Abstract
We have constructed new yeast vectors for targeted integration of desired sequences at the Saccharomyces cerevisiae HO locus. Insertion at HO has been shown to have no effect on yeast growth, and thus these integrations should be neutral. One vector contains the KanMX selectable marker, and integrants can be selected by resistance to G418. The other vector contains the hisG-URA3-hisG cassette, and integrants can be selected by uracil prototrophy. Subsequent growth on 5-FOA permits identification of colonies where recombination between the hisG tandem repeats has led to loss of the URA3 marker and return to uracil auxotrophy. We also describe several new bacterial polylinker vectors derived from pUC21 (ampicillin resistance) and pUK21 (kanamycin resistance).
INTRODUCTION
The budding yeast Saccharomyces cerevisiae is widely used as an experimental organism. Many genetic screens are facilitated by the stable integration of sequences into the yeast genome. For instance, one may want to insert a reporter gene needed for the screen so that it is stably maintained in the absence of growth on selective media and with no variation in copy number. Integration of an additional copy of a wild-type gene before conducting a mutant screen is another use of chromosomal integration. For example, we conducted a pilot screen for mutations that affect repression by LexA-Sin3 and identified numerous mutations in the RPD3 gene (1). We then integrated a second copy of RPD3 at the URA3 locus and thus were able to eliminate this common mutation in a subsequent screen. One might want to integrate a cassette that conditionally expresses a gene, either the wild-type or a dominant negative version, in order to screen for suppressors (2–5).
Integration of a sequence into the yeast genome is often done by cloning a DNA fragment into a Yeast Integrating (YIp) plasmid, such as YIp5 (which has a URA3 marker). Cleavage of the plasmid within the URA3 sequence and transformation into a ura3 strain usually leads to integration of the plasmid at the URA3 locus (6). This integration frequently results in the plasmid sequences being present between two copies of URA3 sequence, one being wild-type URA3 and the other the original mutant ura3 allele.
There are two difficulties with this common integration strategy. The first problem is that it uses up one of the markers, such as ura3, which may be needed for plasmids, libraries or disrupting specific genes. A second problem is that recombination can occur between the tandemly repeated sequences (i.e. URA3 and ura3 flanking the plasmid sequences), leading to loss of the integrated DNA fragment (6). Depending on exactly where recombination occurs, the strain that has excised the YIp plasmid may be URA3 or ura3, and thus continuous selection for uracil prototrophy does not guarantee maintenance of the integrated plasmid.
Loss of the integrated DNA sequences can cause problems for some genetic screens involving selections. For example, suppose one has integrated a YFG1-URA3 reporter gene for a screen in which the promoter of the YFG1 (Your Favorite Gene) gene is driving expression of the URA3 gene. One would mutagenize this strain and select for growth on 5-FOA (a uracil analog that selectively kills cells expressing the Ura3 enzyme; 7) to identify mutations that reduce expression of the YFG1 gene. However, the recombination between the repeats flanking the reporter gene will result in excision of the reporter gene and an undesired 5-FOA-resistant colony. In a second example we have integrated a DNA fragment with a GAL-YFG1* cassette. This fragment expresses the dominant negative YFG1* allele from the inducible GAL1 promoter, and thus the strain is unable to grow on galactose medium. A suppressor screen would be to look for either second site mutations or multicopy plasmids that allow this strain to grow on galactose. However, loss of the GAL-YFG1* cassette through recombination between the flanking repeats would also result in growth on galactose, if YIp-type integration constructs are used.
To overcome these problems we have constructed plasmids that target integration of desired sequences at the HO locus without generation of any tandem duplication, using either the KanMX or hisG-URA3-hisG selectable markers. The hisG-URA3-hisG marker confers uracil prototrophy, but recombination between the repeated hisG sequences results in loss only of the URA3 marker (8). Growth on 5-FOA can be used to select for strains that have returned to uracil auxotrophy. We have selected the HO locus as the target for integration for several reasons. HO encodes an endonuclease that initiates interconversion of the mating-type locus and promotes diploidization of haploid strains (9). The HO locus is not required for growth, and nearly all laboratory strains have a mutation at the HO locus. In fact, Baganz et al. (10) have quantitatively demonstrated that disruption of the HO gene has no effect on growth rate.
