Abstract
The past decade of cancer research has been marked by a growing appreciation of the role of immunity in cancer. Mutations in the tumour genome can cause tumours to express mutant proteins that are tumour specific and not expressed on normal cells (neoantigens). These neoantigens are an attractive immune target because their selective expression on tumours may minimize immune tolerance as well as the risk of autoimmunity. In this Review we discuss the emerging evidence that neoantigens are recognized by the immune system and can be targeted to increase antitumour immunity. We also provide a framework for personalized cancer immunotherapy through the identification and selective targeting of individual tumour neoantigens, and present the potential benefits and obstacles to this approach of targeted immunotherapy.
Recent advances in genome sequencing have revealed that during the process of initiation and progression, cancers acquire tens to thousands of different somatic mutations. Most of these mutations confer no intrinsic growth advantage (passenger mutations) and are often the result of genomic instability within the tumour. A smaller number of cancer mutations interfere with normal cell regulation and help to drive cancer growth and resistance to targeted therapies (driver mutations)1. To date, approximately 140 genes have been identified that can drive tumorigenesis2. However, both driver mutations and passenger mutations can alter amino acid coding sequences, collectively known as nonsynonymous mutations, causing tumours to express mutant proteins that are not expressed by normal cells3,4. These abnormal protein sequences are processed into short peptides (epitopes) and presented on the cell surface in the context of major histocompatibility complex (MHC; also known as human leukocyte antigen (HLA) in humans), thereby becoming recognizable to T cells as foreign antigens (FIG. 1).
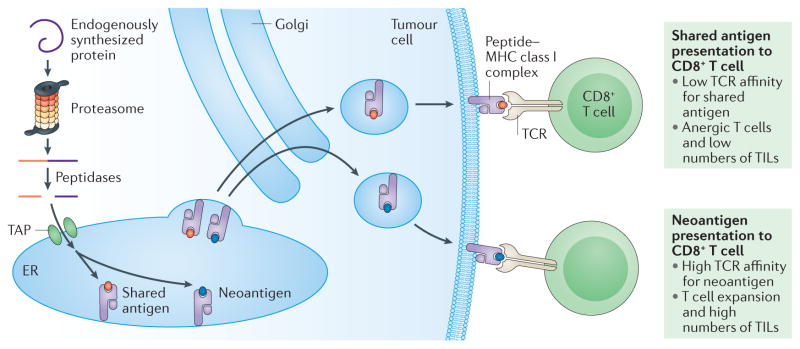
Figure 1. Tumour antigen processing and presentation on MHC class I.
To destroy established cancers, CD8+ T effector cells must recognize antigens displayed by major histocompatibility complex (MHC) class I molecules on tumour cells. This process begins with the ubiquitylation and proteasome degradation of endogenously synthesized proteins in the tumour cell into shorter sequences of 8–11 amino acids. These smaller peptides may undergo further cleavage by peptidases (including aminopeptidases and carboxypeptidases) in the cytosol and also in the endoplasmic reticulum (ER). Peptides enter the ER by way of the transporter associated with antigen processing (TAP) complex. In the ER, these peptides bind with variable affinities to MHC class I. Together, the peptide MHC class I complexes are delivered to the plasma membrane through the Golgi complex, where the peptide can be recognized by CD8+ cytotoxic T cells. Although some T cells do recognize antigens shared between both normal and tumour cells, T cell receptors (TCRs) typically bind with higher affinity to neoantigens23,24, and tumours expressing higher numbers of neoantigens are more likely to induce immune-mediated tumour elimination. The processing and presentation of exogenous antigens to CD4+ T helper cells via MHC class II on professional antigen presenting cells (APCs) follows somewhat similar steps, but MHC class II presents longer sequences of amino acids (11–20 amino acids or longer)118,119. TILs, tumour-infiltrating lymphocytes.
Cancers with a single dominant mutation can often be treated effectively by targeting the dominant driver mutation5. Former American Society for Clinical Oncology (ASCO) president George Sledge has termed such cancers “stupid cancers” for their sensitivity to targeted therapy6. By contrast, “smart cancers” have a much higher number of mutations, multiple simultaneous driver mutations and are less amenable to treatment with traditional targeted therapies6. However, these smart cancers with high mutational burdens often have high expression of aberrant proteins6–8. In the current immunotherapy era, these aberrant proteins are increasingly recognized as opportunities for the immune system to recognize and control tumour growth.
Owing to their selective expression in tumours, the tumour-specific antigens (TSAs) that arise from non-synonymous mutations and other genetic alterations are called neoantigens9 (TABLE 1). In the subset of human tumours with a viral aetiology, as with Merkel cell carcinoma (MCC) associated with the Merkel cell polyomavirus (MCPyV) and cancers of the cervix, oropharynx and other sites associated with the human papillomavirus (HPV), the proteins encoded by viral open reading frames are another type of neoantigen10–12. In addition to TSAs, there are two other broad categories of tumour antigen. Tumour-associated antigens (TAAs) are over-expressed in malignant cells but are also present in normal cells at low levels of expression9. Cancer/testis antigens (CTAs) are expressed by various tumour types and by reproductive tissues (for example, testes, fetal ovaries and trophoblasts) but have limited expression on other normal tissues in the adult and are generally not presented on normal reproductive cells, because these tissues do not express MHC class I molecules13,14.
Table 1.
Broad categories of tumour antigen
| Antigen type | Description | Examples of antigen type | Examples of approved immunotherapies for target antigen |
|---|---|---|---|
| Tumour-specific antigens8,9 |
|
None approved, multiple in clinical development | |
| Tumour-associated antigens9 |
|
||
| Cancer/testis antigens13,14 |
|
None approved, multiple in clinical development |
ALL, acute lymphoblastic leukaemia; HPV, human papillomavirus; MAGE, melanoma-associated antigen; PAP, prostatic acid phosphatase.
Initial attempts to target TSAs, including efforts targeting virus-associated antigens15, mutated growth factor receptors16,17 or mutated KRAS18,19, have provided an early validation of TSAs as tumour antigen targets. However, most neoantigens arise from unique mutations, and technical challenges related to the identification and targeting of neoantigens specific to individual tumours have prevented widespread adoption of an individualized approach. This Review discusses how such limitations are beginning to be overcome through major advancements in genomics and bioinformatics, including massively parallel sequencing (MPS) and epitope prediction algorithms. It is now widely recognized that the development of immune checkpoint inhibitors is transforming cancer care by providing unprecedented clinical benefit for some tumour types20,21. However, novel approaches to immunotherapy are now required to improve on current objective response rates (ORRs) and to extend benefit to all tumour types. In this Review we describe the relationship between neoantigens and sensitivity to current immunotherapies, and also provide a framework for the targeting of individual tumour neoantigens, with the potential to make antitumour immunity more specific, more effective and less toxic.
TSAs, TAAs and CTAs as targets
TSAs, TAAs and CTAs have all been considered as targets for immunotherapy (TABLE 1). Until now, most cancer vaccines have focused on commonly overexpressed TAAs because of the potential to treat many patients with the same therapeutic vaccine. Although CTA targets are also shared between tumours, they are known to be expressed in only a limited number of tumour types, potentially reducing the applicability of CTA-targeted immunotherapy. Unfortunately, most attempts to target TAAs with vaccines have met with limited success22, potentially because TAAs are normal host proteins and are therefore subject to both central and peripheral tolerance mechanisms13. Although some TAA-specific T cells do avoid negative selection, high-affinity T cell receptors (TCRs) for TAAs are preferentially depleted and the range of affinities of the remaining TCRs for TAAs is lower than TCR affinities for typical TSAs and other foreign antigens23,24. T cell cytotoxicity and activation are correlated with TCR binding affinity25, which supports the idea that immune responses against TAAs are less vigorous than those against TSAs.
A separate concern with TAAs and CTAs is the potential for autoimmune toxicities related to immune activation in non-target tissues, because these targets are expressed on normal host cells. Concerns about collateral damage to non-target tissues have received increased attention as modern immunotherapies have become potent enough to induce lethal autoimmunity26. In some cases the immune destruction of normal organs can be managed. For example, the destruction of all CD19-positive B lineage cells (both healthy and malignant) with anti-CD19 chimeric antigen receptor (CAR) T cells in patients with acute lymphoblastic leukaemia (ALL)27,28 can be managed with immunoglobulin replacement, or through reconstitution of B lineage cells with allogeneic stem-cell transplantation. However, in other cases, toxicity from off-target immune activation has led to serious complications. For example, attempts to target the TAA carbonic anhydrase 9 in renal cell carcinoma (RCC) led to severe liver toxicity owing to expression of this antigen in bile duct epithelial cells29, and likewise, an attempt to target ERBB2 (also known as HER2 or NEU) in a patient with a metastatic colon cancer that overexpressed ERBB2 led to rapid respiratory failure and death, probably due to low levels of ERBB2 expression on lung epithelial cells30. Similarly, attempts to use adoptive cell therapy (ACT) to target the CTA melanoma-associated antigen 3 (MAGE-A3) resulted in severe neurological toxicity and death, which was probably related to previously unrecognized expression of MAGE-A family members in the brain31. This case challenges the concept of high CTA specificity. Although CAR T cells have shown considerable promise against some haematological cancers27,28, the ability to treat solid cancers with CAR T cells and other targeted immunotherapies has thus far been severely limited by the lack of suitable TAA and CTA targets.
