Abstract
Hairy cell leukemia (HCL) and hairy cell leukemia-variant (HCL-v) are rare diseases with overlapping clinico-pathological features. We performed flow cytometry analysis (FCM) of 213 cases (169 HCL, 35 HCL-v, 9 splenic marginal zone lymphoma (SMZL)), correlating results with available corresponding clinical and morphological data.
FCM distinguished HCL-v from HCL and SMZL based solely upon expression of four antigens (CD11c, CD25, CD103, CD123) combined with B-cell markers (CD19, CD20, CD22). HCL-v expressed bright CD20, bright CD22, CD11c(100%), CD103(100%), dim(40%) or negative(60%) CD123, and uniformly lacked CD25(100%). HCL expressed bright CD20, bright CD22, bright CD11c, bright CD25, CD103, and bright homogeneous CD123(100%). Aberrant expression of CD5(2%/3%), CD10(12%/3%), CD23(21%/11%), CD38(14%/0%), CD2(2%/9%), CD4(0.5%/0%) and CD13(0.5%/3%), was observed in HCL/HCL-v, respectively. SMZL cases were CD103(−) and CD123(−) except for one case with dim CD123. HCL showed significantly greater marrow infiltration over HCL-v. Prominent nucleoli were observed in most HCL-v but rarely in HCL. A third of HCL and HCL-v marrows were hypocellular or aplastic-appearing. Detection of BRAFV600E mutation and annexin A1 were examined in a subset of cases to further validate FCM diagnostic criteria. HCL-v was negative for both annexin A1 (100%) and BRAFV600E mutation (100%), in contrast to HCL (74% positive for annexin A1; 76% positive for BRAFV600E mutation).
HCL-v is resistant to traditional HCL therapy, making accurate diagnosis imperative. We have defined FCM criteria for differentiation of HCL-v from HCL and SMZL.
Keywords: Hairy Cell Leukemia; Hairy Cell Leukemia Variant; flow cytometry; immunophenotype, HCL, HCL-v, CD11c, CD25, CD103, CD123
Introduction
Hairy Cell Leukemia (HCL) is a rare mature B-cell lymphoproliferative disorder (2% of all lymphoid leukemias) characterized by splenomegaly, cytopenias and distinctive cells with circumferential cytoplasmic projections in the blood and bone marrow [1, 2]. Most patients respond to apoptosis-inducing purine analogs with long-term complete remission [3–6] and refractory patients respond well to immunotoxin therapy [7–9], resulting in a near normal life expectancy [10]. However, other clinical entities mimicking this disease, such as hairy Cell Leukemia variant (HCL-v) and splenic marginal zone lymphoma (SMZL) do not respond to HCL therapies and have a significantly lower survival. Hairy Cell Leukemia variant (HCL-v) is rare, constituting 0.4% of all lymphoid malignancies [1, 11, 12] and is a relatively ill-defined entity. The neoplastic cells also have circumferential cytoplasmic projections but a prominent nucleolus and basophilic cytoplasm are common. Although the patients typically present at an older age with higher numbers of circulating hairy cells than observed in HCL, there is overlap in the spectrum of clinical features [1, 11, 13]; Furthermore the prominent nucleoli characteristic of HCL-v may be absent [14]. HCL-v is a more aggressive disease, with significantly shorter median survival and poor response to purine analogs [1, 5]; however, complete remission has been achieved with rituximab and BL22 [8, 15]. In SMZL leukemic cells may appear morphologically similar to HCL and HCLv but again response to therapy differs. Distinguishing HCL-v from HCL and SMZL can be challenging, as clinical and pathologic findings (splenomegaly, cells with cytoplasmic projections) may overlap [1], but accurate diagnosis is crucial for treatment and prognosis.
Immunophenotypic analysis by multi-parameter flow cytometry (FCM) is essential in establishing the diagnosis of HCL. It is extremely sensitive [16–18], detecting HCL cells at levels of 0.01%. By FCM, HCL expresses CD20 (bright), CD22 (bright), CD11c (bright), CD25, CD103 and CD123 [1, 19–23]. In contrast, HCL-v is reported as typically positive for CD20, CD22, CD11c (bright), CD103 and negative for CD25 and CD123 [2, 11, 13, 20, 21], although evaluation of a large series of HCL-v using currently available diagnostic modalities has not been described, due to its rarity. SMZL cells usually show a nonspecific immunophenotype (negative for CD5, CD10, CD23 or CD103). Nevertheless, variations in antigen expression of CD103, CD25, CD10 and CD23 are reported in HCL [19], potentially causing diagnostic confusion.
We evaluated cases of HCL, HCL-v and SMZL, correlating immunophenotypic and morphologic findings. We propose diagnostic criteria for HCL-v, confirmed in a subset of cases with BRAFV600E mutation analysis and annexin A1 staining. These criteria identify HCL-v from other B-cell lymphoproliferative disorders, including HCL and SMZL.
Design and Methods
Case Selection and Patient Information
A total of 213 patients (169 HCL; 35 HCL-v; 9 SMZL) were evaluated by FCM and reviewed by hematopathologists (M.S.-S. or C.M.Y.) between 1999 and 2011 at the National Cancer Institute, National Institutes of Health, Bethesda, MD. Specimens included peripheral blood and bone marrow aspirates. The morphology from corresponding bone marrow biopsies and peripheral smears was reviewed, as available. All specimens were submitted as part of routine evaluation and screening for protocol eligibility to the Flow Cytometry Unit, Laboratory of Pathology, National Cancer Institute, Bethesda, MD. Treatment status was not available and patients may have had previous therapy. All patients signed institutional review board–approved informed consents for screening and further research evaluations.
Flow Cytometric Immunophenotyping
FCM was performed as previously described [16, 17]. Specimens were acquired with 6-parameter 4-color FC on the FACSCalibur (BD Biosciences) using CellQuest Pro software prior to September 2009, and from September 2009 onward, with 10-parameter 8-color FC on the FACSCanto II (BD Biosciences) using DIVA software (sensitivity of fluorescent detectors monitored using standard beads according to manufacturer’s recommendations). Minimum of 5,000 lymphocytes were acquired per tube. For analysis, lymphocytes, monocytes and granulocytes were examined with forward versus side scatter gating, in conjunction with antigen back-gating for T-cells (CD3), B-cells (CD19), and monocytes (CD14), ensuring that cells of interest were appropriately evaluated. Normal lymphoid cells within the specimens served as internal positive and negative controls for determination of antibody-binding intensity, according to U.S.-Canadian Consensus guidelines [24].