MATERIALS AND METHODS
Plasmid construction
Plasmid pUC21ΔBB was constructed by cleaving pUC21 (11) with BamHI and BglII followed by ligation to delete the fragment between the two sites; the resulting plasmid has neither a BamHI nor a BglII restriction site. Plasmid pUC21-NotI was constructed by inserting the 123 bp NotI polylinker fragment from plasmid pFA6b (12) into pUC21ΔBB that had been cleaved with NotI. A similar strategy was used to construct equivalent vectors with a Kanamycin resistance marker, pUK21ΔBB and pUK21-NotI, starting with plasmid pUK21 (11).
Plasmid HO-poly-HO was constructed in several steps. A 912 bp fragment (HO-L) with sequences from the HO promoter (–2720 to –1814 upstream from the ATG) was made by PCR with primers designed to generate a fragment with HinDIII and BsiWI overhang sequences (see below). This fragment was inserted into HinDIII–BsiWI digested pUC21-NotI. A 507 bp fragment (HO-R) with sequences from the 3′ end of the HO gene (+1199 to +1699 downstream of the ATG) was made by PCR with primers designed to generate a fragment with EcoRI and SfiI overhang sequences (see below). This fragment was inserted into EcoRI–SfiI digested pUC21-NotI. Plasmid HO-poly-KanMX4-HO was constructed by inserting the 1494 bp BsiWI–EcoRI fragment with KanMX4 from plasmid pFA6-KanMX4 (12) into BsiWI–EcoRI digested HO-poly-HO. Plasmid HO-hisG-URA3-hisG-poly-HO was constructed by inserting the 3853 bp BamHI–BglII fragment with the hisG-URA3-hisG cassette (8) into BamHI digested HO-poly-HO.
The PCR amplification of the HO-L was performed using the ‘sticky end PCR cloning’ method (13) using four primers, F852–F855 (Table 1). Briefly, this method involves performing two PCR reactions followed by DNA melting and annealing. PCR with primers F852 and F854 results in a blunt-ended fragment where each end contains 5 bp of either the HinDIII or BsiWI recognition sites. In a separate reaction, PCR with primers F853 and F855 results in a blunt-ended fragment where each end contains 1 bp needed for the HinDIII or BsiWI recognition sites. The two PCR reactions are mixed, melted at 100°C and then slow cooled to allow DNA annealing. This DNA annealing results in four types of DNA products, with different ends. One-quarter of the DNA products have overhangs for cloning into the vector digested with HinDIII and BsiWI. This method eliminates the sometimes problematic step of restriction digestion of DNA products after PCR amplification. The same method was used to prepare the HO-R fragment using primers F856–F859.
Table 1. Primers.
| F852 |
AGC TTA ATT ATC CTG GGC ACG AGT |
| F853 | TAA TTA TCC TGG GCA CGA GT |
| F854 | GAC GCC ATT TTA AGT CCA AAG |
| F855 | GTA CGA CGC CAT TTT AAG TCC AAA G |
| F856 | AAT TCC TGG GGG AAC AAC TTC AC |
| F857 | CCT GGG GGA ACA ACT TCA C |
| F858 | CAT AGG CCA CTG TAA GAT TCC GCC ACA T |
| F859 | AGG CCA CTG TAA GAT TCC GCC ACA T |
RESULTS AND DISCUSSION
Construction of pUC plasmids with different polylinkers
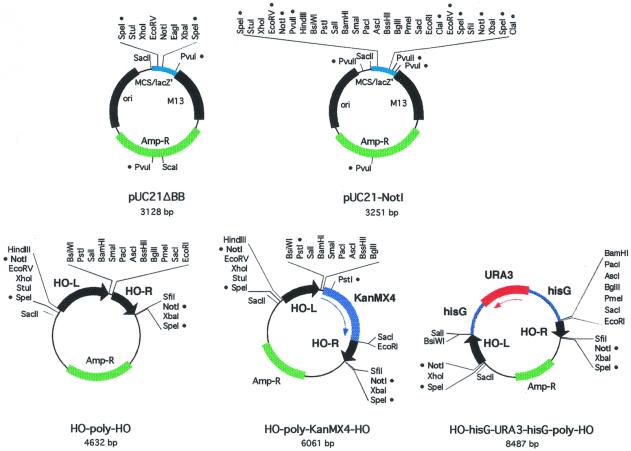
Beginning with plasmid pUC21 (11), we constructed two new plasmids, pUC21ΔBB and pUC21-NotI, with different polylinkers (Fig. 1 and Table 2). The NotI restriction endonuclease has an 8 bp recognition sequence, and thus cuts rarely in genomic DNA. pUC21-NotI has two NotI sites with a large polylinker in between. Thus, it is likely that any fragment inserted in this region can be excised with NotI. We also made two related vectors, pUK21ΔBB and pUK21-NotI, which have Kanamycin resistance markers (Table 2). All of these plasmids maintain a continuous open reading frame through the polylinker and the lacZ’ coding region, and can thus be used for blue/white screening for inserts during cloning. However, all of these plasmids turn blue more slowly than traditional pUC vectors due to a change in the spacing between the –10 and –35 regions of the bacterial promoter driving lacZ’ expression (11).