TSAs are theoretically a more attractive immunotherapy target because they may be recognized as non-self by the host immune system, and are therefore less likely to establish complex immune tolerance mechanisms. Additionally, immunotherapies targeting TSAs should theoretically be less likely to induce autoimmunity, because the target is not expressed on normal cells. Despite these theoretical advantages, T cell responses against TSAs do not ensure clinically detectable anti-tumour activity, as evidenced by the inability of a host to eradicate melanoma despite the presence of high frequencies of cytotoxic T lymphocytes (CTLs) before vaccination32. Furthermore, neoantigen-specific approaches to immunotherapy cannot completely eliminate the risk of autoimmunity, because neoantigen-specific T cells can be cross-reactive with the non-mutated version of the antigen. This is exemplified by cancer-induced auto-immune disease. A recent study identified eight patients with cancer who concurrently developed scleroderma, an autoimmune disease, with detectable autoantibodies against RNA polymerase III subunit RPC1. Although these patients had various malignancies (breast, ovarian and colon), it was demonstrated that most tumours from these patients (six of eight) had specific genetic alterations in POLR3A, the gene that encodes RPC1. Therefore, the neoantigen-specific T cells against mutated POLR3A in the context of a malignancy seem to be driving the autoimmune disease process33.
Neoantigens and the immune response
Evidence for immune system recognition of tumour neoantigens
The immune system has an extraordinary ability to distinguish self from non-self. This inherent property of the immune system gives it the ability to recognize and target non-self antigens on cancer cells to control cancer. In 1943, Gross and colleagues34 were among the first to report that the immune system can recognize and destroy cancer cells. They showed that mice with resected tumours were protected against subsequent re-exposure from the same tumour cells, and that similar protection against tumour cells could be induced by first exposing mice to lethally irradiated tumour cells. However, at the time, the nature of the antigens that enabled the immune system to reject cancers was not clear. In the late 1980s the Boon group35,36 made the important initial observation in a mouse model that antitumour T cells can recognize aberrant peptides derived from tumour-specific mutations. Several years later, somatic mutations were also shown by several separate groups to be a source of neo-antigens recognized by T cells in human tumours37–40. Another important advance in our understanding of TSAs occurred in 2005, when Wölfel and colleagues41 analysed the naturally occurring antitumour T cell response against melanoma in a single patient. They found that the T cells of the patient were reactive against five mutated epitopes resulting from nonsynonymous mutations and that this immunoreactivity against melanoma neoantigens predominated over the response to TAAs. Similarly, the Rosenberg group42 showed that the adoptive transfer of ex vivo-expanded tumour-reactive tumour-infiltrating lymphocytes (TILs) into a patient with melanoma could cause complete tumour regression, and that these T cells reacted to mutated forms of two neoantigens on the melanoma cells. These neoantigen-specific T cells persisted at high levels in the tumour 1 month after transfer. Together, these studies provided support for the role of neoantigens in the naturally occurring antitumour T cell response.
More recent support for neoantigens as important tumour antigens for the human immune system has emerged from studies of novel immune checkpoint inhibitors targeting cytotoxic T lymphocyte-associated protein 4 (CTLA4) and programmed cell death protein 1 (PD1), which are expressed by activated T cells43,44. The remarkable clinical activity of immune checkpoint inhibitors against a wide variety of human cancers has provided indisputable evidence that the immune system can recognize and destroy established cancers20,21. To destroy established cancers in the presence of immune checkpoint inhibitors, T cells must recognize antigens displayed by MHCs on tumour cells (FIG. 1). Theoretically, any class of tumour antigen could be recognized by T cells; however, it is increasingly clear that TSAs serve as the most important tumour antigens in certain cancers where immune checkpoint inhibitors have shown clinical efficacy. For example, TSAs have been demonstrated to be T cell targets of sarcoma-bearing mice treated with immune checkpoint inhibitors, and administration of peptide vaccines incorporating these antigens into mice could achieve similar outcomes to immune checkpoint blockade45. More recently, PD1-expressing neoantigen-specific T cells have been identified in the peripheral blood of patients with melanoma, correlating with the recently documented activity of PD1 inhibitors in this population46.
Tumour somatic mutation frequency and sensitivity to immune checkpoint blockade
Although the full therapeutic potential of immune checkpoint inhibitors remains undefined, it is evident from the available human clinical data that the frequency of somatic mutations within a tumour type, and by extension, the potential for neoantigens within a tumour type, is largely correlated with sensitivity to immune checkpoint inhibitors (FIG. 2). Two tumour types with notably high ORRs to immune checkpoint blockade are melanoma47–51 and non-small-cell lung cancer (NSCLC)52–54. Both cancers are usually formed in the presence of mutagens (ultraviolet light in the case of melanoma and tobacco smoke in the case of NSCLC55), and as a result they have amongst the highest somatic mutation burdens of any cancer type. By comparison, immune checkpoint inhibitors thus far have shown little or no activity in the subset of cancers with lower mutation burdens, such as Ewing sarcoma56 and prostate cancer26,57. The clinical observation that somatic mutations and potential for neoantigens in a tumour type are correlated with objective clinical responses to immune checkpoint inhibitors strongly supports the idea that neoantigens are important immunotherapy tumour antigens.
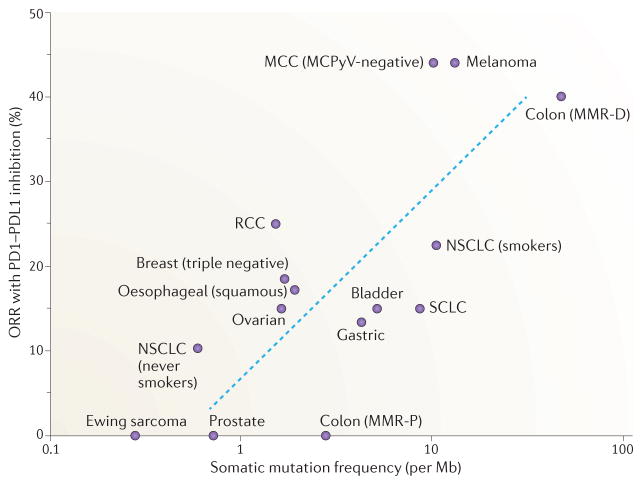
Figure 2. Correlation of tumour somatic mutation frequency with objective response rates to immune checkpoint blockade.
The full potential of programmed cell death protein 1 (PD1)–PD1 ligand 1 (PDL1) checkpoint inhibitors remains undefined; however, from the available clinical data there is a positive correlation between tumour somatic mutation frequency and clinical benefit, measured by the objective response rate (ORR). The ORR is defined in the context of these clinical studies as the proportion of patients who achieved 30% decrease in the sum of the longest diameters of target tumours based on modified Response Evaluation Criteria in Solid Tumours (RECIST)157. Here, we show the somatic mutation frequency (somatic mutations per megabase) and ORR to single-agent PD1 or PDL1 inhibition across multiple solid tumours of non-viral origin for which clinical and sequencing data are available. Tumours with the highest somatic mutation rates (mismatch repair-deficient (MMR-D) colon cancer, Merkel cell polyomavirus (MCPyV)-negative Merkel cell carcinoma (MCC), melanoma and non-small-cell lung cancer (NSCLC) in current or prior tobacco smokers) have amongst the highest ORRs to PD1 PDL1 inhibition (23–44%), whereas tumours with lower mutation rates have demonstrated less frequent responses. These data are obtained from a diverse group of clinical trials conducted at different stages of disease and included are some early-stage exploratory clinical trials, which probably accounts for some of the variability that is observed. However, extrapolating from these data, we can predict the ORR to single-agent PD1–PDL1 blockade for any tumour type with the formula ORR (%) = 0.08 × ln(x)+ 9, where x is the somatic mutation frequency per Mb of DNA. The blue dashed line on the graph represents the line of best fit calculated from the formula. Many PD1–PDL1 inhibitor trials are ongoing and will further clarify the slope of this correlation. MMR-P, mismatch repair proficient; RCC, renal cell carcinoma; SCLC, small-cell lung cancer.
Somatic mutation burden seems to be an important marker of immune response even within a particular tumour type. This was recently illustrated in a series of clinical trials in colorectal cancer. Although most colorectal cancer has DNA mismatch repair proficiency (MMR-P), a small subset of colorectal cancer (<5%) have DNA mismatch repair deficiency (MMR-D), also referred to as microsatellite instability (MSI). As mismatch repair proteins correct errors in DNA replication, colorectal cancers with MMR-D are marked by genomic instability, a remarkably high mutation burden and the potential for high numbers of neoantigens. In clinical trials of PD1 inhibitors in unselected populations of patients with colorectal cancer, little to no activity was observed26,58. However, a clinical trial of the PD1 inhibitor pembrolizumab in a selected population of patients with MMR-D colorectal cancer demonstrated high response rates58. Therefore, within colon cancer, MMR-D predicts responses to immune checkpoint inhibition. A similar correlation between intratumour mutation burden and clinical benefit from immune checkpoint inhibition has been demonstrated in melanomas43 and lung cancers44, providing additional support for the role of tumour mutations and associated neoantigens in the antitumour immune response.
