Summary
Dividing cells called neoblasts contain pluripotent stem cells and drive planarian flatworm regeneration from diverse injuries. A long-standing question is whether neoblasts directly sense and respond to the identity of missing tissues during regeneration. We used the eye to investigate this question. Surprisingly, eye removal was neither sufficient nor necessary for neoblasts to increase eye progenitor production. Neoblasts normally increase eye progenitor production following decapitation, facilitating regeneration. Eye removal alone, however, did not induce this response. Eye regeneration following eye-specific resection resulted from homeostatic rates of eye progenitor production and less cell death in the regenerating eye. Conversely, large head injuries that left eyes intact increased eye progenitor production. Large injuries also non-specifically increased progenitor production for multiple uninjured tissues. We propose a model for eye regeneration in which eye tissue production by planarian stem cells is not directly regulated by the absence of the eye itself.
eTOC blurb
Whether planarian stem cells sense and respond to the absence of specific tissues during regeneration is unclear. LoCascio et al. provide evidence for a mechanism of tissue-specific eye regeneration that does not involve regulation of stem cells by the presence or absence of the eye itself.
Introduction
Regeneration is the replacement of body parts lost to injury, such as organs or appendages, and occurs throughout the animal kingdom (Poss, 2010; Sánchez Alvarado, 2000; Tanaka and Reddien, 2011). How animals respond to the absence of specific tissues following injury to bring about their precise replacement is a central but poorly understood problem in regeneration biology.
Planarians are free-living flatworms that can regenerate from almost any injury, making them a powerful model for the study of animal regeneration (Reddien and Sánchez Alvarado, 2004). Underlying this regenerative ability is a proliferative population of cells called neoblasts that contain pluripotent stem cells (Wagner et al., 2011). Neoblasts constitute the only dividing adult somatic planarian cells and are required for the regeneration and homeostatic maintenance of all differentiated tissues. A remarkable aspect of planarian regeneration is that it is tissue-specific; whether an injury removes an entire section of the body, or specifically ablates a single tissue of virtually any type, the animal replaces precisely those tissues that were lost (Adler et al., 2014; Nishimura et al., 2011; Reddien and Sánchez Alvarado, 2004). One hypothesis to explain this highly specific nature of planarian regeneration is that neoblasts sense the presence and absence of specific tissues after injury, modifying their output in accordance with the identity of missing tissues (Adler and Sánchez Alvarado, 2015; Mangel et al., 2016; Nishimura et al., 2011). However, whether neoblast output is directly regulated by the presence or absence of the specific tissues to be regenerated is unclear.
Planarian eyes present an ideal venue to investigate the mechanistic basis of tissue-specific regeneration in vivo. The paired planarian eyes, which can be formed de novo after head amputation, are simple organs comprised of pigmented optic cup cells and photoreceptor neurons (PRNs) that connect to a bilobed brain. The eyes are discretely located, visible in live animals, and dispensable for viability, making them good targets for specific surgical manipulation. Molecular characterization has identified tissue-specific markers for eye cell types and provided tools for the visualization of eye progenitors during regeneration (Lapan and Reddien, 2011, 2012; Sánchez Alvarado and Newmark, 1999). Previously, we found that head amputation leads to the formation of a large number of specialized neoblasts expressing eye-associated transcription factors. These eye-specialized neoblasts give rise to progenitors that migrate anteriorly, progressively differentiate, and coalesce to form the regenerated eyes (Lapan and Reddien, 2011, 2012). The potential for inducing tissue-specific injuries combined with the ability to observe the cellular stages of eye regeneration presented a unique opportunity to investigate the mechanistic basis of tissue-specific regeneration.
To directly test the hypothesis that neoblasts are regulated by the presence or absence of eye tissue, we examined eye progenitor responses to tissue-specific eye resection and to various large injuries that either removed the eyes or left the eyes uninjured. Surprisingly, our data demonstrate that stem cell-based eye progenitor production is not regulated by the presence or absence of the eye itself. Specific removal of the eye did not impact eye progenitor production. Instead, less cell death occurred in regenerating eyes, allowing them to grow in size despite no specific increase in the rate of eye progenitor production. Such a passive process could fuel regeneration from a myriad of injuries removing different cell types. Eye absence was also not necessary for increased eye progenitor formation. Increased eye progenitor formation was induced whenever large injuries triggered general neoblast proliferation in the body position where eye progenitor specification occurs, regardless of the presence or absence of the eyes. Large injuries also non-specifically increased the production of uninjured pharynx and ventral nerve cord tissue. We propose a “target-blind” progenitor model for planarian eye regeneration, which could apply to many other regenerative contexts, in which stem cells do not respond to the presence or absence of the specific tissue to be regenerated.