Figure 1.
Plasmid maps. Each plasmid map is drawn to scale, with restriction sites indicated. A bullet (•) next to a restriction site indicates that it is not unique.
Table 2. Plasmids.
| Plasmid |
Features |
Marker in Escherichia
coli |
GenBank accession no. |
ATCC number |
| pUK21ΔBB | Bacterial vector | Kan | AF324725 | 87799 |
| pUK21-NotI | Bacterial vector | Kan | AF324726 | 87800 |
| pUC21ΔBB | Bacterial vector | Amp | AF324723 | 87801 |
| pUC21-NotI | Bacterial vector | Amp | AF324724 | 87802 |
| HO-poly-HO | Integrating vector | Amp | AF324727 | 87803 |
| HO-poly-KanMX4-HO | Integrating vector, KanMX | Amp | AF324728 | 87804 |
| HO-hisG-URA3-hisG-poly-HO | Integrating vector, URA3 | Amp | AF324729 | 87805 |
Construction of plasmids for integration at HO
Plasmid HO-poly-HO (Fig. 1 and Table 2) was constructed by inserting two fragments from the HO gene into pUC21-NotI. These two fragments are 912 and 507 bp, respectively, and fragments of this size are very effective at directing homology-mediated recombination (14). These fragments were chosen for the lack of restriction sites present in the polylinker. The integration cassette can be excised by NotI cleavage. If the inserted fragment contains a NotI site there are several other sites in the polylinker that could be used.
Although we have not tested this, one could use this plasmid lacking a selectable marker for targeted integration at HO. One first inserts the URA3 gene into the HO gene, and then transforms this strain with the HO-poly-HO plasmid with an insert. Selection on 5-FOA should identify transformants where ho::URA3 has been replaced by the integrating vector, allowing retention of more available markers.
Plasmids HO-poly-KanMX4-HO and HO-hisG-URA3-hisG-poly-HO (Fig. 1 and Table 2) contain the KanMX4 and URA3 selectable markers, respectively. KanMX, which confers resistance to G418, is rarely used for maintaining YCp or YEp plasmids, and use of this marker here allows the use of other markers (i.e. URA3, HIS3, etc.) for other genetic manipulations. Selection for uracil prototrophy can be used to identify integration of the HO-hisG-URA3-hisG-poly-HO plasmid at the HO locus. Moreover, one can select for cells that have returned to the ura3 state by selection on 5-FOA, identifying yeast in which recombination has occurred between the hisG repeats flanking the URA3 gene (8). Thus, these cells which have the integrated sequences that are stably maintained in the absence of selection can be transformed with a URA3 plasmid.
There are a number of other markers available as cassettes that could be that inserted into the HO-poly-HO plasmid, including nutritional markers such as HIS3, LEU2 or TRP1 (15), as well as dominant drug resistance markers (16).