Although tumour somatic mutation burden is largely correlated with clinical response to currently available immunotherapy agents, the neoantigens resulting from somatic mutations are only one component dictating the rate and quality of immune response to immune checkpoint inhibitors. As a result, the association between potential neoantigens and response to immune checkpoint blockade is not linear (FIG. 2) and does not preclude multiple additional factors influencing responses to immune checkpoint inhibitors. For example, in the study discussed above that correlated intratumour mutation burden in melanoma with response to immune checkpoint blockade, there were multiple patients with high mutational burden who did not benefit from CTLA4 blockade, indicating that mutation burden alone is not sufficient to predict benefit43. One possible explanation for this varied therapeutic benefit may be that not all tumour neoantigens are expressed and/or confer increased MHC class I binding. Another potential explanation is that neoantigens may need to be expressed in every tumour cell in order to be targeted, as patients with NSCLC and melanoma whose tumours contain increased clonal neoantigen burden have improved overall survival and increased response to PD1 and CTLA4 blockade 59. It is also possible that some patients lack T cells that are capable of recognizing the available neoantigen pool, or that capable T cells are present but suppressed by additional or other regulatory mechanisms. Many additional genomic60 and non-genomic features61 have recently been described that also contribute to anti-PD1 response patterns. As demonstrated in FIG. 2, there are also some cancers that respond to immune checkpoint blockade at a higher or lower frequency than would be anticipated from mutation burden alone. For example, RCC, despite having a modest mutation burden, was one of the few cancers to show durable responses to the cytokine interleukin-2 (IL-2), the first effective systemic cancer immunotherapy62. IL-2 induces the growth and proliferation of T cells and natural killer (NK) cells and acts by expanding pre-existing cancer-targeting effector T (Teff) cells63. More recently, RCC also demonstrated high sensitivity to PD1 immunotherapy64. Additional research on the RCC tumour microenvironment (TME) is needed to understand why this tumour type responds disproportionately well to current immunotherapies.
MCC is another cancer that illustrates the complexity of the relationship between tumour antigens and the antitumour immune response. MCC is a relatively rare and aggressive cutaneous neoplasm that can either be caused by MCPyV or can form as a result of somatic mutations resulting from ultraviolet light. MCPyV-positive MCCs generally have a low mutation burden, whereas MCPyV-negative MCCs consistently demonstrate a higher number of ultraviolet light-induced somatic mutations65. If the absolute number of neoantigens singularly mediated sensitivity to immune checkpoint blockade, one would anticipate that MCPyV-positive MCC would show little sensitivity to such therapies. And yet MCPyV-positive MCCs are characterized by a robust lymphoid infiltrate, high PD1 ligand 1 (PDL1) expression and high sensitivity to immunotherapy66,67. In fact, despite having a fraction of the number of somatic mutations, MCPyV-positive MCCs have demonstrated a response rate to PD1 immune checkpoint blockade equivalent to or even higher than that of MCPyV-negative MCC66. In the case of MCPyV-positive MCC, MCPyV DNA is clonally integrated into the tumour genome10, and viral antigens probably serve as TSAs and induce a robust immune response. This supports the idea that neoantigens derived from viruses can serve as strong immune stimulants and may be an attractive target for cancer immunotherapy in some cancers.
It remains unclear what frequency of somatic mutations generate expressed and targetable neoantigens, what percentage of neoantigens are depleted during tumour progression as a result of immunoediting (discussed in detail below) and ultimately what percentage of human cancers contain candidate neoantigens for successful immune targeting. However, there is still reason to believe that tumours with low mutation burdens continue to express neoantigens and will be susceptible to some forms of immune attack, even if clinical responses with single-agent PD1 immune checkpoint inhibitors are rarer in such tumours. For example, medulloblastoma has one of the lowest mutational burdens of any adult cancer type68, yet frequently expresses indoleamine 2,3 dioxygenase 1 (IDO1), an enzyme implicated in immunosuppression, ostensibly to foster immune escape69. Similarly, acute myeloid leukaemia (AML), which has a low mutation burden68, often overexpresses PDL1 (REF. 70) as well as IDO1 (REF. 71). In a separate study, the leukaemic cells from 12 of 15 patients with AML were found to suppress T cell proliferation in vitro by secreting arginase II, an enzyme that actively converts arginine into urea. Arginine is an important modulator of lymphocyte and monocyte function, indicating that leukaemic cells deplete arginine from the TME in an arginase II-dependent manner to create an immunosuppressive microenvironment. Therefore, leukaemic cells in most patients with AML actively modulate the TME, presumably to avoid immune detection72. Although the clinical utility of novel immunotherapies remains undefined in this disease, early studies using the cytokine IL-2 in AML also provide some preliminary clinical evidence that immunotherapy may be successful in tumours with low mutation burdens73,74. Recent work from the Rosenberg group75 further demonstrated that cancers with lower mutational frequencies may be susceptible to immunotherapy. This group successfully identified tumour mutation-specific T cells in 9 of 10 patients with metastatic gastrointestinal tumours, a collection of tumours with relatively modest mutation burden. Collectively, these observations suggest that most adult malignancies have neoantigens, are recognizable by the immune system and are potentially susceptible to immune-targeting interventions. Additional research is needed to determine whether childhood malignancies, which often arise from the dysregulation of developmental pathways and have markedly fewer somatic mutations than adult malignancies76, will also be amenable to therapies enhancing the immune recognition of neoantigens.
In cancers with higher mutation burdens, in which clinical benefit is observed more often with immune checkpoint inhibitors, objective clinical responses still occur in only a minority of patients and complete clinical responses in which all measurable tumours completely disappear are rare47–54. Combinations of immune checkpoint inhibitors may increase response rates, but such combinations are often limited by treatment-related adverse events associated with immune activation in non-target tissues. For example, in the pivotal phase III clinical trial in melanoma combining the PD1 antibody nivolumab with the CTLA4 antibody ipilimumab, 84% of patients receiving this combination required immunomodulatory agents such as topical or systemic steroids to manage adverse immunological events51. Therefore, even in cancers with a high number of mutations, immune checkpoint inhibitors have a relatively small therapeutic index, operating in the narrow space between antitumour immunity and autoimmunity. How to potentiate antitumour immune responses against TSAs in cancers with low or high mutation burdens, without inducing autoimmunity, remains one of the challenges of current immunotherapy research.
Cancer immunoediting and antigen loss
Increasing evidence suggests that the immune system interacts actively with tumour antigens before tumours become clinically discernible. This process has been proposed to include three phases: elimination, equilibrium and escape, known collectively as cancer immunoediting77,78 (FIG. 3).
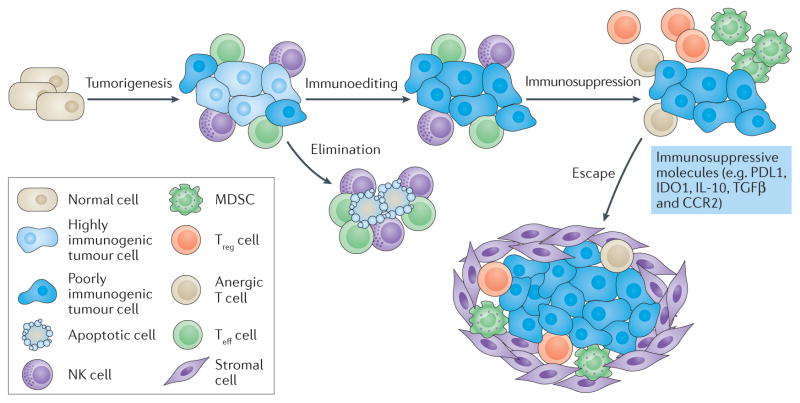
Figure 3. Cancers acquire immune tolerance.
During tumorigenesis, cancers acquire genetic changes that result in the presentation of neoantigens. Innate immune cells and primed adaptive immune cells recognize neoantigens and work together to eliminate the newly formed cancer cells. Cancers that survive the initial elimination phase enter an equilibrium phase in which the tumour remains at a stable size. During this phase, some of the most immunogenic neoantigens are edited out as the immune system selects for less immunogenic disease clones. This process selects for clones with downregulation of antigen processing and presentation on major histocompatibility complex (MHC) class I, as well as clones that have silenced or deleted genes that provide antigens targeted by the immune system. Cancers further avoid immune recognition and destruction through the downregulation of immune activation pathways (including CD137, OX40, CD40 and CD40 ligand (CD40L)), the upregulation of immunosuppressive pathways (including programmed cell death protein 1 ligand 1 (PDL1), PDL2, lymphocyte activation gene 3 (LAG3), T cell immunoglobulin mucin receptor 3 (TIM3), indoleamine-2,3-dioxygenase 1 (IDO1), cytotoxic T lymphocyte-associated antigen 4 (CTLA4)), and changes in expression of cytokines and chemokines or their receptors. Tumours also induce the production of immunosuppressive cytokines (for example, transforming growth factor-β (TGFβ), interleukin-10 (IL-10) and vascular endothelial growth factor (VEGF)) and recruit immune cells that actively mediate tolerance (e.g. myeloid-derived suppressor cells (MDSCs), regulatory T (Treg) cells). Activation of metabolic pathways, such as arginase, adenosine, inducible nitric oxide synthase (iNOS) and glucose consumption by tumours, also leads to immunosuppression in the tumour microenvironment. The remaining T effector (Teff) cells in this immunosuppressive tumour microenvironment become anergic (unresponsive and cannot be activated by antigen) and exhausted. As solid tumours begin to grow, they further impair Teff cells by inducing a stroma, a physical barrier for the immune system that consists of fibroblasts, endothelial cells and other cell types. At this point cancer begins to ‘escape’ the immune system and become clinically evident. CCR2, C-C motif chemokine receptor 2; NK, natural killer. This figure has been adapted from REF. 77, Macmillan Publishing Limited.
The elimination phase refers to elimination of pre-cancerous lesions via innate and adaptive immune mechanisms. This is implied by evidence that lymphocyte-deficient and interferon-γ(IFNγ) receptor (IFNGR)-deficient mice acquire more tumours following exposure to the carcinogen 3′-methylcholanthrene (MCA) when compared with wild-type mice, suggesting that both lymphocytes and IFNγ participate in the tumour elimination process79. IFNα receptor (IFNAR) expression on dendritic cells (DCs) also seems to be essential for tumour elimination80. Tumours are also eliminated by NK cell-induced apoptosis, which is mediated by perforin, granzyme and tumour necrosis factor (TNF)-related apoptosis inducing ligand (TRAIL)81. Expression of other molecules, such as the ligands for the activating NK cell receptor D (NKG2D), by tumours results in tumour eradication82–84.
