Both auxin and CK affect WUS function and the stem cell niche, and the WUS gene expression is regulated by auxin and CK (Laux et al., 1996; Mayer et al., 1998; Gallois et al., 2004; Gordon et al., 2007; Chen et al., 2010; Zhao et al., 2010). WUS also plays an important role in the specification of the stem cell organizing center during de novo shoot regeneration. met1 knockout callus starts greening and regenerating faster than that of WT on shoot induction medium. During de novo shoot regeneration, the DNA methylation status around the WUS gene is gradually reduced, eventually leading to active transcription of WUS gene in the organizing center. WUS gene expression is also controlled by dynamic changes of histone modification levels. Epigenetic marks of active transcription, H3K4me3 and H3Kac9, were increased, whereas the repressive mark, H3K9me2 was reduced, consistent with WUS expression during de novo shoot regeneration. These observations suggest that the WUS-mediated specification of organizing centers seems to be regulated by complex epigenetic mechanisms (Li et al., 2011). In addition, it has been shown that WUS expression is controlled by several epigenetic regulators in the organizing center of the shoot apical meristem (Kaya et al., 2001; Takeda et al., 2004; Kwon et al., 2005). However, the connection between DNA methylation and these epigenetic regulators involved in the WUS expression is currently unclear.
A recent study has linked DNA demethylation to auxin-mediated chromatin opening and gene expression. PINOID (PID) gene encodes an AGC family Ser/Thr protein kinase (Christensen et al., 2000), and controls polarized localization patterns of PINs (Friml et al., 2004). The expression of PID gene is regulated by chromatin loop from PID promoter to APOLO locus that is located 5,148 bp upstream of the PID. APOLO encodes a non-coding APOLO RNA (AUXIN REGULATED PROMOTER LOOP RNA) that influences PID expression directly (Ariel et al., 2014). APOLO RNAi knockdown line displayed a delayed response to gravitropism, a phenotype that is similar to pid knockout mutant (Sukumar et al., 2009; Ariel et al., 2014). Auxin treatment can induce both PID and APOLO expression. Remarkably, the APOLO RNA directly binds to the LHP1 protein, which is associated with genes marked by H3K27me3 in Arabidopsis (Gaudin et al., 2001; Turck et al., 2007; Hennig and Derkacheva, 2009), and this RNA-protein complex is physically associated with the APOLO locus through an LHP-mediated chromatin loop. Interestingly, auxin treatment rapidly reduces the chromatin loop formation and the direct binding of the APOLO RNA and LHP1 protein. Consistent with these findings, a decrease of repressive marks, H3K27me3 and H3K9me2, and accumulation of LHP1 protein at the PID-APOLO locus was observed in response to auxin. Furthermore, the authors showed that active DNA demethyaltion also contributes to the auxin-mediated dynamics of the chromatin loop, indicating the existence of an auxin-mediated complex regulation at the locus (Ariel et al., 2014). The rdd mutant, a triple mutant of ros1, dml2 and dml3 (Penterman et al., 2007), displayed enhanced basal loop formation. Moreover, dynamic changes in both PID and APOLO expression, chromatin loop dynamic and DNA demethylation in response to auxin treatment were significantly altered in triple knockout of the ROS1 family genes (Ariel et al., 2014). Although the significance of auxin inducible active DNA demethylation at the APOLO locus for PID function in the polar localization of PIN2 is still unknown, these observations show that ROS1 family genes-mediated active DNA demethylation plays a role for chromatin loop opening and PID gene expression in response to auxin. The authors proposed an attractive possibility that the chromatin loop formation may affect the accessibility of ARFs TF to the promoter of PID and APOLO for proper auxin responsive expression (Ariel et al., 2014). The function of active DNA methylation in chromatin loop opening seemed to be a very rare event. However, the response to auxin in chromatin loop opening was very quick and dynamic (Ariel et al., 2014). Given the association of auxin action with both ROS1 family-mediated active DNA demethylation and changes in chromatin structures, it would be of interest to determine whether auxin signaling directly regulates ROS1 activity and targeting to the chromatin loop.