Morphologic. Immunohistochemical and Molecular Studies
Paraffin-embedded and H&E stained sections of bone marrow core biopsies, Wright-stained aspirate smears, and peripheral blood blood smears were reviewed from 82 patients diagnosed with HCL (73 male/9 female; 56.8 years, average age) and 14 patients diagnosed with HCL-v (9 male/5 female; 47.8 years, average age). Peripheral blood smears were reviewed from 7 patients diagnosed with SMZL (5 male/2 females; 61.3 years, average age), and an additional 12 patients with HCL-v. Immunohistochemical studies, including annexin A1 staining were performed on formalin-fixed and paraffin-embedded bone marrow core biopsy sections using an automated immunostainer (Ventana Medical Systems, Inc. Tucson, AZ) according to the company’s protocols. Antigen retrieval was performed using Cell Marque's Trilogy (EDTA) in pressure cooker. Antibodies included CD20 (L26, prediluted, Dako, Carpinteria, CA), CD3 (F7.2.38, prediluted, Dako), CD79a (JCB117, prediluted, Dako) and TRAP (9C5, prediluted, Cell Marque, CA). Positive and negative controls were performed with all cases and showed appropriate staining patterns. BRAF mutation detection was performed using a pyrosequencing assay, with PCR primers initially flanking the V600 E mutation hotspot within exon 15 of BRAF, followed by targeted pyrosquencing, as decribed [25].
Results
Clinical Findings
HCL patients were comprised of 144/169 males and 25/169 females (M:F ratio of 5.8:1) with an age range from 17 to 98 years (median, 55 years). HCL-v patients were comprised of 29/35 males and 6/35 females (M:F ratio of 4.8:1) with an age range from 40 to 98 years (median, 65 years). SMZL patients were comprised of 7/9 males and 2/9 females (M:F ratio of 3.5) with an age range from 46 to 87 years (median, 61 years). White blood counts (WBC) from 102 patients were available at the time of initial FCM evaluation (102 HCL; 20 HCL-v; 7 SMZL). The mean WBC count of HCL-v, 74.9 × 109/L (range: 1.8–839.2 ×109/L), was higher in comparison with that of HCL and SMZL, 8.0 × 109/L (range: 0.6–142.0 ×109/L) and 10.5 × 109/L (range 2.2–80.7 ×109/L) respectively, although significant overlap in the range was noted.
Morphology
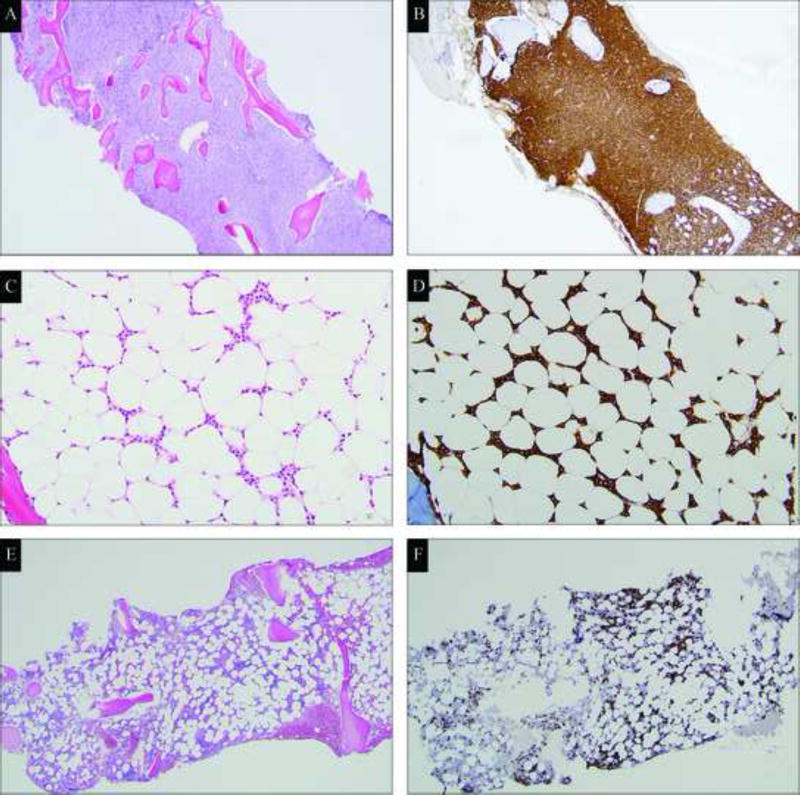
Morphologic findings are summarized in Table 1. HCL patients most frequently demonstrated a hypercellular marrow (44/82 cases, 54%; Figure 1A), with diffuse, infiltrating sheets of neoplastic cells (45/82 cases, 55%; Figure 1B), and markedly suppressed trilineage hematopoiesis (52/82 cases, 63%). Hypocellularity (23/82 cases, 28%; Figure 1C) was observed less frequently and was associated with markedly aplastic areas containing a subtle interstitial infiltrate of neoplastic cells between adipocytes, best revealed by CD20 immunohistochemistry (Figure 1D). In contrast, HCL-v patients presented with hyper-, normo- and hypo-cellular marrows, most frequently with an interstitial infiltration pattern, (7/14 cases, 58%; Figure1E and 1F). Interstitial infiltration with aggregates of neoplastic cells was also observed (5/14, 36%), but diffuse sheets of HCL-v cells was less common in the marrow (2/14, 14%).
Table 1.
Summary of morphologic findings
| HCL | HCL-v | SMZL | |
|---|---|---|---|
| BONE MARROW | |||
| Cellularity | |||
| Hypercellular | 54% (44/82) | 43% (6/14) | N/A |
| Normocellular | 18% (15/82) | 28.5% (4/14) | N/A |
| Hypocellular | 28% (23/82) | 28.5% (4/14) | N/A |
| Infiltration pattern | |||
| Interstitial | 24% (20/82) | 50% (7/14) | N/A |
| Interstitial with aggregates | 21% (17/82) | 36% (5/14) | N/A |
| Sheets | 55% (45/82) | 14% (2/14) | N/A |
| Level of marrow infiltrate | |||
| 75–100% | 73% (60/82) | 21% (3/14) | N/A |
| 25–75% | 22% (18/82) | 50% (7/14) | N/A |
| Less than 25% | 5% (4/82) | 29% (4/14) | N/A |
| Immunostaining for TRAP | |||
| Positive for TRAP | 95% (73/77) | 38% (5/13) | N/A |
| Negative for TRAP | 5% (4/77) | 62% (8/13) | N/A |
| Residual hematopoiesis | |||
| Markedly suppressed | 63% (52/82) | 21% (3/14) | N/A |
| Present | 37% (30/82) | 79% (11/14) | N/A |
| CYTOMORPHOLOGY | |||
| Cytoplasmic projections | |||
| Present | 94% (67/71) | 96% (25/26) | 33% (2/6) |
| Absent | 6% (4/71) | 4% (1/26) | 67% (4/6) |
| Nuclear Shape | |||
| Round | 77% (55/71) | 92% (24/26) | 33% (2/6) |
| Atypical (dumbbell-shape, irregular, grooves) | 23% (16/71) | 8% (2/26) | 67% (4/6) |
| Nucleoli | |||
| Absent | 79% (56/71) | 0% (0/26) | 17% (1/6) |
| Small and inconspicuous | 20% (14/71) | 38% (10/26) | 83% (5/6) |
| Prominent | 1% (1/71) | 62% (16/26) | 0% (0/6) |
HCL: Hairy cell leukemia, HCL-v: Hairy cell leukemia-variant, SMZL: Splenic marginal zone lymphoma, N/A: not available.
Figure 1.