Precancerous lesions that are not eliminated proceed to the equilibrium phase, in which the immune system holds the lesion in check and prevents it from becoming clinically detectable. The equilibrium process has been inferred from clinical observations. For example, two patients with renal failure who received kidney transplants from the same donor were placed on immunosuppressants, and both of these patients subsequently developed melanoma85. It was later discovered that the donor had a history of excised melanoma but had been clinically cancer-free for 15 years. This suggests that the immune system of the donor had maintained a state of equilibrium with the melanoma for many years, keeping the cancer from becoming clinically detectable until it was transplanted into immunosuppressed recipients. Furthermore, mice treated with MCA without overt evidence of tumour formation, subsequently grew tumours after treatment with antibodies blocking adaptive immunity86. This suggests that a robust immune system prevents some precancerous lesions that survive the elimination phase from further growth.
Throughout the equilibrium phase, tumours may undergo a process of editing, in which the most immunostimulatory antigens are lost through the immunoselection of less immunogenic disease clones. This process selects for clones with downregulation of antigen processing and presentation on MHC class I, as well as clones that have silenced or deleted genes that provide antigens targeted by the immune system33,87–90. Evidence for immuno-editing first came from the Boon group91 who showed that tumour escape in the P815 mastocytoma tumour mouse model coincided with the acquisition of tumour clones with antigen loss. The authors also showed that these mice were simultaneously able to eliminate injected tumours formed from clones that had not lost antigens, demonstrating that the host had not become tolerant of the tumour cells but that the tumour had evolved to evade the host immune response. In subsequent experiments, primary MCA-induced sarcomas formed in lymphocyte-deficient mice were rejected when implanted into immunocompetent mice, whereas primary MCA-induced sarcomas formed in wild-type mice continued to grow when implanted into lymphocyte-deficient mice86,92. This suggests that tumours growing in immunodeficient mice are more immunogenic, thus demonstrating that immune system–tumour interactions in early tumorigenesis led to tumour immunoediting and subsequent escape.
Escape refers to how tumours evade immune control and ultimately become clinically discernible. Four mechanisms by which this occurs are: via tumour expression of immunosuppressive molecules and antiphagocytosis signals; via tumour secretion of immunosuppressive factors; via tumour modulation of metabolism; and via recruitment of immune cells that actively mediate tolerance. Tumours can acquire an immunosuppressive phenotype via expression of PDL1, IDO1 or other immuno-suppressive proteins93–95 and expression of CD47 to inhibit phagocyte-dependent clearance96. Factors secreted by tumours to modulate immune function include vascular endothelial growth factor (VEGF), adenosine, IDO1, IL-10, transforming growth factor-β (TGFβ) and galectins97. Activation of metabolic pathways, such as arginase, inducible nitric oxide synthase (iNOS) and glucose consumption by tumours also leads to immunosuppression in the TME and tumour growth72,98,99. Immunosuppressive immune cells such as IDO1-expressing DCs, activated regulatory T (Treg) cells and myeloid-derived suppressor cells (MDSCs) are recruited into the TME, leading to tumour tolerance100,101. For example, tumours induce DCs in the TME to express IDO1 (REF. 102), resulting in Treg cell activation and PD1-dependent T cell suppression by Treg cells100. MDSCs exert their immunosuppressive function via reactive oxygen species, generated by NADPH oxidase 2 (NOX2, encoded by CYBB)103,104, and IDO1-expressing DCs require T cells to have functional general control non-derepressible 2 (GCN2; also known as eIF2AK4), a protein kinase that becomes upregulated in response to amino acid deprivation, in order to exert immunosuppressive function105. Collectively, this suggests that the immunosuppressive TME and tumour growth are promoted by multiple intricate signals involving the tumour, several different immune cells and varied metabolic pathways.
The popular framework of elimination, equilibrium and escape may not occur as originally described in all cases. For example, tolerance may develop earlier, even in the premalignant phase106, and the immune system may fail to have a notable impact on tumour development in some situations107. Furthermore, there is no clear distinction between the various phases in tumour progression, as both equilibrium and escape describe the same biological phenomenon of reduced immunogenicity and antigenicity. However, the hypothesis of tumour elimination, equilibrium and escape illustrates the principle that tumours orchestrate a complex signalling network to induce immune tolerance. Neoantigen-directed immunotherapies seek to overcome the immunosuppressive mechanisms that have been established by tumours, by targeting the neoantigens that have not been eliminated during this process.
A framework for targeting neoantigens
Although neoantigens are appealing targets for cancer immunotherapies, most neoantigens arise from unique mutations and are not shared between different patients. Thus, neoantigen-directed immunotherapy will probably need to be personalized. Until recently, the discovery of neoantigens relied on classic screening approaches that were highly labour intensive and impractical on a large scale. However, with the advent of relatively inexpensive next-generation sequencing, it is now possible to systematically predict cancer neoantigens for individual patients7,8 (FIG. 4).
Figure 4. A framework for identifying and targeting tumour neoantigens.
Whole-exome sequencing is carried out on tumour cells and matched normal tissue to identify the somatic mutations expressed in the tumour cells. Software algorithms designed to predict major histocompatibility complex (MHC)/human leukocyte antigen (HLA) binding affinity are then used to prioritize which predicted neoantigens are most attractive for immune targeting. Attributes that may be considered during the selection of a neoantigen target are listed. Next, neoantigen vaccines or cellular therapies are used to target the predicted neoantigens. Such therapies should be combined with immune checkpoint inhibitors and other immune-based therapies to overcome immunosuppressive mechanisms in the tumour microenvironment that inhibit neoantigen-specific immune responses. A2AR, adenosine A2A receptor; ACT, adoptive cell therapy; B7H3, also known as CD276; CD137, also known as 4-1BB; CD40L, CD40 ligand; CTLA4, cytotoxic T lymphocyte-associated antigen 4; GITR, glucocorticoid-induced tumour necrosis factor receptor family related protein; ICOS, inducible T cell co stimulator; IDO1, indoleamine-2,3-dioxygenase 1; LAG3, lymphocyte activation gene 3; MDSCs, myeloid derived suppressor cells; PDL1, programmed cell death 1 ligand 1; TIM3, T cell immunoglobulin mucin receptor 3.
Approaches to predicting and prioritizing neo−antigens
Tumour-specific somatic mutations can be identified by comparing DNA sequences isolated from tumour and matched normal host cells using MPS2,3,75. Of the approximately 3 billion nucleotides in the human genome, only a small percentage (30 Mb or 1%) of these nucleotides code for known expressed genes (the exome)108,109. Therefore, sequencing only the functional exome can significantly reduce costs and minimize the complexity of the analysis. Detailed information describing whole-exome sequencing can be found elsewhere108,109. Briefly, the process begins with extraction of DNA from host and tumour cells and breakdown of the DNA into fragments. Coding fragments of DNA are sequestered with artificial DNA or RNA baits that are complementary to the targeted DNA, and the noncoding sequences are discarded. The coding sequences are then amplified, sequenced and analysed using a reference genome. The DNA mutations can subsequently be constructed into an altered sequence of amino acids by matching each DNA codon with its corresponding amino acid, providing the initial data required to prioritize tumour neoantigens.
Multiple approaches have been used to prioritize these candidate neoantigens. Dedicated software algorithms have been developed that attempt to predict how these aberrant proteins will be processed into smaller peptides through proteasomal cleavage110 and which peptides will be transported into the endoplasmic reticulum by transporter associated with antigen processing (TAP)111,112 (FIG. 1). However, such algorithms are not reliable at present, and for this reason algorithms based on proteosomal processing and TAP transport are not a preferred approach. Currently, the best established and most promising tool for identifying T cell neoepitopes are software algorithms designed to predict MHC/HLA binding affinity113–115. However, there are some challenges with using current MHC/HLA binding predictions. First, human tumour cells, like normal cells, can express up to six MHC class I molecules and binding affinities cannot be predicted for all MHC class I molecules. Second, as current approaches rely on whole-exome sequencing, they can reliably predict only neoantigens arising from point mutations and insertions and/or deletions of 8–10 nucleotides or smaller present in the exome116. Therefore, they cannot predict certain neo-antigens arising from long insertion and/or deletion variants, splicing aberrations, gene fusions, epigenetic alterations and post-translational modifications such as phosphorylation or deglycosylation. Lastly, predicting binding affinities for MHC class II molecules has proved more difficult, as these molecules present longer sequences of amino acids (11–20 amino acids, or even longer, as the peptide-binding groove is open, unlike that of MHC class I) than MHC class I molecules (8–11 amino acids)117,118. As a result, the predictive performance is generally not as reliable119. The need for improved predictive algorithms for MHC class II molecules, which present exogenous antigens to CD4+ T cells, has received renewed attention because of recent evidence that CD4+ T cells recognize a higher number of neoantigens than was previously appreciated and, moreover, can generate potent antitumour activity120,121.