Results
Planarian eyes exhibit tissue-specific regeneration
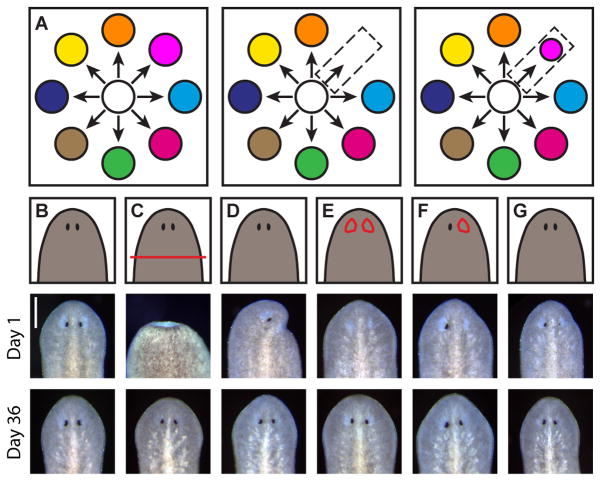
How regeneration occurs following removal of specific tissues is poorly understood (Figure 1A). To address this problem we developed tissue-specific surgical manipulations to partially or fully resect one or both of the planarian eyes (Figures 1B–1G and S1A–S1C). In all cases, the injured or absent eye returned, representing the regeneration of an entire organ following its specific removal (Figures 1B–1G). We therefore utilized these tissue-specific surgical strategies in combination with various large injuries to seek the mechanistic basis of tissue-specific regeneration.
Figure 1. Planarian eyes exhibit tissue-specific regeneration.
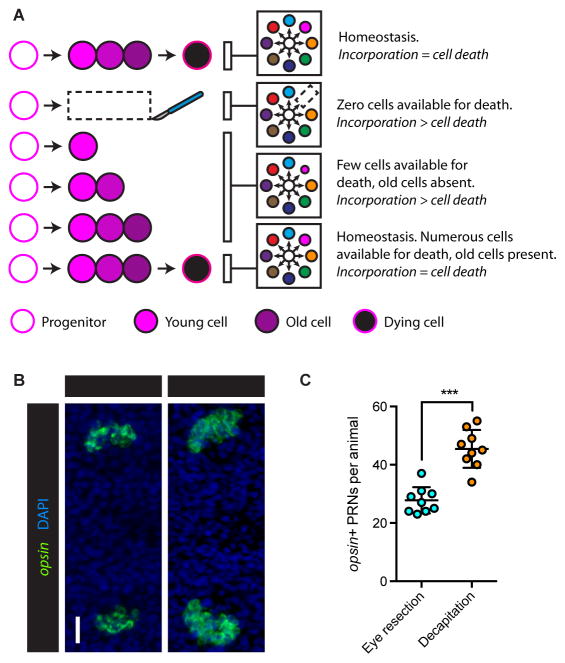
(A) Tissue-specific regeneration schematic. White circle represents the neoblast population, colored circles represent distinct differentiated tissues. Neoblasts produce differentiated tissue types (left). Tissue-specific injury (center) is followed by tissue-specific regeneration (right).
(B–G) Cartoons (top) and live images 1 day (center) and 36 days (bottom) after no injury (B), decapitation (C), half decapitation (D), bilateral eye resection (E), right eye resection (F), and partial right eye resection (G). Scale bar, 200 μm. See also Figures S1A–S1C.