Hypercellular marrow with diffuse infiltrate of hairy cell leukemia cells (A, hematoxylin-eosin stain; B, CD20 immunostain, magnification ×400); Hypocellular marrow with interstitial infiltrate of hairy cell leukemia cells (C, hematoxylin-eosin stain, magnification ×1000; D, CD20 immunostain, magnification ×400); Hypocellular marrow with interstitial infiltrate of hairy cell leukemia variant cells (E, hematoxylin-eosin stain; F, CD20 immunostain, magnification ×400).
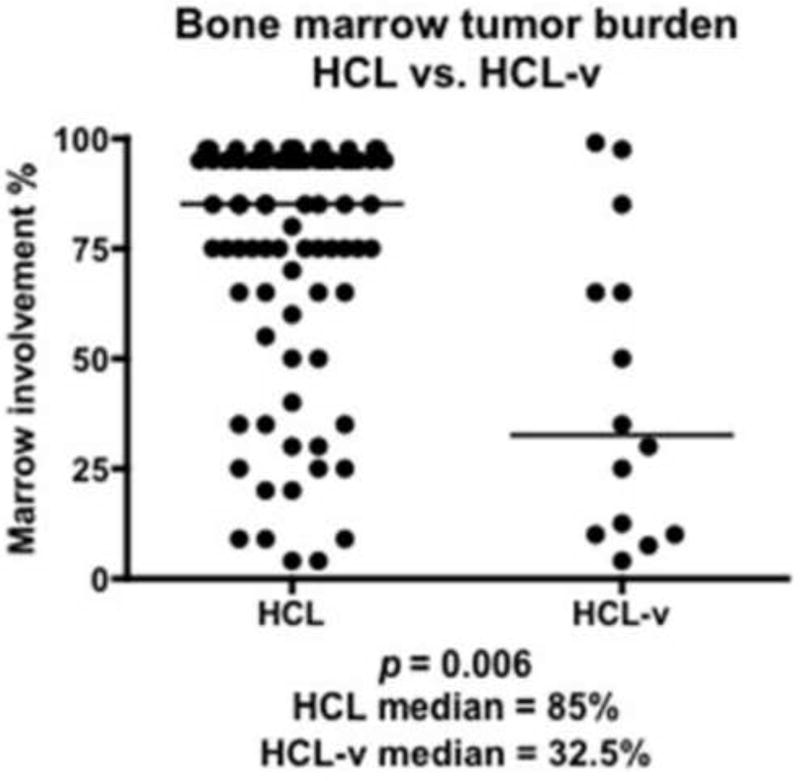
The percentage of marrow infiltration was significantly higher (p=0.0066) in HCL (85% median involvement; 28.4 SD) versus HCL-v (32.5% median involvement; 34.3 SD, Figure 2). We observed that 73% of HCL and only 17% of HCL-v cases showed >75% bone marrow involvement. A greater proportion of HCL-v cases exhibited significant residual hematopoiesis (11/14, 79%) compared to HCL. TRAP immunohistochemical staining was detected in 95% of HCL and 38% of HCL-v cases; HCL cells tended to have stronger, more diffuse staining, while HCL-v cells usually showed weaker staining for TRAP.
Figure 2.
Difference in marrow involvement (% of marrow cellularity) between HCL and HCL-v marrow biopsies. Marrow involvement was 2.6 times as extensive in HCL and HCL-v (p=0.0066, Mann-Whitney).
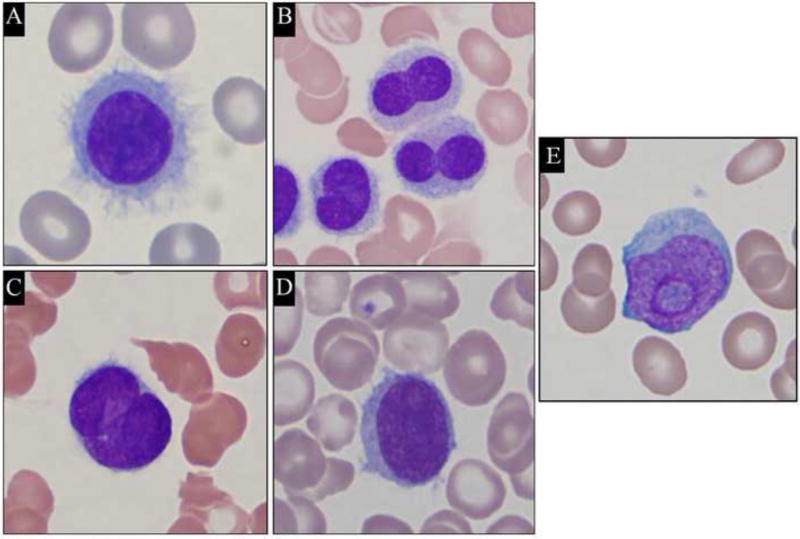
Cytological features of HCL, HCL-v and SMZL cells are summarized in Table 1. Cytoplasmic projections were present in 94% of HCL (Figure 3A–C) and 96% of HCL-v (Figure 3D), but only in 33% (2/6 cases) of SMZL cases. In HCL, 23% of cases (16/71 cases, Figure3B and 3C) exhibited nuclear atypia (homogeneous chromatin, nuclear grooves, or Dumbbell-shaped nuclei). These features were seen in only 8% (2/26 cases) of HCL-v. SMZL demonstrated irregular nuclear contours and coarse chromatin in 67% (4/6 cases). Prominent nucleoli were observed in 63% (16/26 cases) of HCL-v (Figure 3D–E), but in only 1 HCL case (1/71) and none of the SMZL cases (0/6). Small and inconspicuous nucleoli were detected in 38% HCL-v (10/26 cases), 20% HCL (14/71 cases), and in 83% SMZL (5/6 cases).
Figure 3.
Representative images of cytological features of hairy cell leukemia and hairy cell leukemia variant cells in the peripheral blood and bone marrow aspirates. (A, B and C) Hairy cell leukemia cells (Wright's Stain, magnification ×1000); (D and E) Hairy cell leukemia variant cells (Wright's Stain, magnification ×1000).