Using current MHC/HLA binding prediction algorithms, a significant fraction of mutations have been predicted to generate peptides that will bind to MHC class I molecules. In 2008, Segal et al.3 predicted that the average human breast or colon cancer would carry 40–60 neoantigens and that approximately 10% of somatic mutations generate peptides that bind to MHC class I molecules. This estimation does not take into account certain factors that might limit peptide generation, transport and MHC binding, such as processing by the proteasome and by peptidases110–112. But how many of these predicted neoantigens are bona fide neo-antigens that can be recognized by patients’ T cells? The precise answer is not known, but it is clear that in most cases only a small fraction of predicted neoepitopes are recognized by T cells from the patient122,123. For example, Robbins et al.124 predicted 229 tumour-specific neoepitopes across three patients with melanoma using NetMHC, an MHC class I binding prediction algorithm. However, T cell immune responses were detected to only 11 (4.8%) of these neoepitopes. Despite these limitations, selection based on predicted binding affinity does seem to enrich for true neoepitopes that can be recognized by T cells from the patient. For example, Fritsch et al.125 and van Buuren et al.126 retrospectively applied a prediction algorithm to try to predict naturally recognized tumour neoepitopes that are associated with immune responses. Both groups found that most neoepitopes were successfully predicted to have strong or moderate binding affinity using current prediction algorithms. These studies suggest that predictive algorithms demonstrate high sensitivity (and therefore would not be expected to overlook many neoepitopes); however, the specificity of these algorithms, that is, their accuracy in correctly identifying bona fide neoantigens that can be recognized by the T cells of the patient, is difficult to assess. The simultaneous use of multiple algorithms averaged together into a single score to prioritize neoantigens might improve the specificity of available algorithms.
Several other tools have been explored to improve the reliability of available algorithms. One explanation for the low success rate of available MHC class I binding prediction algorithms such as NetMHC is that most neoepitopes will have non-mutated versions that also bind to MHC class I molecules. Therefore, T cells that would potentially react to the neoepitopes may be depleted or tolerized by the non-mutated counterpart of the antigen. To select for neoantigens with peptide-binding determinants that are most dissimilar from their non-mutated counterparts and are therefore more likely to be recognized by the patient’s T cells, Duan et al.127 recently created a novel algorithm that they named the “differential agretopicity index” (DAI; where agretopicity is the ability of a peptide to bind to MHC) that subtracts the NetMHC scores of the non-mutated form of the neoepitope from the corresponding NetMHC scores of the predicted neoepitope. Interestingly, whereas Fritsch et al.125 suggest that neoepitopes are more commonly generated from mutations in residues in exposed regions of the peptides (these TCR-exposed regions are crucial for the interaction with the TCR), Duan et al.127 found that amino acid substitutions of residues that anchor peptides within MHC molecules make the biggest contributions (both positive and negative) to the DAI. A separate tool for validating presented neoantigens has been to confirm the presentation of predicted epitopes using mass spectrometry. In a recent report of this approach, only a small fraction of predicted high-binding peptides were confirmed by mass spectrometry122. This indicates that the sensitivity of the peptide purification and mass spectrometry is not sufficient to detect certain antigens. Moreover, reliance on mass spectrometry is probably too impractical for clinical use.
Interestingly, Carreno et al.123 observed a much higher hit rate than other reported studies. This study has key differences compared with others. First, this group used a more complicated predictive approach and included physical peptide-binding studies. Therefore, they confirmed that the peptides they used bound to their predicted HLA class I molecules. Second, they immunized the three patients with advanced melanoma with seven selected peptides. This is unique in that no other study has used lymphocytes from vaccinated individuals. In each of the three patients, they saw T cells specific for one of the seven peptides before vaccination and three of the seven peptides after vaccination. More specifically, they found that even for neoepitopes that had pre-existing T cell responses, the T cell repertoire expanded after vaccination and that for neoepitopes that were undetectable before vaccination T cell responses were induced. This study demonstrates that vaccination can induce neoantigen-specific T cell responses that do not occur naturally. Thus, neoantigens can be divided into two broad categories: ‘dominant’ neo-antigens that spontaneously induce immune responses and ‘subdominant’ neoantigens that do not naturally induce immune responses but instead require immunization to induce a response against them. At this point, as most studies have not focused on vaccinated patients, it remains somewhat unclear how common subdominant neoantigens are or how useful they are as tumour antigen targets; the work of Carreno et al.123 proves only that they exist. However, if the dominant neoantigens of a tumour are eliminated as a result of immunoediting, subdominant neoantigens may provide an attractive target for immunotherapies, as long as they are processed and presented by HLA molecules on the surface of tumour cells.
Therapeutic approaches for targeting predicted neo−antigens
Multiple approaches for targeting personalized neoantigens have been proposed, but the two approaches that are best established are ACT using neoantigen-specific T cell products and personalized vaccines encoding predicted neoantigens. Antibody-based therapies, including bispecific T cell-engaging antibodies128–130 and CAR T cells27,28,131,132, are also potent immunotherapies with remarkable therapeutic activity against certain TAAs. Although such therapies are currently used only to target extracellular proteins, they could theoretically be designed to target intracellular tumour antigens presented on the cell surface in the context of MHC by specifically engaging the peptide–MHC complex on the cell surface133. However, the process of antibody design is currently too impractical and costly for widespread use targeting individual tumour neoantigens.
In 2014, the Rosenberg group121 provided proof of principle for neoantigen-specific ACT by successfully inducing partial regression of a metastatic cholangiocarcinoma by infusing neoantigen-specific CD4+ T cells against a neoantigen identified by whole-exome sequencing. The cholangiocarcinoma of this patient had only 26 nonsynonymous mutations, again supporting the idea that tumours with modest numbers of neoantigens are susceptible to neoantigen-specific immunotherapy approaches. More recently, the Rosenberg group18 successfully treated a patient with KRAS-mutant colorectal cancer with an enriched population of CD8+ T cells that reacted to the specific KRAS mutation expressed by the colorectal cancer. The patient had a partial response upon therapy that lasted approximately 9 months, at which point one of the lung metastases of the patient began to increase in size. Upon resection of the single progressing lung lesion, it was revealed that the progressing lesion contained the targeted KRAS mutation but had acquired a defect in antigen presentation, thereby providing a direct mechanism of immune evasion by the progressing lesion. Although autologous T cell transfer has shown promise, recent work from Strønen et al.134 suggests that ‘outsourced’ exogenous T cells obtained from healthy blood donors, rather than endogenous patient T cells, may broaden neoantigen-specific T cell reactivity and enable the targeting of neoantigens that have not been recognized by the patient’s own immune system. This study showed that T cells from healthy donors were able to specifically recognize 11 of 57 predicted neoepitopes across three patients, supporting the idea that T cell repertoires from healthy donors can provide a rich source of T cells and can enable personalized immunotherapy, independently of endogenous immune function.
Personalized vaccines, designed to present the neo-antigen or neoantigens coupled to an adjuvant to prime and activate DCs, could also be used to selectively target predicted neoantigens. Although multiple therapeutic vaccines have entered clinical development, most have failed in isolation to demonstrate objective clinical responses when administered to humans with established tumours. At this point, a survival benefit has conclusively been demonstrated in humans for only one therapeutic vaccine, sipuleucel-T, an autologous DC-based vaccine approved for the treatment of castration-resistant metastatic prostate cancer135. Many therapeutic cancer vaccines probably failed because of flawed antigen targets (such as antigens with low immunogenicity, low specificity or low expression level on tumour cells136) or weak adjuvants. Novel adjuvants that activate specific innate immune responses, including Toll-like receptor (TLR) and stimulator of interferon genes (STING) pathways, to enhance antigen-specific immune responses are under clinical development137,138. However, the failure of so many therapeutic cancer vaccines may also imply a barrier to antitumour immunity, or tolerance, that in most cases cannot be overcome with vaccination alone22. We and others have shown that antitumour vaccines can increase tumour-infiltrating CD8+ Teff cells, but these cells produce IFNγ, leading to upregulation of the PD1–PDL1 pathway and other immunosuppressive pathways139,140. Thus, the upregulation of immunosuppressive regulatory mechanisms by cancer vaccines may be one reason why many cancer vaccines in isolation have been unsuccessful in demonstrating objective clinical responses in clinical trials and why cancer vaccines need to be re-examined in combination with immune checkpoint inhibitors or other therapies that modify the TME. Multiple preclinical models have demonstrated the potential for synergy between antitumour vaccines and immune checkpoint inhibitors141–144, and clinical trials combining therapeutic vaccines with immune checkpoint inhibitors are now ongoing.
It remains unclear which immune-based therapy or combinations of immune-based therapies will be most effective in combination with neoantigen-specific vaccines or ACT in overcoming the immunosuppressive mechanisms within the TME and enhancing immune responses. Two classes of immune checkpoint inhibitor are approved for clinical use at this time (inhibitors of the CTLA4 and PD1–PDL1 pathways); however, multiple additional immune checkpoint inhibitors are in clinical development (see Topalian et al.145). In addition to traditional immune checkpoints, many other negative feedback loops within the TME have recently been identified, and multiple novel immunotherapies are now in clinical development alone or in combination with traditional immune checkpoint inhibitors. Examples of other TME targets that may synergize with neoantigen-specific therapies and immune checkpoint inhibitors include inhibitors of IDO1 (REF. 146), adenosine signalling through the adenosine A2A receptor (A2AR)147 and other metabolic signalling pathways97, tumour extracellular matrix and stromal inhibitors148 and MEK inhibitors149. Clinical trials are needed to identify the most effective combinations of immune-based therapies for each tumour type. Eventually, it may become possible to personalize not only the neoantigen-targeted therapy, but also the selection of other immunotherapies targeting the TME based on tumour type or even on an individual patient basis. However, this type of personalized approach to immunotherapy may require improved biomarkers of response beyond those such as PDL1 expression patterns that are in clinical use today150. In summary, neoantigen-specific therapy may be significantly enhanced by combining such agents with immune checkpoint inhibitors, immune adjuvants and TME modulators, with the eventual goal of converting all cancers to be responsive to immunotherapy.