Eye absence is not sufficient to increase eye progenitor production
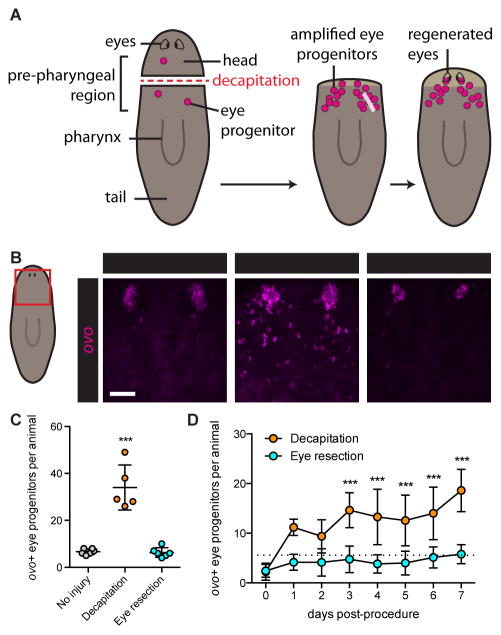
Previously, we found that head amputation leads to increased neoblast-derived eye progenitor numbers (eye progenitor amplification), facilitating eye regeneration (Figure 2A) (Lapan and Reddien, 2011, 2012). If eye progenitor amplification following decapitation involved neoblasts responding to eye absence, then eye removal alone should also induce eye progenitor amplification. We therefore assessed whether eye resection was sufficient to increase eye progenitor numbers above basal levels found in uninjured animals. As expected, three days after injury, eye progenitor numbers were increased in response to decapitation. Surprisingly, however, eye progenitor numbers were not increased after eye resection (Figures 2B and 2C), despite the fact that eyes regenerated following this injury (Figure 1E). We also quantified eye progenitors every day for one week following injury. This time window allows substantial regeneration, including of a functional head and eyes, with animals capable of feeding and negative phototaxis. Eye-resected animals did not show elevated progenitor numbers at any of the eight time points quantified, whereas decapitated animals exhibited elevated progenitor numbers from Day 3 – 7 (Figure 2D). Quantification with a semi-automated computer protocol yielded similar results (Figures S1D–S1H). ovo RNAi animals did not regenerate eyes following eye resection, indicating that the amplification of an ovo− progenitor population does not contribute to eye regeneration in this context (Figure S1I). We conclude that eye absence alone is not sufficient to induce eye progenitor amplification.
Figure 2. Eye absence is not sufficient to induce eye progenitor amplification.
(A) Schematic of eye regeneration following decapitation. Decapitation (left) leads to amplification of ovo+ eye progenitors (center), which migrate anteriorly and coalesce to form regenerating eyes (right).
(B) Fluorescence in situ hybridization (FISH) with ovo RNA probe, 3 days after indicated surgeries. Maximum intensity projections. Red box on cartoon indicates displayed region. Asterisks mark presumptive eyes, identifiable as coalesced ovo+ cells anterior to progenitors. Scale bar, 50 μm.
(C and D) ovo+ eye progenitor numbers 3 days (C) and 0 to 7 days (D) post-surgery. Data represented as mean ± SD. Dots reflect eye progenitor numbers for individual animals for (C), means for (D). In (D), dotted line reflects mean of uninjured animals on Day 0. Decapitation but not eye resection resulted in significantly elevated eye progenitor numbers in comparison to uninjured controls. n≥4 animals per condition. Statistical significance assessed with respect to uninjured animals by one-way ANOVA (***p<0.001). See also Figures S1D–S1I.
Eye absence is not sufficient to induce a tissue-specific increase in progenitor incorporation
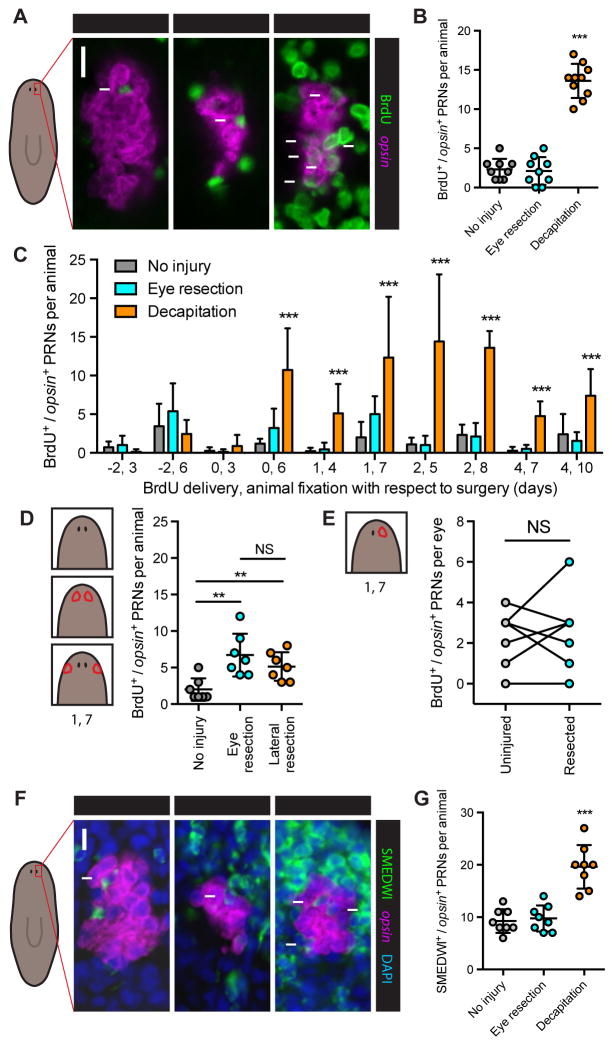
Although eye resection did not increase eye progenitor numbers, a tissue-specific increase in eye progenitor incorporation could in principle drive regeneration in this context. For instance, eye progenitors might only survive and incorporate into eyes that are disproportionately small or absent. More generally, if neoblasts respond to eye absence in a tissue-specific manner, then an eye-specific increase in progenitor incorporation should occur following eye resection. To assess progenitor incorporation rate, BrdU was utilized to label neoblasts (Newmark and Sánchez Alvarado, 2000) and the number of BrdU+/opsin+ PRNs was quantified 6 days later. Whereas decapitation resulted in an increased rate of new PRN formation from neoblasts, eye resection alone did not (Figures 3A and 3B).