Efficiency of integration at HO
Several experiments were performed to determine the efficiency of integration at HO. Strain DY131, which has a HO-lacZ reporter integrated at the HO locus, was transformed with NotI cleaved HO-poly-KanMX4-HO and G418 resistant colonies were identified. The parent strain expresses lacZ from the HO promoter and this strain is blue on the chromogenic substrate X-gal. In contrast, integration of HO-poly-KanMX4-HO at the HO locus will eliminate the lacZ gene, and these colonies should be white on X-gal. We tested 14 transformants and found that all were white, showing that integration was very efficient. A similar experiment was performed with the HO-hisG-URA3-hisG-poly-HO plasmid, and we found that seven of eight Ura+ transformants were white.
In another test of the HO-poly-KanMX4-HO integrating vector, three separate genes were cloned into the polylinker. The plasmid was digested with NotI and the 10.6 kb NotI fragment was transformed into yeast. Ten independent G418-resistant colonies were examined, and for all 10 isolates PCR analysis demonstrated that the upstream and downstream endpoints of the integration were as predicted. For four of these 10 isolates, further PCR analysis was performed to characterize the internal sequences of the integrating fragment. The results showed that correct integration had occurred for all four. We conclude that integration is an efficient process.
Finally, we wish to draw attention to the ‘sticky end PCR cloning’ method (13) that we used in preparing these vectors (see Materials and Methods). One might have a fragment that one wishes to insert into these vectors, but there is a problem if the insert contains restriction sites for all of the sites in the polylinker. The sticky end PCR cloning method eliminates this problem, as there is no need to cleave the ends of the DNA after PCR amplification.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the NIH to D.J.S. and by grants from the NIH and ACS to J.M.S.
DDBJ/EMBL/GenBank accession nos AF324723–9
References
- 1.Dorland S., Deegenaars,M.L. and Stillman,D.J. (2000) Roles for the Saccharomyces cerevisiae SDS3, CBK1 and HYM1 genes in transcriptional repression by SIN3. Genetics, 154, 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burke D., Gasdaska,P. and Hartwell,L. (1989) Dominant effects of tubulin overexpression in Saccharomyces cerevisiae. Mol. Cell Biol., 9, 1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akada R., Yamamoto,J. and Yamashita,I. (1997) Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol. Gen. Genet., 254, 267–274. [DOI] [PubMed] [Google Scholar]
- 4.Liu H., Krizek,J. and Bretscher,A. (1992) Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics, 132, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandrock T.M., Brower,S.M., Toenjes,K.A. and Adams,A.E. (1999) Suppressor analysis of fimbrin (Sac6p) overexpression in yeast. Genetics, 151, 1287–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothstein R. (1991) Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol., 194, 281–302. [DOI] [PubMed] [Google Scholar]
- 7.Boeke J.D., LaCroute,F. and Fink,G.R. (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet., 197, 345–346. [DOI] [PubMed] [Google Scholar]
- 8.Alani E., Cao,L. and Kleckner,N. (1987) A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics, 116, 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herskowitz I., Rine,J. and Strathern,J. (1992) Mating-type determination and mating-type interconversion in Saccharomyces cerevisiae. In Jones,E.W., Pringle,J.R. and Broach,J.R. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Press, Plainview, NY, pp. 583–656.
- 10.Baganz F., Hayes,A., Marren,D., Gardner,D.C. and Oliver,S.G. (1997) Suitability of replacement markers for functional analysis studies in Saccharomyces cerevisiae. Yeast, 13, 1563–1573. [DOI] [PubMed] [Google Scholar]
- 11.Vieira J. and Messing,J. (1991) New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene, 100, 189–194. [DOI] [PubMed] [Google Scholar]
- 12.Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 13.Zeng G. (1998) Sticky-end PCR: new method for subcloning. Biotechniques, 25, 206–208. [DOI] [PubMed] [Google Scholar]
- 14.Ma H., Kunes,S., Schatz,P.J. and Botstein,D. (1987) Plasmid construction by homologous recombination in yeast. Gene, 58, 201–216. [DOI] [PubMed] [Google Scholar]
- 15.Jones J.S. and Prakash,L. (1990) Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast, 6, 363–366. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein A.L. and McCusker,J.H. (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast, 15, 1541–1553. [DOI] [PubMed] [Google Scholar]