Perspectives for clinical translation
In the past decade, there has been an astounding growth in our understanding of the role of immunity in cancer and a rapid clinical uptake of novel immunotherapies in the care of cancer patients. By specifically targeting tumour neoantigens, it may be possible to maintain or improve on the antitumour effects of current immunotherapies while minimizing off-target toxicities. Therefore, the targeting of tumour neoantigens may be more specific, more effective and less toxic than other cancer immunotherapy approaches. However, clinical trials are needed to demonstrate the superiority of this approach and to rule out unanticipated autoimmune side effects. Many challenges persist, and several key questions remain unanswered.
One practical concern that must be addressed before such therapies could enter mainstream use will be that of cost. Although personalized neoantigen-directed immunotherapies may already be technically feasible, this approach represents a massive undertaking for the care of a single patient, at a time when there is an increasing focus on value in cancer care. Because the approach described here will be customized for an individual patient, it is intrinsically more expensive than off-the-shelf pharmaceutical drugs developed for larger groups of patients. However, technologies for neoantigen sequencing and interpretation are improving rapidly. The cost of whole-exome sequencing has already fallen from several hundred million US dollars for the first human genome as part of the Human Genome Project, to approximately $10 million in 2007 and to less than $1,000 today. There is reason to hope that the entire process of neoantigen identification and targeting could eventually become scaled and automated to reduce costs. Additionally, if this approach results in durable clinical responses, the value of this therapy, a measure that incorporates not only cost but also efficacy151, would probably be acceptable. Increased funding for personalized medicine and institutional infrastructures to accommodate personalized cancer therapies will help to accelerate the development of such approaches. A separate challenge is that regulatory agencies may be unprepared to evaluate neoantigen-directed therapies, because each personalized neoantigen therapy could theoretically carry its own risks and benefits. This approach is a significant departure from the current model of drug development, and regulatory processes are not currently tailored to individualized therapies.
Tumour heterogeneity also presents a clinical challenge. A single core tumour biopsy may underestimate the total number of antigen clones present in the tumour152, and treatments with antigen-directed therapies can select for tumour clonal variants that lack the target antigen and can therefore escape the directed immune response. This is exemplified by recent clinical trials of CAR T cells or bispecific antibodies targeting CD19 in ALL. In these trials, many patients who initially responded to such therapies eventually relapsed with CD19-negative ALL28,89,90,130. Therefore, targeting a single antigen may lead to relapsed disease via tumour evasion of immune control, which as a consequence restores immune tolerance. The main mechanisms of acquired tumour resistance to immune checkpoint inhibitors have only recently been investigated. In some cases, acquired defects in pathways involved in IFN-receptor signalling or antigen presentation may explain disease relapse in patients who initially respond to such therapies88. Somewhat paradoxically, we anticipate that tumours with higher levels of genomic instability and high mutation burdens may be the most likely to initially respond to immune checkpoint inhibitors, but would also be expected to display more tumour heterogeneity and have a greater propensity to acquire resistance to targeted immunotherapies. The idea that cancer can acquire resistance to targeted therapies through the clonal evolution of resistant variants of disease is not new. For example, it has been recognized for many decades that selective pressure on oestrogen-positive breast cancers with anti-oestrogen therapies can result in loss of sensitivity to anti-oestrogens153 and more recently that cancers can acquire resistance to tyrosine kinase inhibitors154. These observations have led to the development of novel inhibitors that overcome resistance to primary therapy and combinations of therapies that reduce the chance of resistance (as with the addition of palbociclib to reduce resistance to endocrine therapy in breast cancer155 or the addition of the MEK inhibitor cobimetinib to the BRAF inhibitor vemurafenib in BRAF-mutant melanoma156). Similar approaches to combination therapy, through the concomitant targeting of multiple neoantigens may eventually be needed to prevent resistance to neoantigen-directed immunotherapy. Targeting of tumour driver mutations that are essential for carcinogenesis, whenever feasible to do so, may also improve the chance of a durable clinical response. Finally, the presumption that tumour heterogeneity increases as tumours progress presents a strong biological argument for treating patients as early in the course of their disease as possible, when tumour heterogeneity is at its lowest.
In conclusion, the best approach for predicting neo-antigens is unknown. So far, the most useful tool for identifying T cell neoepitopes are algorithms designed to predict MHC class I/HLA binding affinity. However, improvements in current prediction algorithms are needed to predict certain types of neoantigen, including those that form MHC class II antigens, as well as proteins arising from gene fusions and neoantigens arising from errors in translation. Clinical trials will also need to establish the best platform for delivering neoantigen-specific immunotherapy. Finally, to maximize immunity, we will need novel approaches to overcome immunosuppressive mechanisms in the TME that can inhibit neoantigen-specific immune responses. We anticipate steady progress in all of these areas as we build on our current understanding of the immune system and the mechanisms that occur in tumours to subvert the immune system and to escape detection and destruction.
Acknowledgments
The authors gratefully acknowledge support from the Bloomberg–Kimmel Institute for Cancer Immunotherapy and the Skip Viragh Center for Pancreatic Cancer Research and Clinical Care at Johns Hopkins University, Baltimore, Maryland, USA.
Glossary
- Passenger mutations
Mutations that have no effect on the fitness of the cell, and are therefore not implicated in oncogenesis.
- Driver mutations
Mutations that cause a selective advantage to a cell clone and are therefore causally implicated in oncogenesis.
- Nonsynonymous mutations
Point mutations and missense mutations that alter the amino acid sequence of proteins.
- Major histocompatibility complex (MHC)
Cell surface proteins that bind to short sequences of amino acids (epitopes) and display them on the cell surface for recognition by the immune system. MHC class I is present on the surface of virtually all nucleated cells including tumour cells and presents intracellular antigens to CD8+ T cells; MHC class II is present on antigen presenting cells and presents exogenous antigens to CD4+ T helper cells.
- Neoantigens
Mutations in the tumour genome can cause tumours to express mutant proteins that are tumour specific and not expressed on normal cells — referred to as neoantigens.
- Objective response rates (ORRs)
ORR is a common efficacy end point used in clinical trials of cancer therapies in solid tumours, usually defined as the percentage of patients who experience at least a 30% decrease in tumour diameter on an imaging scan.
- Central and peripheral tolerance
A state in which immune cells are unresponsive to antigens as a result of clonal deletion of autoreactive B cells and T cells, as well as the dampening of potentially autoreactive B cells and T cells through regulatory immune cells and subsequent downregulation of costimulatory molecules.
- Chimeric antigen receptor (CAR) T cells
T cells that are modified to express an antigen recognition domain of a specific antibody fused to an intracellular signalling domain.
- Adoptive cell therapy (ACT)
An immunotherapy treatment in which antitumour lymphocytes are identified, expanded in vitro and then infused into a patient with cancer. The administered antitumour lymphocytes can be autologous (patient’s own) or allogeneic (donor).
- Microsatellite instability (MSI)
Increased propensity for changes in microsatellite (short tandem repeat) sequences resulting from defects in DNA mismatch repair.
- Immunogenic
The ability to induce an immune response.
- Adjuvant
An immunostimulatory agent designed to enhance the immune response to a vaccine.
Footnotes
Competing interests statement
The authors declare competing interests: see Web version for details.