Figure 3. Increased eye progenitor incorporation does not contribute to eye-specific regeneration.
(A) BrdU immunofluorescence (IF) together with FISH for opsin. Scale bar, 10 μm.
(B–E), BrdU/opsin double-positive PRNs per animal or per eye after surgeries indicated in text or cartoons. (B) BrdU delivery Day 2, animal fixation Day 8. Decapitated but not eye-resected animals had significantly more BrdU+ PRNs than uninjured animals. (C) BrdU delivery-fixation intervals denoted on x-axis as day of delivery, day of fixation. Asterisks indicate significant increase above uninjured condition for that interval. Day 2–8 interval data is also shown in (B). PRN incorporation is higher in eye-resected animals than uninjured controls for Day 0–6 (p=0.0235) and Day 1–7 (p=0.0120) intervals by Student’s t-test. n≥5 animals per condition. (D) BrdU delivery Day 1, fixation Day 7. n=7 animals per condition. (E) BrdU delivery Day 1, fixation Day 7. n=7 animals. See also Figure S2. Data are represented as mean ± SD. Dots represent values from individual animals. In (B) and (C), statistical significance was assessed by one-way ANOVA comparing eye-resected and decapitated animals to uninjured animals for same interval, in (D) by Student’s t-test, and in (E) by paired Student’s t-test (**p<0.01, ***p<0.001); NS, not significant.
(F) SMEDWI IF with FISH for opsin and DAPI labeling, 5 days after indicated surgeries. Scale bar, 10 μm.
(G) SMEDWI/opsin double-positive PRNs per animal 5 days after indicated surgeries. n=8 animals per condition.
To exclude the possibility that we failed to observe an increase in PRN incorporation following eye resection because of the specific timing of our experiment, we systematically varied the timing of BrdU delivery and animal fixation with respect to surgery in uninjured, eye-resected and decapitated animals (Figure 3C). In most cases decapitated animals had significantly more BrdU+/opsin+ cells than did uninjured animals. Conversely, in most cases no difference between eye-resected animals and uninjured controls was observed. A modest increase in PRN incorporation was observed in eye-resected animals only for the Day 0–6 and Day 1–7 delivery-fixation intervals. To determine whether this effect was specifically a consequence of eye absence, we resected a similar amount of tissue from a region lateral to the eyes and used the Day 1–7 delivery-fixation interval to assess PRN incorporation. Tissue resection lateral to the eye resulted in a similar increase in PRN incorporation (Figure 3D). These data suggest that this modest, transient increase in progenitor incorporation is a generic consequence of injury-induced proliferation, rather than eye absence. This is consistent with a global wave of mitosis previously described to occur in planarians following any small injury (Baguñà, 1976; Wenemoser and Reddien, 2010). Tissue-specific regeneration also occurred in the case of single eye removal, enabling paired comparisons of regenerating and non-regenerating eyes within the same individuals (Figure 1F). Uninjured and regenerating eyes had similar incorporation rates, regardless of the delivery-fixation interval (Figures 3E and S2).