Flow Cytometric Immunophenotyping
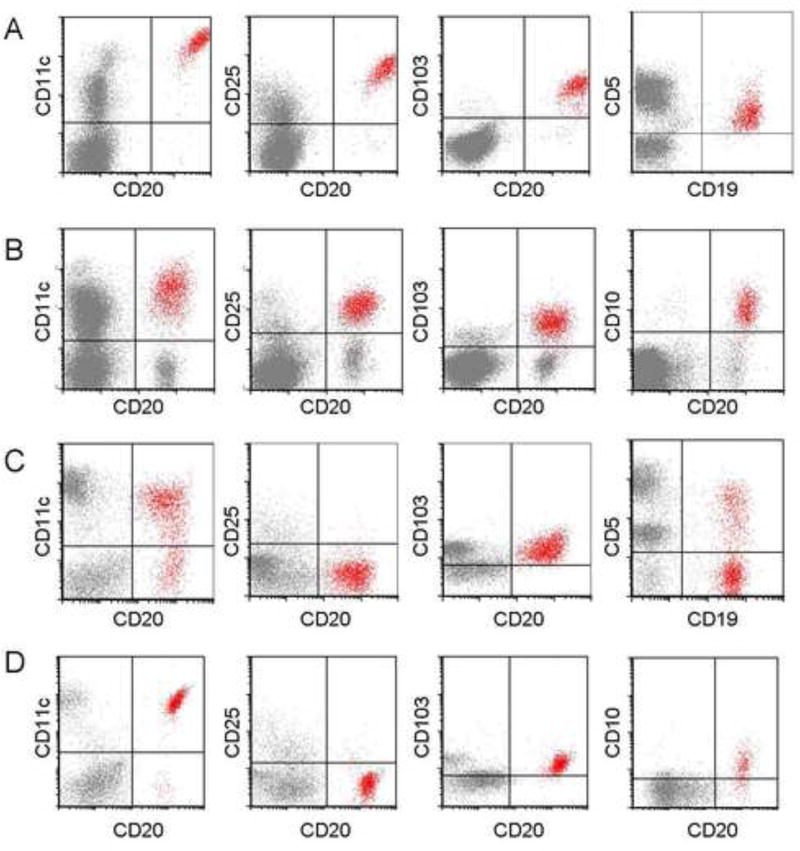
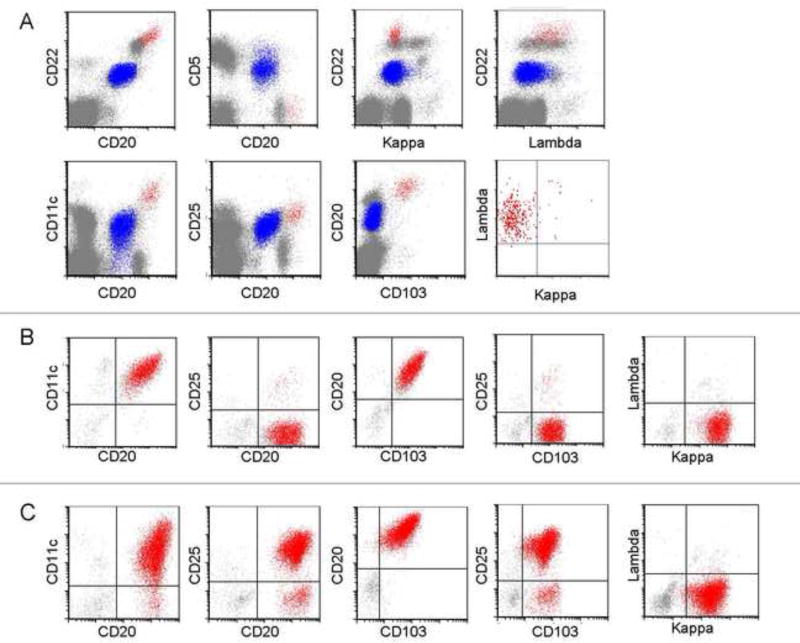
FCM findings are summarized in Table 2. The utility of a subset of these antigens (e.g., CD11c, CD25, CD103, and CD123) to distinguish between HCL-v, HCL and SMZL are examined in Table 3. In HCL, 163/169 cases showed detectable surface immunoglobulin light chain restriction (81 kappa, 82 lambda). Light chain expression was not definitively demonstrated in 6/169 HCL due to obscuring cytophillic antibody, or technically suboptimal anti-light chain staining. The HCL cells in almost all patients were positive for pan-B-cell antigens: CD19 (99%), CD20 (99%), and CD22 (100%). The expression of CD22 and CD20 were consistently brighter than that of normal B-cells. All HCL cells showed expression of the following antigens: CD11c (100%), CD25 (100%), CD103 (100%) and CD123 (100%). The intensity of the expression of CD11c, CD25 and CD123 were consistently bright. Aberrant antigen expression included expression of CD5 (2%, Figure 4A), CD10 (12%, Figure 4B), CD23 (21%), CD2 (2%), CD4 (0.5%) and CD13 (0.5%). The aberrant expression of CD23 was usually weak, with variable intensity. No case showed simultaneous expression of CD5 and CD23. CD38 expression was uncommon, observed in only 14% of cases.
Table 2.
Immunophenotypic comparison between HCL, HCL-v and SMZL
| Antigen | HCL | HCL-v | SMZL |
|---|---|---|---|
| CD19 | 99% (168/169) | 100% (35/35) | 100% (9/9) |
| 2% (4/169)@ | 3% (1/35)@ | ||
| 0.6% (1/169)¶ | 3% (1/35)¶ | ||
|
| |||
| CD20 | 99% (168/169) | 100% (35/35) | 89% (8/9) |
| 88% (148/169)@ | 94% (33/35)@ | ||
|
| |||
| CD22 | 100% (169/169) | 100% (35/35) | 100% (9/9) |
| 96% (162/169)@ | 97% (34/35)@ | 11% (1/9)@ | |
|
| |||
| CD11c | 99% (167/169) | 100% (35/35) | 67% (6/9)¶ |
| 91% (154/169)@ | 63% (22/35)@ | ||
|
| |||
| CD25 | 100% (169/169) | 0% (0/35) | 22% (2/9)¶ |
| 98% (165/169)@ | 0% (0/9)@ | ||
|
| |||
| CD103 | 100% (169/169) | 100% (35/35) | 0% (0/9) |
| 3% (1/35)* | |||
|
| |||
| CD123 | 100% (114/114)@ | 40% (8/20)¶ | 25% (1/4)¶ |
|
| |||
| CD5 | 2% (4/169) | 3% (1/35) | 22% (2/9)¶ |
|
| |||
| CD10 | 12% (21/169) | 3% (1/35) | 0% (0/8) |
|
| |||
| CD23 | 21% (36/169)* | 14% (5/35)* | 38% (3/8) |
|
| |||
| CD38 | 14% (8/58) | 0% (0/35) | 22% (2/9) |
|
| |||
| CD2 | 2% (3/169) | 9% (3/35) | ND |
|
| |||
| CD4 | 0.5% (1/169) | 0% (0/35) | ND |
|
| |||
| CD13 | 0.5% (1/169) | 3% (1/35) | ND |
|
| |||
| Kappa | 48% (81/169) | 49% (17/35) | 67% (6/9) |
|
| |||
| Lambda | 48% (82/169) | 40% (14/35) | 33% (3/9) |
|
| |||
| Monophasic# | 4% (6/169) | 11% (4/35) | ND |
Bright expression;
Dim expression;
Heterogeneous/partial expression
Monophasic without apparent surface light chain expression
ND: Not Done, HCL: Hairy cell leukemia, HCL-v: Hairy cell leukemia-variant, SMZL: splenic marginal zone lymphoma
Table 3.
Evaluation of Antigens to Distinguish HCL, HCL-v and SMZL
| Antigen | Comparison | P value* |
|---|---|---|
| CD11c | HCL (167/169) vs. SMZL (6/9) | p=0.0009 |
| HCL-v (35/35) vs. SMZL (6/9) | p =0.006 | |
|
| ||
| CD11c (bright) | HCL (154/169) vs. HCL-v (22/35) | p =0.00008 |
|
| ||
| CD25 | HCL (169/169) vs. HCL-v (0/35) | p =3 × 10−40 |
| HCL (169/169) vs. SMZL (2/9) | p =4 × 10−11 | |
| HCL-v (0/35) vs. SMZL (2/9) | p =0.04 | |
|
| ||
| CD25 (bright) | HCL (165/169) vs. SMZL (0/9) | p =2 × 10−12 |
|
| ||
| CD103 | HCL (169/169) vs. SMZL (0/9) | p =2 × 10−15 |
| HCL-v (35/35) vs. SMZL (0/9) | p =1 × 10−9 | |
|
| ||
| CD123 | HCL (114/114) vs. HCL-v (8/20) | p =3 × 10−12 |
| HCL-v (8/20) vs. SMZL (1/4) | p =1 | |
| HCL (114/114) vs. SMZL (1/4) | p =1.5 × 10−5 | |
Fisher’s Exact Test
Figure 4.