References
- 1.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segal NH, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68:889–892. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 4.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 5.O’Brien SG, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348:994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 6.Sledge GW. The challenge and promise of the genomic era. J Clin Oncol. 2012;30:203–209. doi: 10.1200/JCO.2011.39.0088. [DOI] [PubMed] [Google Scholar]
- 7.Lu YC, Robbins PF. Cancer immunotherapy targeting neoantigens. Semin Immunol. 2016;28:22–27. doi: 10.1016/j.smim.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gubin MM, et al. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest. 2015;125:3413–3421. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ward JP, Gubin MM, Schreiber RD. The role of neoantigens in naturally occurring and therapeutically induced immune responses to cancer. Adv Immunol. 2016;130:25–74. doi: 10.1016/bs.ai.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillison ML. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 12.Walboomers JMM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 13.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014;14:135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 14.Simpson AJG, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 15.Trimble CL, et al. Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6 and E7 proteins for cervical intraepithelial neoplasia 2/3: a randomised, double-blind, placebo-controlled phase 2b trial. Lancet. 2015;386:2078–2088. doi: 10.1016/S0140-6736(15)00239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nedergaard MK, Hedegaard CJ, Poulsen HS. Targeting the epidermal growth factor receptor in solid tumor malignancies. BioDrugs. 2012;26:83–99. doi: 10.2165/11599760-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 17.Sampson JH, et al. Immunologic escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma. J Clin Oncol. 2010;28:4722–4729. doi: 10.1200/JCO.2010.28.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran E, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med. 2016;375:2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wedén S, et al. Long-term follow-up of patients with resected pancreatic cancer following vaccination against mutant K-ras. Int J Cancer. 2011;128:1120–1128. doi: 10.1002/ijc.25449. [DOI] [PubMed] [Google Scholar]
- 20.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, et al. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125:3384–3391. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melero I, et al. Therapeutic vaccines for cancer: an overview of clinical trials. Nat Rev Clin Oncol. 2014;11:509–524. doi: 10.1038/nrclinonc.2014.111. [DOI] [PubMed] [Google Scholar]
- 23.Stone JD, Harris DT, Kranz DM. TCR affinity for p/MHC formed by tumor antigens that are self-proteins: impact on efficacy and toxicity. Curr Opin Immunol. 2015;33:16–22. doi: 10.1016/j.coi.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aleksic M, et al. Different affinity windows for virus and cancer-specific T-cell receptors: implications for therapeutic strategies. Eur J Immunol. 2012;42:3174–3179. doi: 10.1002/eji.201242606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian S, Maile R, Collins EJ, Frelinger JA. CD8+ T cell activation is governed by TCR-peptide/MHC affinity, not dissociation rate. J Immunol. 2007;179:2952–2960. doi: 10.4049/jimmunol.179.5.2952. [DOI] [PubMed] [Google Scholar]
- 26.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grupp SA, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maude SL, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamers CH, et al. Treatment of metastatic renal cell carcinoma with CAIX CAR-engineered T cells: clinical evaluation and management of on-target toxicity. Mol Ther. 2013;21:904–912. doi: 10.1038/mt.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan RA, et al. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther. 2010;18:843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan RA, et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J Immunother. 2013;36:133–151. doi: 10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Germeau C, et al. High frequency of antitumor T cells in the blood of melanoma patients before and after vaccination with tumor antigens. J Exp Med. 2005;201:241–248. doi: 10.1084/jem.20041379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joseph CG, et al. Association of the autoimmune disease scleroderma with an immunologic response to cancer. Science. 2014;343:152–157. doi: 10.1126/science.1246886. This study showed that even neoantigen-specific T cells against a mutated tumour protein can be cross-reactive with the non-mutated version of the antigen and can induce autoimmunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross L. Intradermal immunization of C3H mice against a sarcoma that originated in an animal of the same line. Cancer Res. 1943;3:326–333. [Google Scholar]
- 35.De Plaen E, et al. Immunogenic (tum−) variants of mouse tumor P815: cloning of the gene of tum-antigen P91A and identification of the tum-mutation. Immunology. 1988;85:2274–2278. doi: 10.1073/pnas.85.7.2274. This study demonstrated that antitumour T cells can recognize aberrant peptides derived from tumour-specific mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lurquin C, et al. Structure of the gene of tum− transplantation antigen P91A: the mutated exon encodes a peptide recognized with Ld by cytolytic T cells. Cell. 1989;58:293–303. doi: 10.1016/0092-8674(89)90844-1. [DOI] [PubMed] [Google Scholar]
- 37.Monach PA, Meredith SC, Siegel CT, Schreiber H. A unique tumor antigen produced by a single amino acid substitution. Immunity. 1995;2:45–59. doi: 10.1016/1074-7613(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 38.Coulie PG, et al. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc Natl Acad Sci USA. 1995;92:7976–7980. doi: 10.1073/pnas.92.17.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wölfel T, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. This study analysed the naturally occurring antitumour T cell response against melanoma in a patient to demonstrate that immunoreactivity against neoantigens predominates over the response to TAAs. [DOI] [PubMed] [Google Scholar]
- 40.Zhou X, et al. Diverse CD8+ T-cell responses to renal cell carcinoma antigens in patients treated with an autologous granulocyte–macrophage colony-stimulating factor gene-transduced renal tumor cell vaccine. Cancer Res. 2005;65:1079–1088. [PubMed] [Google Scholar]
- 41.Lennerz V, et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci USA. 2005;102:16013–16018. doi: 10.1073/pnas.0500090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J, Dudley ME, Rosenberg SA, Robbins PF. Persistence of multiple tumor-specific T-cell clones is associated with complete tumor regression in a melanoma patient receiving adoptive cell transfer therapy. J Immunother. 2005;28:53–62. doi: 10.1097/00002371-200501000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rizvi NA, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gros A, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22:433–438. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dummer R, et al. A randomized controlled comparison of pembrolizumab and chemotherapy in patients with ipilimumab-refractory melanoma. J Transl Med. 2015;13:O5. [Google Scholar]
- 48.Robert C, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2014;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 49.Robert C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 50.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larkin J, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borghaei H, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sundar R, Cho BC, Brahmer JR, Soo RA. Nivolumab in NSCLC: latest evidence and clinical potential. Ther Adv Med Oncol. 2015;7:85–96. doi: 10.1177/1758834014567470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garon EB, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 55.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abdul-Hassan Tawbi H, et al. Safety and efficacy of PD-1 blockade using pembrolizumab in patients with advanced soft tissue (STS) and bone sarcomas (BS): results of SARC028 — a multicenter phase II study. J Clin Oncol. 2016;34 abstr. 11006. [Google Scholar]
- 57.Kwon ED, et al. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 2014;15:700–712. doi: 10.1016/S1470-2045(14)70189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. This paper provided support for the role of somatic mutations in the response to immune checkpoint inhibitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hugo W, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenberg SA. Interleukin 2 for patients with renal cancer. Nat Clin Pract Oncol. 2007;4:497. doi: 10.1038/ncponc0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosenberg SA. IL-2: the first effective immunotherapy for human cancer. J Immunol. 2014;192:5451–5458. doi: 10.4049/jimmunol.1490019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Motzer RJ, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goh G, et al. Mutational landscape of MCPyV-positive and MCPyV-negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget. 2016;7:3403–3415. doi: 10.18632/oncotarget.6494. This study showed that in a subset of MCCs with a viral aetiology, neoantigens derived from viral proteins can serve as strong immune stimulants and may be an attractive target for cancer immunotherapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nghiem PT, et al. PD-1 blockade with pembrolizumab in advanced Merkel-cell carcinoma. N Engl J Med. 2016;374:2542–2552. doi: 10.1056/NEJMoa1603702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lipson EJ, et al. PD-L1 expression in the Merkel cell carcinoma microenvironment: association with inflammation, Merkel cell polyomavirus and overall survival. Cancer Immunol Res. 2013;1:54–63. doi: 10.1158/2326-6066.CIR-13-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Folgiero V, et al. IDO1 involvement in mTOR pathway: a molecular mechanism of resistance to mTOR targeting in medulloblastoma. Oncotarget. 2016;7:52900–52911. doi: 10.18632/oncotarget.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L, Gajewski TF, Kline J. PD-1/PD-L1 interactions inhibit antitumor immune responses in a murine acute myeloid leukemia model. Blood. 2009;114:1545–1552. doi: 10.1182/blood-2009-03-206672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fukuno K, et al. Expression of indoleamine 2,3-dioxygenase in leukemic cells indicates an unfavorable prognosis in acute myeloid leukemia patients with intermediate-risk cytogenetics. Leuk Lymphoma. 2015;56:1398–1405. doi: 10.3109/10428194.2014.953150. [DOI] [PubMed] [Google Scholar]
- 72.Mussai F, et al. Acute myeloid leukemia creates an arginase-dependent immunosuppressive microenvironment. Blood. 2013;122:749–758. doi: 10.1182/blood-2013-01-480129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stone RM, et al. Low dose interleukin-2 following intensification therapy with high dose cytarabine for acute myelogenous leukemia in first complete remission. Am J Hematol. 2008;83:771–777. doi: 10.1002/ajh.21253. [DOI] [PubMed] [Google Scholar]
- 74.Meloni G, et al. Interleukin-2 may induce prolonged remissions in advanced acute myelogenous leukemia. Blood. 1994;84:2158–2163. [PubMed] [Google Scholar]
- 75.Tran E, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350:1387–1390. doi: 10.1126/science.aad1253. This study supports the idea that most tumours have neoantigens that are recognizable by the immune system and are potentially susceptible to immune-targeting interventions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 78.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 79.Shankaran V, et al. IFNγ and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 80.Diamond MS, et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Screpanti V, Wallin RPA, Grandien A, Ljunggren HG. Impact of FASL-induced apoptosis in the elimination of tumor cells by NK cells. Mol Immunol. 2005;42:495–499. doi: 10.1016/j.molimm.2004.07.033. [DOI] [PubMed] [Google Scholar]
- 82.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev. 2010;235:267–285. doi: 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Diefenbach A, Jensen ER, Jamieson AM, Raulet DH. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 2001;413:165–171. doi: 10.1038/35093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases — elimination, equilibrium and escape. Curr Opin Immunol. 2014;27:16–25. doi: 10.1016/j.coi.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.MacKie RM, Reid R, Junor B. Fatal melanoma transferred in a donated kidney 16 years after melanoma surgery. N Engl J Med. 2003;348:567–568. doi: 10.1056/NEJM200302063480620. [DOI] [PubMed] [Google Scholar]