We also used perdurance of the neoblast protein SMEDWI-1 (Guo et al., 2006; Reddien et al., 2005; Scimone et al., 2010) as a marker of newly differentiated PRNs, providing a second direct readout of progenitor incorporation rate into the eye. Again, whereas animals regenerating from decapitation showed elevated numbers of SMEDWI+/opsin+ PRNs, animals regenerating from eye resection did not (Figures 3F and 3G). We conclude that tissue-specific eye regeneration, following eye resection, is accomplished in the absence of a tissue-specific neoblast response.
Regenerating eyes exhibit less cell death than uninjured eyes
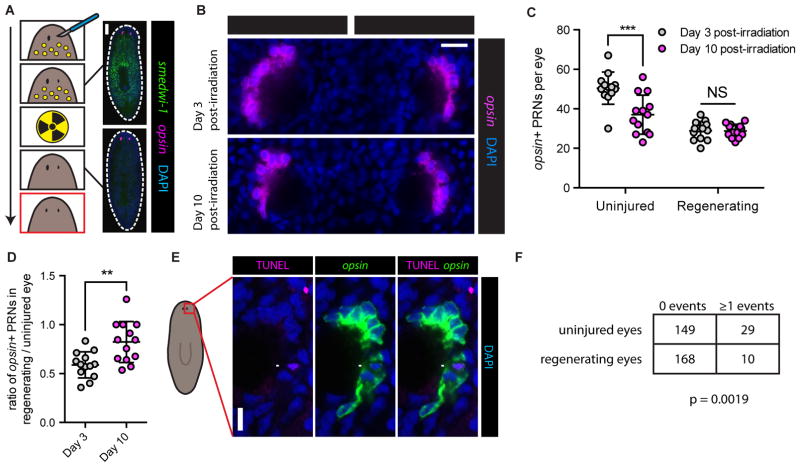
How are the eyes specifically regenerated following eye resection if their absence is not sensed by neoblasts, and there is no specific alteration in their progenitor production or incorporation rates? For instance, in animals with only one resected eye, the intact eye and the regenerating eye have the same rate of progenitor incorporation (Figures 1F and 3E). Thus, how does growth occur only on the injured side? Because the size of a tissue remains constant when cell production and cell death are in equilibrium, alteration in either process can affect tissue size. Therefore, if the rate of eye progenitor incorporation remains constant following eye resection, then the rate of cell loss in regenerating eyes (defined as total cell loss events per eye per unit time) must be lower in the regenerating eye in order to facilitate net growth. To test this prediction, we sought to compare rates of cell loss in uninjured and regenerating eyes (Figure 4A). Animals underwent right eye resection, leaving left eyes uninjured. Right eyes were allowed to partially regenerate for eight days. Animals were then irradiated with 6,000 rads, a procedure that specifically and rapidly eliminates neoblasts (Figure 4A) (Dubois, 1949) and neoblast-derived eye progenitors (Lapan and Reddien, 2011), but that has no detectable effect on differentiated planarian tissues (Guo et al., 2006; Reddien et al., 2005; Wagner et al., 2012; Wagner et al., 2011). Because neoblasts are the only dividing planarian cells and the sole source of new differentiated tissue (Newmark and Sánchez Alvarado, 2000), subsequent alterations in the number of eye cells could be attributed to eye cell loss. We therefore predicted that the right eyes, which were regenerating at the time of irradiation, would decrease in size more slowly than the uninjured left eyes. To quantify eye size we counted the total number of PRNs per eye (Figure S3A). As predicted, uninjured left eyes significantly decreased in size from Day 3 to Day 10 post-irradiation. Uninjured eyes also decreased in size following irradiation when the contralateral eye was not injured, indicating that this effect was not a consequence of contralateral eye absence (Figure S3B). In contrast to the uninjured left eyes, regenerating right eyes did not significantly decrease in size, indicating that less cell loss occurred in the regenerating eye during this interval (Figures 4B and 4C). Consistent with these observations, the intra-animal PRN number difference between uninjured and regenerating eyes was decreased from Day 3 to Day 10 post-irradiation (Figure S3C). The ratio of PRNs in the regenerating to uninjured eye was increased over this time interval, also demonstrating proportionally less cell loss in the regenerating eye (Figure 4D). We also used FISH combined with whole-mount TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling) (Pellettieri et al., 2010) to observe apoptotic cell death events in intact and regenerating PRNs (Figures 4E and S3D). Apoptosis accounts for a small fraction of the lifetime of a cell, making TUNEL+/opsin+ PRNs rare. We therefore analyzed >350 eyes that were uninjured or regenerating from eye resection. A greater proportion of uninjured eyes contained TUNEL+/opsin+ PRNs than did regenerating eyes (Figure 4F). Taken together, our data indicate that regenerating eyes specifically exhibit a decrease in the rate of cell death (cell death events per eye per unit time), facilitating net growth without a specific increase in eye progenitor incorporation.