Conclusions
In the past decade, extensive genetic mutant studied have revealed that epigenetic controls contribute to the regulation of plant hormonal signaling/action in various developmental and physiological processes. These findings give us exciting prospects of considering the unexpected complex and layered but ordered regulatory mechanisms modulating hormone actions at the epigenetic levels in addition to biochemical and cellular levels of post-transcriptional controls. Remarkably, auxin signaling is modulated by all of epigenetic modification machineries discussed in this review. Moreover, some clear lines of evidence reveal the existence of auxin-linked epigenetic loop (Figure 2B). It would be interesting to see if these observations can be extended to other plant hormones. In this review, we discussed the involvement of different epigenetic modifications in the hormone actions, however, as we discussed in the section of DNA methylation/demethylation, these regulatory mechanisms do not exist in solitary in the regulatory network, but are interconnected. In addition, given these epigenetic modifications are reversible reactions, it is likely that these modifications happen simultaneously at the same locus, thus may affect each other synergistically or antagonistically, in response to numerous external or internal signals.
Plant hormones often display crosstalk with other plant hormones, and these signals are integrated as gene regulatory networks. We discussed that some plant hormones, such as auxin, GA and BR can modulate epigenomes. The competing or coordinating rewriting of epigenomes might be one crosstalk point among different plant hormones. In addition, the common critical regulatory factors in multiple plant hormones signaling such as PKL and TPL could be key factors to reveal the new paradoxes of plant hormone crosstalks. The studies of conventional plant hormone signaling pathways are well advanced in Arabidopsis. This advance should aid in our future studies aimed at understanding how these conventional hormone signaling pathways are integrated into the new layers of hormone regulations at various epigenetic levels.
Short summary.
This review focuses on recent works on the interrelationship between mechanisms of epigenetic control and the actions of plant hormone.
Acknowledgments
We thank Irene Lavegi for her critical comments on and helpful editing of this manuscript. The work is supported by funds from Fujian Agriculture and Forestry University, by grants from US National Institutes of Health Grants R01GM070795 and R01GM059138 and by the Chinese Academy of Sciences to J.-K.Z., and by grant from the US National Institute of General Medical Sciences (R01GM081451 and R01GM100130) to Z.Y.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Ahringer J. NuRD and SIN3: histone deacetylase complexes in development. Trends in Genetics. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- Alinsug MV, Yu CW, Wu K. Phylogenetic analysis. Subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plant. BMC Plant Biol. 2009;9:37. doi: 10.1186/1471-2229-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR. Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci U S A. 2003;100:2992–2997. doi: 10.1073/pnas.0438070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, Zhang S, Ecker JR, Guo H. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell. 2010;22:2384–2401. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzola JM, Sieberer T, Ortbauer M, Butt H, Korbei B, Weinhofer I, Müllner AE, Luschnig C. Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc Nat Acad Sci USA. 2010;107:10308–10313. doi: 10.1073/pnas.0913918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archacki R, Buszewicz D, Sarnowski TJ, Sarnowska E, Rolicka AT, Tohge T, Fernie AR, Jikumaru Y, Kotlinski M, Iwanicka-Nowicka R, Kalisiak K, Patryn J, Halibart-Puzio J, Kamiya Y, Davis SJ, Koblowska MK, Jerzmanowski A. BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in arabidopsis. PLoS One. 2013;8:e58588. doi: 10.1371/journal.pone.0058588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, Crespi M. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell. 2014;55:383–396. doi: 10.1016/j.molcel.2014.06.011. [DOI] [PubMed] [Google Scholar]
- Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. Micro RNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Benhamed M, Bertrand C, Servet C, Zhou DX. Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell. 2006;18:2893–2903. doi: 10.1105/tpc.106.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M, Martin-Magniette ML, Taconnat L, Bitton F, Servet C, De Clercq R, De Meyer B, Buysschaert C, Rombauts S, Villarroel R, Aubourg S, Beynon J, Bhalerao RP, Coupland G, Gruissem W, Menke FL, Weisshaar B, Renou JP, Zhou DX, Hilson P. Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 2008;56:493–504. doi: 10.1111/j.1365-313X.2008.03606.x. [DOI] [PubMed] [Google Scholar]
- Bergonzi S, Albani MC, Ver Loren, van Themaat E, Nordström KJ, Wang R, Schneeberger K, Moerland PD, Coupland G. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science. 2013;340:1094–1097. doi: 10.1126/science.1234116. [DOI] [PubMed] [Google Scholar]
- Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol. 2014;65:473–503. doi: 10.1146/annurev-arplant-050213-035728. [DOI] [PubMed] [Google Scholar]
- Bowen AJ, Gonzalez D, Mullins JG, Bhatt AM, Martinez A, Conlan RS. PAH-domain-specific interactions of the Arabidopsis transcription coregulator SIN3-LIKE1 (SNL1) with telomere-binding protein 1 and ALWAYS EARLY2 Myb-DNA binding factors. J Mol Biol. 2010;395:937–949. doi: 10.1016/j.jmb.2009.11.065. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, Campilho A, Sebastian J, Bowman JL, Helariutta Y, Benfey PN. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 2012;158:423–438. doi: 10.1104/pp.111.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Tian Roles of dynamic and reversible histone acethylation in plant development and polyploidy. Biochem Biophys Acta. 2007;1769:29–307. doi: 10.1016/j.bbaexp.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-T, Wu K. Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signaling & Behavior. 2010;5:1318–1320. doi: 10.4161/psb.5.10.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-T, Luo M, Wang Y-Y, Wu K. Involvement od Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J Exp Bot. 2010;61:3345–3353. doi: 10.1093/jxb/erq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Bao ML, Sun YZ, Yang YJ, Xu XH, Wang JH, Han N, Bian HW, Zhu MY. Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol Biol. 2011;77:619–629. doi: 10.1007/s11103-011-9838-1. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhang H, Jablonowski D, Zhou X, Ren X, Hong X, Schaffrath R, Zhu JK, Gong Z. Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol Cell Biol. 2006;26:6902–6912. doi: 10.1128/MCB.00433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC. Pattern formation via small RNA mobility. Genes Dev. 2009;23:549–554. doi: 10.1101/gad.1770009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Dagenais N, Chory J, Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell. 2000;100:469–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Cryderman DE, Grade SK, Li Y, Fanti L, Pimpinelli S, Wallrath LL. Role of Drosophila HP1 in euchromatic gene expression. Dev Dyn. 2005;232:767–774. doi: 10.1002/dvdy.20310. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Galinha C, Fletcher AG, Grigg SP, Molnar A, Willemsen V, Scheres B, Sabatini S, Baulcombe D, Maini PK, Tsiantis M. A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Curr Biol. 2012;22:1699–1704. doi: 10.1016/j.cub.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;26:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Ding Y, Avramova Z, Fromm M. The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J. 2011;66:735–744. doi: 10.1111/j.1365-313X.2011.04534.x. [DOI] [PubMed] [Google Scholar]
- Ding Y, Mou Z. Elongator and its epigenetic role in plant development and responses to abiotic and biotic stresses. Front Plant Sci. 2015;6:296. doi: 10.3389/fpls.2015.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci U S A. 2012;109:E2183–91. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Han SK, Kim HJ, Wu MF, Steiner E, Birnbaum KD, Hong JC, Eshed Y, Wagner D. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev Cell. 2013;24:438–445. doi: 10.1016/j.devcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emes RD, Ponting CP. A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum Mol Genet. 2001;15:2813–2820. doi: 10.1093/hmg/10.24.2813. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell. 1999;99:199–209. doi: 10.1016/s0092-8674(00)81651-7. [DOI] [PubMed] [Google Scholar]