- 86.Koebel CM, et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–917. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 87.Khong HT, Restifo NP. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat Immunol. 2002;3:999–1005. doi: 10.1038/ni1102-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zaretsky JM, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yannakou CK, Came N, Bajel AR, Juneja S. CD19 negative relapse in B-ALL treated with blinatumomab therapy: avoiding the trap. Blood. 2015;126:4983–4983. [Google Scholar]
- 90.Gardner R, et al. Acquisition of a CD19 negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T cell therapy. Blood. 2016;127:2406–2410. doi: 10.1182/blood-2015-08-665547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Uyttenhove BC, Maryanski J, Boon T. Escape of mouse mastocytoma. J Exp Med. 1983;157:1040–1052. doi: 10.1084/jem.157.3.1040. This work showed for the first time that as tumours progress, they lose immunogenicity and antigenicity through the immunoselection of clones expressing weaker antigens, a process that has been termed immunoediting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matsushita H, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Uyttenhove C, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 94.Iwai Y, et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci USA. 2002;99:12293–12297. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tseng D, et al. Anti-CD47 antibody-mediated phagocytosis of cancer by macrophages primes an effective antitumor T-cell response. Proc Natl Acad Sci USA. 2013;110:11103–11108. doi: 10.1073/pnas.1305569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pardoll D. Cancer and the immune system: basic concepts and targets for intervention. Semin Oncol. 2015;42:523–538. doi: 10.1053/j.seminoncol.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Singer K, Gottfried E, Kreutz M, Mackensen A. Suppression of T-cell responses by tumor metabolites. Cancer Immunol Immunother. 2011;60:425–431. doi: 10.1007/s00262-010-0967-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chang CH, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sharma MD, et al. Plasmacytoid dendritic cells from mouse tumor-draining lymph nodes directly activate mature Tregs via indoleamine 2,3-dioxygenase. J Clin Invest. 2007;117:2570–2582. doi: 10.1172/JCI31911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Serafini P, Borrello I, Bronte V. Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Semin Cancer Biol. 2006;16:53–65. doi: 10.1016/j.semcancer.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 102.Munn DH, et al. Expression of indoleamine 2,3-dioxygenase by plasmacytoid dendritic cells in tumor-draining lymph nodes. J Clin Invest. 2004;114:280–290. doi: 10.1172/JCI21583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mao Y, Poschke I, Kiessling R. Tumour-induced immune suppression: role of inflammatory mediators released by myelomonocytic cells. J Intern Med. 2014;276:154–170. doi: 10.1111/joim.12229. [DOI] [PubMed] [Google Scholar]
- 104.Corzo CA, et al. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol. 2009;182:5693–5701. doi: 10.4049/jimmunol.0900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Munn DH, et al. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity. 2005;22:633–642. doi: 10.1016/j.immuni.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 106.Willimsky G, et al. Immunogenicity of premalignant lesions is the primary cause of general cytotoxic T lymphocyte unresponsiveness. J Exp Med. 2008;205:1687–1700. doi: 10.1084/jem.20072016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kammertoens T, Qin Z, Briesemeister D, Bendelac A, Blankenstein T. B-cells and IL-4 promote methylcholanthrene-induced carcinogenesis but there is no evidence for a role of T/NKT-cells and their effector molecules (Fas-ligand, TNF-α, perforin) Int J Cancer. 2012;131:1499–1508. doi: 10.1002/ijc.27411. [DOI] [PubMed] [Google Scholar]
- 108.Choi M, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nielsen M, Lundegaard C, Lund O, Keşmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57:33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- 111.Larsen MV, et al. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Peters B, Bulik S, Tampe R, Van Endert PM, Holzhütter HG. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J Immunol. 2003;171:1741–1749. doi: 10.4049/jimmunol.171.4.1741. [DOI] [PubMed] [Google Scholar]
- 113.Nielsen M, Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8:33. doi: 10.1186/s13073-016-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nielsen M, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS ONE. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rammensee HG, Bachmann J, Emmerich NPN, Bachor OA, Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 116.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370:2418–2425. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 117.Bjorkman PJ, et al. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 118.Babbitt BP, Allen PM, Matsueda G, Haber E, Unanue ER. Binding of immunogenic peptides to Ia histocompatibility molecules. Nature. 1985;317:359–361. doi: 10.1038/317359a0. [DOI] [PubMed] [Google Scholar]
- 119.Nielsen M, Lund O, Buus S, Lundegaard C. MHC class II epitope predictive algorithms. Immunology. 2010;130:319–328. doi: 10.1111/j.1365-2567.2010.03268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kreiter S, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;9:641–645. doi: 10.1126/science.1251102. This case report demonstrated proof of principle for treating cancer with neoantigen-directed immunotherapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 123.Carreno BM, et al. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348:803–808. doi: 10.1126/science.aaa3828. This study demonstrated that vaccination can induce neoantigen-specific T cell responses that are not naturally occurring. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Robbins PF, et al. Mining exomic sequencing data to identify mutaded antigens recognized by adoptively transferred tumor-reactive cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fritsch EF, et al. HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol Res. 2014;2:522–529. doi: 10.1158/2326-6066.CIR-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Van Buuren MM, Calis JJ, Schumacher TN. High sensitivity of cancer exome-based CD8 T cell neo-antigen identification. Oncoimmunology. 2014;3:e28836. doi: 10.4161/onci.28836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Duan F, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med. 2014;211:2231–2248. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bargou R, et al. Tumor regression in cancer patients by very low doses of a T cell-engaging antibody. Science. 2008;321:974–977. doi: 10.1126/science.1158545. [DOI] [PubMed] [Google Scholar]
- 129.Baeuerle PA, Reinhardt C. Bispecific T-cell engaging antibodies for cancer therapy. Cancer Res. 2009;69:4941–4944. doi: 10.1158/0008-5472.CAN-09-0547. [DOI] [PubMed] [Google Scholar]
- 130.Topp MS, et al. Safety and activity of blinatumomab for adult patients with relapsed or refractory B-precursor acute lymphoblastic leukaemia: a multicentre, single-arm, phase 2 study. Lancet Oncol. 2015;16:57–66. doi: 10.1016/S1470-2045(14)71170-2. [DOI] [PubMed] [Google Scholar]
- 131.Gross G, Waks T, Eshhar Z. Expression of immunoglobulin-T-cell receptor chimeric molecules as functional receptors with antibody-type specificity. Proc Natl Acad Sci USA. 1989;86:10024–10028. doi: 10.1073/pnas.86.24.10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dao T, et al. Therapeutic bispecific T-cell engager antibody targeting the intracellular oncoprotein WT1. Nat Biotechnol. 2015;33:1079–1086. doi: 10.1038/nbt.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Strønen E, et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. 2016;352:1337–1341. doi: 10.1126/science.aaf2288. [DOI] [PubMed] [Google Scholar]
- 135.Kantoff PW, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363:411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 136.Cheever MA, et al. The prioritization of cancer antigens: a National Cancer Institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15:5323–5337. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Krieg AM. Therapeutic potential of Toll-like receptor 9 activation. Nat Rev Drug Discov. 2006;5:471–484. doi: 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- 138.Baird JR, et al. Radiotherapy combined with novel STING-targeting oligonucleotides results in regression of established tumors. Cancer Res. 2016;76:50–61. doi: 10.1158/0008-5472.CAN-14-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lutz ER, et al. Immunotherapy converts nonimmunogenic pancreatic tumors into immunogenic foci of immune regulation. Cancer Immunol Res. 2014;2:616–631. doi: 10.1158/2326-6066.CIR-14-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Taube JM, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Soares KC, et al. PD-1/PD-L1 blockade together with vaccine therapy facilitates effector T-cell infiltration into pancreatic tumors. J Immunother. 2015;38:1–11. doi: 10.1097/CJI.0000000000000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li B, et al. Anti-programmed death-1 synergizes with granulocyte macrophage colony-stimulating factor-secreting tumor cell immunotherapy providing therapeutic benefit to mice with established tumors. Clin Cancer Res. 2009;15:1623–1634. doi: 10.1158/1078-0432.CCR-08-1825. [DOI] [PubMed] [Google Scholar]
- 143.Fu J, et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med. 2015;7:283ra52. doi: 10.1126/scitranslmed.aaa4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Duraiswamy J, Kaluza KM, Freeman GJ, Coukos G. Dual blockade of PD-1 and CTLA-4 combined with tumor vaccine effectively restores T-cell rejection function in tumors. Cancer Res. 2013;73:3591–3603. doi: 10.1158/0008-5472.CAN-12-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Spranger S, et al. Mechanism of tumor rejection with doublets of CTLA-4, PD-1/PD-L1, or IDO blockade involves restored IL-2 production and proliferation of CD8+ T cells directly within the tumor microenvironment. J Immunother Cancer. 2014;2:3. doi: 10.1186/2051-1426-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Leone RD, Lo YC, Powell JD. A2aR antagonists: next generation checkpoint blockade for cancer immunotherapy. Comput Struct Biotechnol J. 2015;13:265–272. doi: 10.1016/j.csbj.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Provenzano PP, et al. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ebert PJR, et al. MAP kinase inhibition promotes T cell and anti-tumor activity in combination with PD-L1 checkpoint blockade. Immunity. 2016;44:609–621. doi: 10.1016/j.immuni.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 150.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 151.Bach PB, Pearson SD. Payer and policy maker steps to support value-based pricing for drugs. JAMA. 2015;314:2503–2504. doi: 10.1001/jama.2015.16843. [DOI] [PubMed] [Google Scholar]
- 152.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 154.Quintás-Cardama A, et al. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood. 2009;113:1619–1630. doi: 10.1182/blood-2008-03-144790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Turner NC, et al. Palbociclib in hormone-receptor-positive advanced breast cancer. N Engl J Med. 2015;373:209–219. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 156.Larkin J, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 157.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 158.Slamon DJ, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 159.Le DT, et al. A live-attenuated Listeria vaccine (ANZ-100) and a live-attenuated Listeria vaccine expressing mesothelin (CRS-207) for advanced cancers: phase I studies of safety and immune induction. Clin Cancer Res. 2012;18:858–868. doi: 10.1158/1078-0432.CCR-11-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Argani P, et al. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE) Clin Cancer Res. 2001;7:3862–3868. [PubMed] [Google Scholar]
- 161.Thomas AM, et al. Mesothelin-specific CD8+ T cell responses provide evidence of in vivo cross-priming by antigen-presenting cells in vaccinated pancreatic cancer patients. J Exp Med. 2004;200:297–306. doi: 10.1084/jem.20031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chomez P, et al. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- 163.Gnjatic S, et al. NY-ESO-1: review of an immunogenic tumor antigen. Adv Cancer Res. 2006;95:1–30. doi: 10.1016/S0065-230X(06)95001-5. [DOI] [PubMed] [Google Scholar]