Figure 4. Regenerating eyes exhibit less cell death than uninjured eyes.
(A) Experimental schematic and prediction for comparing PRN loss in uninjured and regenerating eyes after neoblast ablation by irradiation (6,000 rads). Arrow represents time, yellow circles represent neoblasts. Bottom panel (red outline) is predicted outcome if there is less cell death in the regenerating eye. Panels on right display FISH for smedwi-1 and opsin with DAPI labeling, pre-irradiation and three days post-irradiation. smedwi-1+ neoblasts are absent three days post-irradiation. Dashed white line denotes animal boundary. Scale bar, 200 μm.
(B) Representative opsin FISH with DAPI labeling, 3 and 10 days post-irradiation. Scale bar, 20 μm.
(C) Total PRNs per eye, 3 and 10 days post-irradiation. Decreased PRN number was detected in uninjured but not regenerating eyes. Data represented as mean ± SD. Dots represent PRN counts for individual eyes. Statistical significance assessed by Student’s t-test (***p<0.001). See also Figures S3A–S3C.
(D) Intra-animal ratio of PRNs in regenerating/intact eyes increased from Day 3 to Day 10 post-irradiation. Data represented as mean ± SD. Dots represent ratios from individual animals. Statistical significance assessed by Student’s t-test (**p<0.01).
(E) Combined TUNEL and FISH for opsin with DAPI labeling in an uninjured eye. Arrow indicates TUNEL+/opsin+ PRN. Scale bar, 10 μm.
(F) Table indicating number of uninjured or regenerating eyes 10 days post-surgery that contained either 0 or ≥1 TUNEL+/opsin+ PRNs. A greater proportion of uninjured eyes contained TUNEL+/opsin+ PRNs than regenerating eyes. Statistical significance assessed by Fisher’s exact test (p=0.0019).
A passive model for tissue-specific eye regeneration
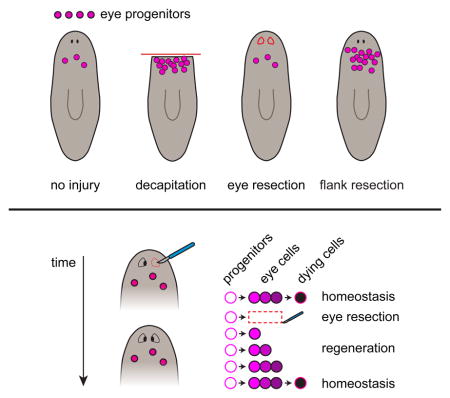
Based on these findings, we propose a simple, passive model for tissue-specific eye regeneration (Figure 5A), in which the rate of eye cell production by progenitors remains constant. Following eye resection, there are initially zero cells available for cell death, allowing any addition of new cells to result in net growth. In the following days, fewer cells are available for cell death than during homeostasis, with no old PRNs present. Net growth thus continues because the rate of cell death is less than the rate of incorporation, as a passive consequence of the emergent properties of a regenerating eye. As sufficient numbers of PRNs accumulate and age, the rate of cell death per eye once again matches the rate of incorporation events per eye, resulting in homeostatic eye size. This model allows tissue-specific eye regeneration to occur, while not requiring neoblasts to specifically interpret or respond to the absence of the eye.
Figure 5. A passive model for tissue-specific eye regeneration.
(A) A passive model for tissue-specific eye regeneration with constant progenitor incorporation. Time represented top to bottom. For panels, white circle represents neoblasts, colored circles represent distinct differentiated tissues, as in Figure 1A.
(B) Maximum intensity projections of FISH for opsin with DAPI labeling, seven days after eye resection or decapitation. Anterior facing left. Scale bar, 20 μm.
(C) Total PRNs per animal seven days post-surgery. Decapitated animals had more PRNs than eye-resected animals. Data represented as mean ± SD. Dots represent PRN counts from individual animals. Statistical significance assessed by Student’s t-test (***p<0.001). See also Figures S4 and S5.