(A) Hairy cell leukemia cells with expression of CD5. (B) Hairy cell leukemia cells with expression of CD10. (C) Hairy cell leukemia variant cells with expression of CD5. (D) Hairy cell leukemia variant cells with expression of CD10. Analysis shows mononuclear cells, as determined by FSC and SSC characteristics, with HCL or HCL-v cells gated as red events.
In HCL-v, 31/35 cases showed detectable surface immunoglobulin light chain restriction (17 kappa, 14 lambda); in 4/35 cases kappa or lambda light chain expression was not definitively determined (due to obscuring cytophillic antibody, or technically suboptimal anti-light chain staining). All HCL-v cases were positive for B-cell antigens: CD19 (100%), CD20 (100%), and CD22 (100%) as well as CD11c (100%) and CD103 (100% positive with 1/35 or 3% only partial positivity). Expression of CD20 and CD22 were consistently bright. The pattern of CD11c expression in HCL-v differed slightly from HCL; the majority (63%), but not all HCL-v, showed bright CD11c expression, whereas bright CD11c was a distinguishing feature of HCL, present in 91% of cases. All HCL-v cases were negative for CD25 (0%). CD123, when present in HCL-v (40%), was characteristically dim. Aberrant antigen expression included CD5 (3%, Figure 4C), CD10 (3%, Figure 4D), CD23 (14%), CD2 (9%), and CD13 (3%). Similarly to HCL, CD23 expression was usually heterogeneous in HCL-v; furthermore, none of the HCL-v cases showed simultaneous expression of CD5 and CD23.
All 9 SMZL cases showed surface immunoglobulin light chain restriction (6 kappa, 3 lambda). SMZL cells in all patients were positive for the B-cell antigens: CD19 (100%), CD20 (100%, one case post anti-CD20 therapy not included) and CD22 (100%). In sharp contrast to HCL and HCL-v, the expression of CD20 and CD22 was of moderate intensity in SMZL. Similarly, CD11c, when positive (67%) was dim. All SMZL cases were negative for CD103 and CD10, and largely negative for CD25 and CD123. In the 2/9 SMZL cases (22%) that were CD25 positive, the intensity of CD25 expression was dim. Similarly, dim expression of CD123 was observed in only 1 case of SMZL. Expression of CD5 (22%), CD23 (38%), or CD38 (22%) was noted in a minority of SMZL cases.
Interestingly, in 3 cases of HCL, 2 simultaneously occurring clonal B-cell processes were immunophenotypically identified (Figure 5). One case of HCL, (lambda restricted) occurred concurrently with a minor B-cell clone immunophenotypically resembling chronic lymphocytic leukemia (expressing CD19, dim CD20, dim CD22, CD11c, CD25, CD5, CD23, and kappa surface light chain, Figure 5A). The other two HCL cases, also lambda restricted, were accompanied by a minor population of kappa restricted B-cells with a non-diagnostic/non-specific immunophenotype (expressing CD19, CD20, CD22; negative for CD5, CD10, CD25 and CD103).
Figure 5.
(A) flow cytometry analysis demonstrating coexistent hairy cell leukemia and monoclonal B-cells consistent with chronic lymphocytic leukemia. Analysis shows mononuclear cells, as determined by FSC and SSC characteristics, with HCL cells gated as red events, and CLL cells gated as blue events. The chronic lymphocytic leukemia cells expressed dim CD20, dim CD22, dim CD11c, moderate CD25 and CD5, and showed dim kappa light chain restriction. (B) The hairy cell leukemia cells comprised approximately 0.55% of the cells, and were positive for CD20 (bright), CD11c (bright), CD25 and CD103. The hairy cell leukemia cells were lambda light chain restricted when gated on bright CD11c positive B-cells; (B–C) Flow cytometry analysis demonstrating a case of HCL-v with an apparent phenotypic change. Analysis shows mononuclear cells, as determined by FSC and SSC characteristics, with HCL-v cells gated as red events. (B) The hairy cell leukemia variant cells were negative for CD25. However, a small, minor subpopulation was positive for CD25. (C) Two years later, the neoplastic cells appeared to have acquired expression of CD25, but may actually represent an expansion of the minor CD25 positive population at diagnosis (B).
One notable case of HCL-v showed the characteristic expression of bright CD20, bright CD22, CD11c, CD103 and absence of CD25 at the time of initial diagnosis (Figure 5B); however, two years later (post treatment), all of the patient’s neoplastic cells acquired CD25 expression (Figure 5C) and appeared to resemble classic HCL. Closer examination of the initial immunophenotypic data revealed that although most of the HCL-v cells were initially negative for CD25, there was a minor subset of CD25-positive cells present at that time. Although the mechanism for the expansion of this CD25-positive population is unknown, we speculate that it was not as susceptible to treatment as the CD25-negative neoplastic cells.
Based on our data, we proposed that HCL-v be defined as: CD19(+), bright CD20(+), bright CD22(+), CD103(+), CD25(−), CD11c(+) or bright(+), with dim or negative CD123. We proposed that HCL be defined as: CD19(+), bright CD20(+), bright CD22(+), CD103(+), CD25(+), CD11c bright(+) and CD123 bright and homogeneously(+). We validated this against a series of 69 cases (14 HCL-v, 55 HCL, based upon diagnostic FCM immunophenotype) with BRAF V600E mutation analysis (Table 4). All HCL-v cases were negative for the BRAFV600E mutation (14/14, 100%). In HCL, 42/55 (76%) were positive for the BRAFV600E mutation while 13/55 (24%) were negative. Additionally, we validated our criteria with annexin A1 staining (Table 4) in a series of 27 cases (8 HCL-v, 19 HCL. based upon diagnostic FCM immunophenotype). All HCL-v (8/8, 100%) were negative for annexin A1 staining. Of HCL cases, 14/19 (74%) were positive for annexin A1 and 5/19 (26%) were negative.
Table 4.