- Exner V, Aichinger E, Shu H, Wildhaber T, Alfarano P, Caflisch A, Gruissem W, Köhler C, Hennig L. The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development. PLoS One. 2009;4(4):e5335. doi: 10.1371/journal.pone.0005335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 1993;21:2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, Palme K. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell. 2002;108:661–673. doi: 10.1016/s0092-8674(02)00656-6. [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature. 2002;415:806–809. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- Friml J. Auxin transport - shaping the plant. Curr Opin Plant Biol. 2003;6:7–12. doi: 10.1016/s1369526602000031. [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, Hooykaas PJ, Palme K, Offringa R. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Taniguchi N, Tasaka M. PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J. 2006;48:380–389. doi: 10.1111/j.1365-313X.2006.02882.x. [DOI] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Fukazawa J, Teramurac H, Murakoshic S, Nasunoc K, Nishidac N, Itoa T, Yoshidac M, Kamiya Y, Yamaguchi S, Takahashia Yohsuke. DELLAs Function as Coactivators of GAI-ASSOCIATED FACTOR1 in Regulation of Gibberellin Homeostasis and Signaling in Arabidopsis. Plant Cell. 2014;26:2920–2938. doi: 10.1105/tpc.114.125690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- Furuta K, Kubo M, Sano K, Demura T, Fukuda H, Liu YG, Shibata D, Kakimoto T. The CKH2/PKL chromatin remodeling factor negatively regulates cytokinin responses in Arabidopsis calli. Plant Cell Physiol. 2011;52:618–628. doi: 10.1093/pcp/pcr022. [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- Gallois JL, Nora FR, Mizukami Y, Sablowski R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 18:375–380. doi: 10.1101/gad.291204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing transacting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Gaudin V, Libault M, Pouteau S, Juul T, Zhao G, Lefebvre D, Grandjean O. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development. 2001;128:4847–458. doi: 10.1242/dev.128.23.4847. [DOI] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez D, Bowen AJ, Carroll TS, Conlan RS. The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol. 2007;27:5306–5315. doi: 10.1128/MCB.01912-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM. Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development. 2007;134:3539–3548. doi: 10.1242/dev.010298. [DOI] [PubMed] [Google Scholar]
- Grzenda A, Lomberk G, Zhang JS, Urrutia R. Sin3: master scaffold and transcriptional corepressor. Biochim Biophys Acta. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci. 2012;190:82–88. doi: 10.1016/j.plantsci.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. Combined networks regulating seed maturation. Trends Plant Sci. 2007;12:294–300. doi: 10.1016/j.tplants.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Baurle I, Kientz M, Jü rgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Henderson JT, Li HC, Rider SD, Mordhorst AP, Romero-Severson J, Cheng JC, Robey J, Sung ZR, de Vries SC, Ogas J. PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol. 2004;134:995–1005. doi: 10.1104/pp.103.030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–24. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Hennig L, Derkacheva M. Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 2009;25:414–423. doi: 10.1016/j.tig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Hollender C, Liu ZC. Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol. 2008;50:875–885. doi: 10.1111/j.1744-7909.2008.00704.x. [DOI] [PubMed] [Google Scholar]
- Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutiérrez-Nava M, Poethig SR. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. Two-component circuitry in Arabidopsis signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- Jaeger G, Solano R, Goossens A. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Jing Y, Lin R. PICKLE is a repressor in seedling de-etiolation pathway. Plant Signal Behav. 2013;8:e25026-1-3. doi: 10.4161/psb.25026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci U S A. 2005;102:12449–1254. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–142. doi: 10.1016/s0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;26:446–4451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Kim JM, To TK, Ishida J, Morosawa T, Kawashima M, Matsui A, Toyoda T, Kimura H, Shinozaki K, Seki M. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:1580–1589. doi: 10.1093/pcp/pcn133. [DOI] [PubMed] [Google Scholar]
- Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- Kornet N, Scheres B. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell. 2009;21:1070–1079. doi: 10.1105/tpc.108.065300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CS, Chen C, Wagner D. WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 2005;19:992–1003. doi: 10.1101/gad.1276305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D. Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 2011;7:e1002040. doi: 10.1371/journal.pgen.1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acethylase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Liu H, Cheng ZJ, Su YH, Han HN, Zhang Y, Zhang XS. DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet. 2011;7:e1002243. doi: 10.1371/journal.pgen.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A, Leubner-Metzger G. Beyond gibberellins and abscisic acid: how ethylene and jasmonates control seed germination. Plant Cell Rep. 2012;31:253–270. doi: 10.1007/s00299-011-1180-1. [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Woody S, Poethig S, Meyerowitz EM, Barton MK. Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development. 2002;129:2297–2306. doi: 10.1242/dev.129.12.2797. [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312:1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Pavangadkar KA, Thomashow MF, Triezenberg SJ. Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochem Biophys Acta. 2006;1759:69–79. doi: 10.1016/j.bbaexp.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JRS, Shaller GE. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17:3007–3018. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–76. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. 2010;22:1104–1117. doi: 10.1105/tpc.109.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- Meng Y, Chen D, Ma X, Mao C, Cao J, Wu P, Chen M. Mechanisms of microRNA-mediated auxin signaling inferred from the rice mutant osaxr. Plant Signal Behav. 2010;5:252–254. doi: 10.4161/psb.5.3.10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahigashi K, Jasencakova Z, Schubert I, Goto K. The Arabidopsis heterochromatin protein1 homolog (TERMINAL FLOWER2) silences genes within the euchromatic region but not genes positioned in heterochromatin. Plant Cell Physiol. 2005;46:1747–1756. doi: 10.1093/pcp/pci195. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S. Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 2003;133:1843–1853. doi: 10.1104/pp.103.030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Nelissen H, Fleury D, Bruno L, Robles P, De Veylder L, Traas J, Micol JL, Van Montagu M, Inzé D, Van Lijsebettens M. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc Natl Acad Sci U S A. 2005;102:7754–7759. doi: 10.1073/pnas.0502600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Cheng JC, Sung ZR, Somerville C. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science. 1997;277:91–94. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:13839–1344. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Ryu H, Hwang I, Wang ZY. TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat Commun. 2014;18(5):4140. doi: 10.1038/ncomms5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci U S A. 2003;100:2987–2991. doi: 10.1073/pnas.0437936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. Analysis of histone acetylase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, García-Casado G, Witters E, Inzé D, Long JA, De Jaeger G, Solano R, Goossens A. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci U S A. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of transacting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez AC, Goossens A. Jasmonate signalling: a copycat of auxin signalling? Plant Cell Environ. 2013;36:2071–2084. doi: 10.1111/pce.12121. [DOI] [PubMed] [Google Scholar]
- Perruc E, Kinoshita N, Lopez-Molina L. The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 2007;52:927–936. doi: 10.1111/j.1365-313X.2007.03288.x. [DOI] [PubMed] [Google Scholar]
- Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S, Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Miki D, Zhang H, Liu Y, Zhang X, Tang K, Kan Y, La H, Li X, Li S, Zhu X, Shi X, Zhang K, Pontes O, Chen X, Liu R, Gong Z, Zhu JK. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336:1445–1448. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao M, Zhao Z, Song Y, Liu Z, Cao L, Yu Y, Li S, Xiang F. Proper regeneration from in vitro cultured Arabidopsis thaliana requires the microRNA-directed action of an auxin response factor. Plant J. 2012;71:14–22. doi: 10.1111/j.1365-313X.2012.04944.x. [DOI] [PubMed] [Google Scholar]
- Qiao M, Xiang F. A set of Arabidopsis thaliana miRNAs involve shoot regeneration in vitro. Plant Signal Behav. 2013;8:e23479-1-3. doi: 10.4161/psb.23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Rizzardi K, Landberg K, Nilsson L, Ljung K, Sundås-Larsson A. TFL2/LHP1 is involved in auxin biosynthesis through positive regulation of YUCCA genes. Plant J. 2011;65:897–906. doi: 10.1111/j.1365-313X.2010.04470.x. [DOI] [PubMed] [Google Scholar]
- Ryu H, Cho H, Bae W, Hwang I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat Commun. 2014;18(5):4138. doi: 10.1038/ncomms5138. [DOI] [PubMed] [Google Scholar]
- Saez A, Rodrigues A, Santiago J, Rubio S, Rodriguez PL. HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell. 2008;20:2972–2988. doi: 10.1105/tpc.107.056705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowska EA, Rolicka AT, Bucior E, Cwiek P, Tohge T, Fernie AR, Jikumaru Y, Kamiya Y, Franzen R, Schmelzer E, Porri A, Sacharowski S, Gratkowska DM, Zugaj DL, Taff A, Zalewska A, Archacki R, Davis SJ, Coupland G, Koncz C, Jerzmanowski A, Sarnowski TJ. DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiol. 2013;163:305–317. doi: 10.1104/pp.113.223933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A. Arabidopsis ARR1 is a transcription factor for genes immediately responsive to cytokinins. Science. 2001;294:1519–1521. doi: 10.1126/science.1065201. [DOI] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell. 2015;27:9–19. doi: 10.1105/tpc.114.133744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- Servet C, Conde e Silva N, Zhou DX. Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol Plant. 2010;3:670–677. doi: 10.1093/mp/ssq018. [DOI] [PubMed] [Google Scholar]
- Sieberer T, Hauser MT, Seifert GJ, Luschnig C. PROPORZ1, a putative Arabidopsis transcriptional adaptor protein, mediates auxin and cytokinin signals in the control of cell proliferation. Curr Biol. 2003;13:837–842. doi: 10.1016/s0960-9822(03)00327-0. [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;13:1555–1566. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci U S A. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell. 2005;17:2384–2396. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar P, Edwards KS, Rahman A, Delong A, Muday GK. PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol. 2009;150:722–735. doi: 10.1104/pp.108.131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol. 2004;55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol. 2011;21:R338–R345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- Takeda S, Tadele Z, Hofmann I, Probst AV, Angelis KJ, Kaya H, Araki T, Mengiste T, Mittelsten Scheid O, Shibahara K, Scheel D, Paszkowski J. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 2004;18:782–793. doi: 10.1101/gad.295404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A. ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol. 2007;48:263–277. doi: 10.1093/pcp/pcl063. [DOI] [PubMed] [Google Scholar]
- Tajima Y, Imamura A, Kiba T, Amano Y, Yamashino T, Mizuno T. Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol. 2004;45:28–39. doi: 10.1093/pcp/pcg154. [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF (COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- Tian L, Chen ZJ. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Nat Acad Sci USA. 2001;98:200–205. doi: 10.1073/pnas.011347998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Wang J, Fong MP, Chen M, Cao H, Gelvin SB, Chen ZJ. Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics. 2003;165:399–409. doi: 10.1093/genetics/165.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Fong MP, Wang JJ, Wei NE, Jiang H, Doerge RW, Chen ZJ. Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics. 2005;169:337–345. doi: 10.1534/genetics.104.033142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci U S A. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2010;107:4477–4482. doi: 10.1073/pnas.0909571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachonasios KE, Thomashow MF, Triezenberg SJ. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell. 2003;15:626–638. doi: 10.1105/tpc.007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- Wang JW, Park MY, Wang LJ, Koo Y, Chen XY, Weigel D, Poethig RS. miRNA control of vegetative phase change in trees. PLoS Genet. 2011;7:e1002012. doi: 10.1371/journal.pgen.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Cao H, Sun Y, Li X, Chen F, Carles A, Li Y, Ding M, Zhang C, Deng X, Soppe WJ, Liu YX. Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell. 2013;25:149–166. doi: 10.1105/tpc.112.108191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Sauer M, Meurette O, Friml J, Ljung K, Sandberg G, Hooykaas P, Offringa R. Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell. 2005;17:2517–2526. doi: 10.1105/tpc.105.034637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiste C, Dröge-Laser W. The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat Commun. 2014;27(5):3883. doi: 10.1038/ncomms4883. [DOI] [PubMed] [Google Scholar]
- Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133:4211–4218. doi: 10.1242/dev.02602. [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Custard KD, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL. DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell. 2006;18:805–814. doi: 10.1105/tpc.105.038836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in transacting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2005;102:12984–1299. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Miki D, Zheng Z, Ma J, Wang J, Yang Z, Dong J, Zhu JK. Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat Commun. 2014;5(5):4062. doi: 10.1038/ncomms5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19:2470–2483. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Li C, Zhao L, Gao S4, Lu J, Zhao M, Chen CY, Liu X, Luo M, Cui Y, Yang C, Wu K. The Arabidopsis SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA Targets Directly to PINs and Is Required for Root Stem Cell Niche Maintenance. Plant Cell. 2015;27:1670–1680. doi: 10.1105/tpc.15.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Yamashino T, Amano Y, Tajima Y, Imamura A, Sakakibara H, Mizuno T. Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 2007;48:84–96. doi: 10.1093/pcp/pcl040. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol. 2007;14:869–871. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, Yang H, Xu Y, Kim SH, Fujioka S, Lin WH, Chong K, Lu T, Wang ZY. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2009;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu JK. Seeing the forest for the trees: a wide perspective on RNA-directed DNA methylation. Genes Dev. 2012;26:1769–73. doi: 10.1101/gad.200410.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Jing Y, Jiang Z, Lin R. The Chromatin-Remodeling Factor PICKLE Integrates Brassinosteroid and Gibberellin Signaling during Skotomorphogenic Growth in Arabidopsis. Plant Cell. 2014;26:2472–2485. doi: 10.1105/tpc.113.121848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TQ, Lian H, Tang H, Dolezal K, Zhou CM, Yu S, Chen JH, Chen Q, Liu H, Ljung K, Wang JW. An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell. 2015;27:349–360. doi: 10.1105/tpc.114.135186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU. Hormonal control of the shoot stem-cell niche. Nature. 2010;465:1089–1092. doi: 10.1038/nature09126. [DOI] [PubMed] [Google Scholar]
- Zhong S, Zhao M, Shi T, Shi H, An F, Zhao Q, Guo H. EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci U S A. 2009;106:21431–21436. doi: 10.1073/pnas.0907670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Zhang L, Duan J, Miki B, Wu K. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell. 2005;17:1196–1204. doi: 10.1105/tpc.104.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CM, Zhang TQ, Wang X, Yu S, Lian H, Tang H, Feng ZY, Zozomova-Lihová J, Wang JW. Molecular basis of age-dependent vernalization in Cardamine flexuosa. Science. 2013;340:1097–1100. doi: 10.1126/science.1234340. [DOI] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- Zhu YX. The epigenetic involvement in plant hormone signaling. Chinese Science Bulletin. 2010;55:2198–2203. [Google Scholar]
- Zhu Z, An F, Feng Y, Li P, Xue L, A M, Jiang Z, Kim JM, To TK, Li W, Zhang X, Yu Q, Dong Z, Chen WQ, Seki M, Zhou JM, Guo H. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]