Consistent with this passive model, the rate of eye de-growth following irradiation was similar to the rate of growth following eye resection, indicating that both incorporation and cell death occur at appreciable rates during homeostasis (Figure S4A). Furthermore, unlike the case for regenerating eyes described above, partially resected eyes (Figure 1G) decreased in size at a rate comparable to larger, uninjured eyes following irradiation (Figures S4B and S4C). This is consistent with cell age contributing to PRN death, because unlike regenerating eyes, partially resected eyes are not exclusively composed of young cells. The result also indicates that an active size-sensing mechanism is unable to suppress death in partially resected eyes. Our passive model for tissue-specific eye regeneration also predicts that eyes regenerate following resection, but slowly. Indeed, eye-resected animals regenerated eyes more slowly than did decapitated animals, despite the fact that decapitated animals were significantly smaller than their eye-resected counterparts because of surgery and had to regenerate not only the eyes but also all other cell types of an entire head (Figures 5B, 5C, S5A and S5B).
Eye progenitor amplification is associated with wounds that induce proliferation in the location of eye progenitor specification
Our data indicate that eye progenitor amplification is not a consequence of eye removal. We therefore sought to explore how large injuries induce eye progenitor amplification if eye absence is not regulating this process. Tail removal did not increase eye progenitor numbers in uninjured heads (Figure 6A), indicating that the response does not simply occur after any wound removing a large amount of tissue. Anterior incisions made in the same location as decapitation, but that did not remove the head, also failed to significantly increase eye progenitors (Figure 6A), indicating that eye progenitor amplification requires anterior wounds that remove tissue. Injuries removing substantial tissue (such as amputation) differ from wounds that do not remove tissue (such as incisions) in multiple ways. Importantly, amputations but not incisions elicit a sustained increase in neoblast proliferation that is localized near the wound site (Wenemoser and Reddien, 2010). Accordingly, decapitation increased proliferation in the pre-pharyngeal region (assessed with phosphorylated histone H3 (H3P) immunofluorescence), whereas eye resection, tail amputation, and anterior incisions did not (Figure 6B). Specification of eye progenitors occurs in a spatially restricted manner within the head and pre-pharyngeal region (Lapan and Reddien, 2011, 2012). We therefore hypothesized that eye progenitor amplification occurs as a consequence of generic induction of proliferation in the body position where eye progenitors are specified by large wounds, rather than as a consequence of eye absence.
Figure 6. Eye progenitor amplification is associated with wounds that induce proliferation in the location of eye progenitor specification.
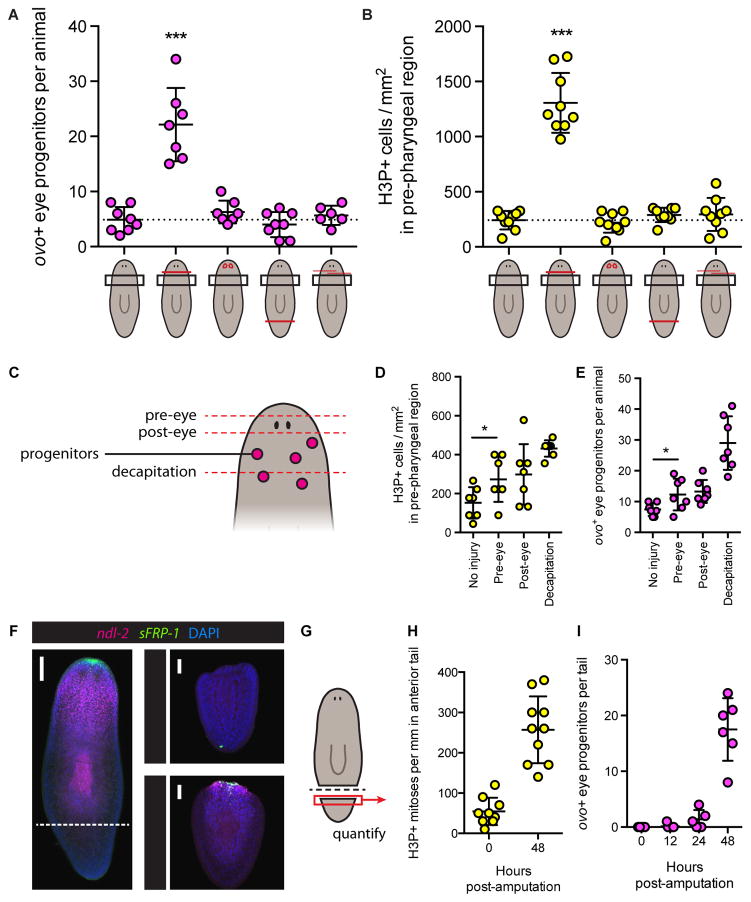
(A–B) ovo+ eye progenitors per animal four days post-surgery (A) and H3P+ mitotic cells per mm2 in pre-pharyngeal region two days post-surgery (B). Cartoons depict surgeries with red lines, left to right: no surgery, decapitation, eye resection, tail amputation, anterior incisions. Black boxes represent quantification area. Only decapitation led to elevated eye progenitor numbers and increased density of mitotic cells. Data represented as mean ± SD. Dotted lines indicate mean of uninjured animals. Dots represent values from individual animals. Significance assessed with respect to uninjured animals by one-way ANOVA (***p<0.001).