Evaluation of BRAFV600E Mutation Status and Annexin A1 staining in HCL and HCL-v.
| Diagnosis | HCL | HCL-v | |
|---|---|---|---|
| BRAF V600E | Mutation | 42/55 (76%) | 0/14 (0%) |
| Wild-type | 13/55 (24%) | 14/14 (100%) | |
| Annexin A1 | Positive | 14/19 (74%) | 0/8 (0%) |
| Negative | 5/19 (26%) | 8/8 (100%) | |
Discussion
HCL, though uncommon, is a well-recognized entity; however, HCL-v is extremely rare and less well-defined, and some of its distinguishing features remain controversial. The focus of this study was to characterize the diagnostic features of HCL-v that distinguish it from HCL and SMZL utilizing our unique and unusually large collection of HCL and HCL-v cases (169 HCL, 35 HCL-v). Morphologic, immunophenotypic, cytologic, and clinical features were reviewed. Our HCL cases exhibited significantly greater marrow involvement compared to HCL-v, with 55% demonstrating sheets of neoplastic cells. This is in contrast with the previously described subtle, diffuse bone marrow infiltration of HCL [2], Although our observations may be due to a skewing of the patient population at our institution, presenting at a later point in the their disease state, it is important to note that extensive marrow infiltration with sheets and aggregates of cells can occur in this disease. We noted an inverse relationship between peripheral blood and marrow involvement in HCL and HCL-v, with higher levels of infiltrate in HCL-involved bone marrow when compared to HCL-v (Figure 2), and higher blood involvement in HCL-v compared to HCL. There was significant overlap in the WBC range in HCL (0.6–142.0 ×109/L) and HCL-v (1.8–839.2 ×109/L). A markedly aplastic pattern was observed occasionally in both HCL and HCL-v cases (Figure 1C–D). Some cases were submitted with a working diagnosis of aplastic anemia, only to have HCL discovered on the biopsy; therefore, patients with cytopenias or marrow biopsies with an aplastic appearance should be evaluated for HCL or HCL-v. HCL showed greater TRAP positivity (95%) with increased staining intensity when compared to HCL-v, which was generally TRAP negative (62%); however, the overlap between HCL and HCL-v, diminished the utility of the TRAP stain.
Cytoplasmic projections were associated with the majority of HCL and HCL-v (Table 1), but less so with SMZL. Nuclear grooves and Dumbbell-shaped nuclei were observed in SMZL and a subset of HCL, while the overwhelming majority of HCL-v exhibited a round nucleus (92%). Prominent nucleoli were associated with HCL-v (62%). Some HCL-v cases (38%) exhibited small, inconspicuous nucleoli (consistent with previous report, [11]); however, no HCL-v case lacked nucleoli. One notable case of HCL-v (Figure 3E) exhibiting a single large nucleolus, but lacking prominent cytoplasmic projections, was a deceptive mimicker of a prolymphocyte at the time of initial diagnosis (hence, the historical synonym of prolymphocytic variant of HCL for cases of HCL-v [2, 26]).
Although CD103 has the highest specificity for HCL and HCL-v [21], it is reportedly rarely expressed in other B-cell lymphoproliferative disorders, including SMZL [20]. Previous studies show CD103 expression in all [20, 22, 27], or nearly all cases (94%, 33/35) of HCL [19]. In HCL-v, CD103 expression varies greatly in the literature, from 36–100% [1, 11, 12, 14, 21]. Here, CD103 was reliably expressed in all HCL (100%) and HCL-v (100%), consistent with our earlier work [21]. Generally, SMZL is CD103 negative, although one report describes CD103 expression as high as 40% in SMZL [28]. We find CD103 clearly distinguishes HCL and HCL-v from SMZL; all of our SMZL cases were CD103 negative.
Bright CD25 expression is a classic feature of HCL; conversely, HCL-v usually lacks CD25. Matutes, et. al. reported occasional expression of CD25 in HCL-v specimens (less than 10%,[11, 12]. However, in our series, CD25 was reliably positive in all HCL and negative in all HCL-v, consistent with recent studies [14, 19, 29, 30]. We find CD25 reliably differentiates HCL from HCL-v (100% sensitivity and specificity).
CD123 is a relatively new marker for evaluation of B-cell lymphoproliferative disorders with hairy/villous cytomorphology. Del Giudice, et al reported CD123 expression in 95% (22/23) of HCL cases but only in 9% (1/11) of cases with HCL-v [31]. Similarly, Muñoz, et al found 100% (6/6) of HCL cases with bright CD123 expression and no CD123 positive HCL-v cases (0%) [32]. Recently, Venkataraman, et al showed CD123 in 100% of HCL cases and weakly in 40% of HCL-v cases [21]. Our study confirms the universal expression of CD123 in HCL (100%), with a characteristically bright and homogeneous pattern. CD123 expression was observed in a minor proportion HCL-v (40%), and when present, was dim. CD123 was dimly expressed in only 1/4 SMZL cases. We conclude that bright, homogeneous expression of CD123 is highly specific for HCL, consistent with our earlier work [21].
Based on our study of 169 HCL, 35 HCL-v, and 9 SMZL, we propose the use of the 4 core antigens, CD11c, CD25, CD103 and CD123, along with common B-cell antigens (CD19, CD20 and CD22) as a clinical standard in FCM evaluation where the differential diagnosis includes HCL, HCL-v, and SMZL (e.g. when cells with villous cytoplasmic projections are present). The expression of CD19, bright CD20, bright CD22, bright CD11c, bright CD25, CD103 and bright, homogeneous CD123 are diagnostic of HCL. The expression of bright CD20, bright CD22, CD11c and CD103, but absent CD25 and dim to absent CD123 are indicative of HCL-v. Absent CD103, weak to moderate expression of CD11c, and dim to absent CD123 would favor SMZL or other splenic B-cell lymphoma within the appropriate clinico-pathologic and cytomorphologic context. Our diagnostic FCM criteria based upon CD11c, CD19, CD20 and CD22, CD25, CD103 and CD123 were validated by BRAFV600E mutation analysis and annexin A1 data in a subset of patients, as HCL-v is characterized by a lack of annexin A1 staining and BRAFV600E mutation observed in HCL [25, 33, 34]. Of the cases classified as HCL-v by our criteria, all were negative for annexin A1; furthermore, none had a detectable BRAFV600E mutation. Criteria for FCM diagnosis of HCL and HCL-v are especially important in evaluation of peripheral blood. Annexin A1 is evaluated by inmmunohistochemistry; as such, it is not generally applicable to blood. Furthermore, routine screening by FCM is within the scope of most clinical laboratories, whereas detection of BRAFV600E mutations is currently not widely available.
Use of a limited antibody panel may result in misdiagnosis in select cases, due to aberrant antigen expression. CD5, CD10, and CD23 are generally negative in HCL and HCL-v; however, variations exist. Previous studies report none, [19, 31] or minimal CD5 expression (2% [20] and 5% [35]) in HCL. In HCL-v, CD5 is generally negative [1, 14, 26], with Del Giudice, et al reporting expression in 9% (1/11 cases) [31]. We found CD5 was rarely expressed in HCL (2%) and HCL-v (3%), consistent with previous reports. CD23 expression is generally negative in both HCL [23] and HCL-v [12, 23, 31], with some exceptions (HCL: 6% [20] and 17% [19] ; HCL-v: 3% [11]). Our CD23 expression in HCL (21%) and HCL-v (12%) is slightly higher than in previous reports. Reported CD10 expression in HCL ranges from 4–26% [7, 11, 19, 20, 27, 36, 37], and our CD10 detection in 12% of HCL is within this range. In HCL-v, CD10 is usually negative [14, 20], with some exceptions (15%, [11]), including our study, which identified CD10 expression in 3% of HCL-v. We also identified occasional aberrant expression of CD2, CD4, CD13 and CD38 in both HCL and HCL-v. Three patients exhibited a second, concurrent monoclonal B-cell population, in addition to the presence of HCL. In one patient, the second clonal population was consistent with CLL. In two other patients, the second clonal population had a non-specific immunophenotype with expression of B-cell antigens, but negative for CD25, CD103, CD5 and CD10. The light chain expression between the two monoclonal B-cell populations was different in all three patients, indicating different clonal origins of the two populations. Simultaneously occurring HCL and CLL has been reported [38, 39] and molecular studies suggest different clonal origins [39]. A limited antibody panel could have resulted in failure to detect HCL in these patients.
In summary, diagnosis of HCL-v can be difficult but is critical as HCL-v is responsive to rituximab and BL22 immunotoxin therapy, but refractory to the purine analogs that are so effective in HCL. This study highlights the importance of recognizing the overlap in the spectrum of presentation of HCL and HCL-v in peripheral blood and bone marrow. We demonstrated that HCL-v and HCL are clearly defined by FCM. While both HCL and HCL-v characteristically express CD19, bright CD20, bright CD22, CD103 and CD11c (moderate or bright), HCL has bright CD25 and CD123 while HCL-v lacks CD25 and CD123 is negative or or dim). We propose a panel containing 4 HCL/HCLv antigens (CD11c, CD25, CD103, CD123) in conjunction with common B-cell antigens (CD19, CD20 and CD22) as part of the standard FCM work-up of these diseases. We hope to improve the identification of HCL-v, prevent its misdiagnosis, and facilitate the initiation of appropriate therapy for patients with this rare and treatable lymphoproliferative disorder.
Acknowledgments
The authors wish to thank Catharine McCoy, Linda Weaver, Gregory Jasper, and Christine Aguhar at the NCI flow cytometry laboratory for graciously providing FCM access, and technical expertise. We are grateful to Jaime Hahn for assistance with retrieving slides.
This work was supported in part by the Intramural Research Program of the NIH, NCI.
Footnotes
Authorship and Disclosures
HS and MG retrieved and reviewed FCM data. KRC performed immunohistochemical evaluation. KRC and PRT reviewed the bone marrow and peripheral blood morphology. RJK collected, organized and reviewed the data. KRC and RJK provided statistical analysis. HS, KRC, MG, MSS and CMY wrote the paper. CMY was the principal investigator and takes primary responsibility for the paper.
Competing interests: the authors have no competing interests.
References
- 1.Sharpe RW, Bethel KJ. Hairy cell leukemia: diagnostic pathology. Hematol Oncol Clin North Am. 2006;20:1023–49. doi: 10.1016/j.hoc.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. 4. Lyon: IARC Press; 2008. [Google Scholar]
- 3.Tallman MS, Hakimian D, Variakojis D, Koslow D, Sisney GA, Rademaker AW, et al. A single cycle of 2-chlorodeoxyadenosine results in complete remission in the majority of patients with hairy cell leukemia. Blood. 1992;80:2203–9. [PubMed] [Google Scholar]
- 4.Jehn U, Bartl R, Dietzfelbinger H, Vehling-Kaiser U, Wolf-Hornung B, Hill W, et al. Long-term outcome of hairy cell leukemia treated with 2-chlorodeoxyadenosine. Ann Hematol. 1999;78:139–44. doi: 10.1007/s002770050490. [DOI] [PubMed] [Google Scholar]
- 5.Robak T, Blasinska-Morawiec M, Blonski J, Hellmann A, Halaburda K, Konopka L, et al. 2-chlorodeoxyadenosine (cladribine) in the treatment of hairy cell leukemia and hairy cell leukemia variant: 7-year experience in Poland. Eur J Haematol. 1999;62:49–56. doi: 10.1111/j.1600-0609.1999.tb01114.x. [DOI] [PubMed] [Google Scholar]
- 6.Tallman MS, Hakimian D, Rademaker AW, Zanzig C, Wollins E, Rose E, et al. Relapse of hairy cell leukemia after 2-chlorodeoxyadenosine: long-term follow-up of the Northwestern University experience. Blood. 1996;88:1954–9. [PubMed] [Google Scholar]
- 7.Robbins DH, Margulies I, Stetler-Stevenson M, Kreitman RJ. Hairy cell leukemia, a B-cell neoplasm that is particularly sensitive to the cytotoxic effect of anti-Tac(Fv)-PE38 (LMB-2) Clin Cancer Res. 2000;6:693–700. [PubMed] [Google Scholar]
- 8.Kreitman RJ, Wilson WH, Bergeron K, Raggio M, Stetler-Stevenson M, FitzGerald DJ, et al. Efficacy of the anti-CD22 recombinant immunotoxin BL22 in chemotherapy-resistant hairy-cell leukemia. N Engl J Med. 2001;345:241–7. doi: 10.1056/NEJM200107263450402. [DOI] [PubMed] [Google Scholar]
- 9.Kreitman RJ, Stetler-Stevenson M, Margulies I, Noel P, Fitzgerald DJ, Wilson WH, et al. Phase II trial of recombinant immunotoxin RFB4(dsFv)-PE38 (BL22) in patients with hairy cell leukemia. J Clin Oncol. 2009;27:2983–90. doi: 10.1200/JCO.2008.20.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grever MR. How I treat hairy cell leukemia. Blood. 2010;115:21–8. doi: 10.1182/blood-2009-06-195370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matutes E, Wotherspoon A, Catovsky D. The variant form of hairy-cell leukaemia. Best Pract Res Clin Haematol. 2003;16:41–56. doi: 10.1016/s1521-6926(02)00086-5. [DOI] [PubMed] [Google Scholar]
- 12.Matutes E, Wotherspoon A, Brito-Babapulle V, Catovsky D. The natural history and clinico-pathological features of the variant form of hairy cell leukemia. Leukemia. 2001;15:184–6. doi: 10.1038/sj.leu.2401999. [DOI] [PubMed] [Google Scholar]
- 13.de Totero D, Tazzari PL, Lauria F, Raspadori D, di Celle PF, Carbone A, et al. Phenotypic analysis of hairy cell leukemia: "variant" cases express the interleukin-2 receptor beta chain, but not the alpha chain (CD25) Blood. 1993;82:528–35. [PubMed] [Google Scholar]
- 14.Cessna MH, Hartung L, Tripp S, Perkins SL, Bahler DW. Hairy cell leukemia variant: fact or fiction. Am J Clin Pathol. 2005;123:132–8. doi: 10.1309/8qytyq1clqmhq9cl. [DOI] [PubMed] [Google Scholar]
- 15.Narat S, Gandla J, Dogan A, Mehta A. Successful treatment of hairy cell leukemia variant with rituximab. Leuk Lymphoma. 2005;46:1229–32. doi: 10.1080/10428190500083433. [DOI] [PubMed] [Google Scholar]
- 16.Sausville JE, Salloum RG, Sorbara L, Kingma DW, Raffeld M, Kreitman RJ, et al. Minimal residual disease detection in hairy cell leukemia. Comparison of flow cytometric immunophenotyping with clonal analysis using consensus primer polymerase chain reaction for the heavy chain gene. Am J Clin Pathol. 2003;119:213–7. doi: 10.1309/G629-9513-NGLC-UB1K. [DOI] [PubMed] [Google Scholar]
- 17.Shao H, Yuan CM, Xi L, Raffeld M, Morris JC, Janik JE, et al. Minimal residual disease detection by flow cytometry in adult T-cell leukemia/lymphoma. Am J Clin Pathol. 2010;133:592–601. doi: 10.1309/AJCPS1K0OHLJYWWV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravandi F, O'Brien S, Jorgensen J, Pierce S, Faderl S, Ferrajoli A, et al. Phase 2 study of cladribine followed by rituximab in patients with hairy cell leukemia. Blood. 2011;118:3818–23. doi: 10.1182/blood-2011-04-351502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YH, Tallman MS, Goolsby C, Peterson L. Immunophenotypic variations in hairy cell leukemia. Am J Clin Pathol. 2006;125:251–9. doi: 10.1309/PMQX-VY61-9Q8Y-43AR. [DOI] [PubMed] [Google Scholar]
- 20.Dong HY, Weisberger J, Liu Z, Tugulea S. Immunophenotypic analysis of CD103+ B-lymphoproliferative disorders: hairy cell leukemia and its mimics. Am J Clin Pathol. 2009;131:586–95. doi: 10.1309/AJCPL13YDUHFKPJU. [DOI] [PubMed] [Google Scholar]
- 21.Venkataraman G, Aguhar C, Kreitman RJ, Yuan CM, Stetler-Stevenson M. Characteristic CD103 and CD123 Expression Pattern Defines Hairy Cell Leukemia: Usefulness of CD123 and CD103 in the Diagnosis of Mature B-Cell Lymphoproliferative Disorders. Am J Clin Pathol. 2011;136:625–30. doi: 10.1309/AJCPKUM9J4IXCWEU. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassan IB, Hagberg H, Sundstrom C. Immunophenotype of hairy-cell leukemia. Eur J Haematol. 1990;45:172–6. doi: 10.1111/j.1600-0609.1990.tb00447.x. [DOI] [PubMed] [Google Scholar]
- 23.Matutes E. Immunophenotyping and differential diagnosis of hairy cell leukemia. Hematol Oncol Clin North Am. 2006;20:1051–63. doi: 10.1016/j.hoc.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Borowitz MJ, Bray R, Gascoyne R, Melnick S, Parker JW, Picker L, et al. U.S.-Canadian Consensus recommendations on the immunophenotypic analysis of hematologic neoplasia by flow cytometry: data analysis and interpretation. Cytometry. 1997;30:236–44. doi: 10.1002/(sici)1097-0320(19971015)30:5<236::aid-cyto4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 25.Xi L, Arons E, Navarro W, Calvo KR, Stetler-Stevenson M, Raffeld M, et al. Both variant and IGHV4-34-expressing hairy cell leukemia lack the BRAF V600E mutation. Blood. 2012;119:3330–2. doi: 10.1182/blood-2011-09-379339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunphy CH, Petruska PJ. Atypical prolymphocytic variant of hairy-cell leukemia: case report and review of the literature. Am J Hematol. 1996;53:121–5. doi: 10.1002/(SICI)1096-8652(199610)53:2<121::AID-AJH11>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 27.Juliusson G, Lenkei R, Liliemark J. Flow cytometry of blood and bone marrow cells from patients with hairy cell leukemia: phenotype of hairy cells and lymphocyte subsets after treatment with 2-chlorodeoxyadenosine. Blood. 1994;83:3672–81. [PubMed] [Google Scholar]
- 28.Ocio EM, Hernandez JM, Mateo G, Sanchez ML, Gonzalez B, Vidriales B, et al. Immunophenotypic and cytogenetic comparison of Waldenstrom's macroglobulinemia with splenic marginal zone lymphoma. Clin Lymphoma. 2005;5:241–5. doi: 10.3816/clm.2005.n.007. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh YC, Chang ST, Chuang SS, Lu CL, Tsao CJ, Lin CN, et al. Hairy cell leukemia and variant in Taiwan: report of a variant case and literature review. Int J Clin Exp Pathol. 2011;4:183–9. [PMC free article] [PubMed] [Google Scholar]
- 30.Kao HW, Dunn P, Kuo MC, Shih LY, Lin TL, Wu JH, et al. Classical hairy cell leukemia and its variant: a 17-year retrospective survey in Taiwan Chinese. Acta Haematol. 2011;126:186–93. doi: 10.1159/000328887. [DOI] [PubMed] [Google Scholar]
- 31.Del Giudice I, Matutes E, Morilla R, Morilla A, Owusu-Ankomah K, Rafiq F, et al. The diagnostic value of CD123 in B-cell disorders with hairy or villous lymphocytes. Haematologica. 2004;89:303–8. [PubMed] [Google Scholar]
- 32.Munoz L, Nomdedeu JF, Lopez O, Carnicer MJ, Bellido M, Aventin A, et al. Interleukin-3 receptor alpha chain (CD123) is widely expressed in hematologic malignancies. Haematologica. 2001;86:1261–9. [PubMed] [Google Scholar]
- 33.Falini B, Tiacci E, Liso A, Basso K, Sabattini E, Pacini R, et al. Simple diagnostic assay for hairy cell leukaemia by immunocytochemical detection of annexin A1 (ANXA1) Lancet. 2004;363:1869–70. doi: 10.1016/S0140-6736(04)16356-3. [DOI] [PubMed] [Google Scholar]
- 34.Tiacci E, Schiavoni G, Forconi F, Santi A, Trentin L, Ambrosetti A, et al. Simple genetic diagnosis of hairy cell leukemia by sensitive detection of the BRAF-V600E mutation. Blood. 2012;119:192–5. doi: 10.1182/blood-2011-08-371179. [DOI] [PubMed] [Google Scholar]
- 35.Goodman GR, Bethel KJ, Saven A. Hairy cell leukemia: an update. Curr Opin Hematol. 2003;10:258–66. doi: 10.1097/00062752-200307000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Jasionowski TM, Hartung L, Greenwood JH, Perkins SL, Bahler DW. Analysis of CD10+ hairy cell leukemia. Am J Clin Pathol. 2003;120:228–35. doi: 10.1309/QVJD-31TE-G9UJ-18BQ. [DOI] [PubMed] [Google Scholar]
- 37.Kaleem Z, White G, Vollmer RT. Critical analysis and diagnostic usefulness of limited immunophenotyping of B-cell non-Hodgkin lymphomas by flow cytometry. Am J Clin Pathol. 2001;115:136–42. doi: 10.1309/8B6V-16DJ-UMTB-6LVX. [DOI] [PubMed] [Google Scholar]
- 38.Sokol L, Agosti SJ. Simultaneous manifestation of chronic lymphocytic leukemia (CLL) and hairy cell leukemia (HCL) Am J Hematol. 2004;75:107–9. doi: 10.1002/ajh.10459. [DOI] [PubMed] [Google Scholar]
- 39.Gine E, Bosch F, Villamor N, Rozman M, Colomer D, Lopez-Guillermo A, et al. Simultaneous diagnosis of hairy cell leukemia and chronic lymphocytic leukemia/small lymphocytic lymphoma: a frequent association? Leukemia. 2002;16:1454–9. doi: 10.1038/sj.leu.2402553. [DOI] [PubMed] [Google Scholar]