(C) Cartoon depicting amputation planes (dashed red lines) for surgeries in (D) and (E).
(D–E) H3P+ mitotic cells per mm2 (D) and ovo+ eye progenitors per animal (E) in pre-pharyngeal region following indicated surgeries. Data represented as mean ± SD. Dots represent values from individual animals. Pre-eye amputation increased mitotic density and eye progenitor numbers, as assessed by Student’s t-test (*p<0.05). See also Figure S6A.
(F) Maximum intensity projection of FISH for ndl-2 and sFRP-1 with DAPI labeling in uninjured animal (left) and amputated tails (right), 0 and 48 hours post-amputation (HPA). Dotted line indicates amputation plane. ndl-2 and sFRP-1 are restored in amputated tails by 48 HPA. Numbers in bottom right of tail insets indicate 6 out of 6 animals displayed the expression patterns shown. Scale bars, 100 μm.
(G) Cartoon indicating region quantified in amputated tails for (H) and (I).
(H–I) H3P+ mitotic cells per mm in anterior of tail (H) and ovo+ cells per tail (I), at indicated hours post-amputation. Data represented as mean ± SD. Dots represent values from individual tails.
To explore this possibility, we assessed neoblast proliferation and eye progenitor amplification in the pre-pharyngeal region following wounds that removed progressively increasing amounts of anterior tissue. Animals were left uninjured, underwent transverse amputation just anterior to the eyes, just posterior to the eyes, or full decapitation (Figure 6C). Neoblast proliferation in the pre-pharyngeal region increased proportionately with the amount of anterior tissue removed (Figure 6D) and eye progenitor amplification also occurred, closely paralleling the degree of neoblast proliferation (Figure 6E). Importantly, the difference in eye progenitor numbers between pre-eye and post-eye amputations was very small, despite the fact that one injury type left the eyes intact while the other completely removed them. The fact that eye progenitor numbers increased following amputation of the anterior head tip, an injury that did not remove eyes or eye progenitors, also demonstrates that eye absence is not required for increased eye progenitor production (Figures 6E and S6A).
Amputated body fragments, such as tails, initially lack the region where eye progenitors are specified and yet they produce large numbers of eye progenitors de novo and regenerate eyes. Positional information is required for maintenance of the planarian adult body plan and proper regeneration (Reddien, 2011). Regional expression gradients of patterning molecules (such as Wnt, BMP, FGFRL/ndl) exist in planarian body wall muscle (Scimone et al., 2016; Witchley et al., 2013), and the pattern of expression of these molecules is restored early in the process of regeneration. In tails, anterior patterning molecules reappear by 48 hours post-amputation (Figure 6F) (Gurley et al., 2010; Petersen and Reddien, 2009), coinciding with increased neoblast proliferation (Figure 6H) (Wenemoser and Reddien, 2010) and the location of de novo eye progenitor amplification (Figure 6I). Therefore, similar to the case of decapitated animals, eye progenitor amplification in tail fragments involves significant tissue removal and induction of sustained neoblast proliferation in a region coinciding with the location of eye progenitor specification (the anterior-facing wound expressing head patterning molecules). We propose that it is not the absence of eyes promoting eye progenitor amplification in either case.
Eye absence is not required for eye progenitor amplification
We utilized additional injuries to further test predictions of the hypothesis that eye progenitor amplification caused by large wounds is not the consequence of eye absence itself. We resected large lateral flanks in the pre-pharyngeal region, while leaving the eyes uninjured (Figures 7A and 7B). Similarly to decapitation, lateral flank resection induced a missing tissue response involving a large increase in neoblast proliferation in the location of eye progenitor specification. These flank injuries caused amplification of eye progenitors (Figure 7C–E). Flank resection also increased the incorporation rate of eye progenitors into uninjured eyes (Figure 7F). We conclude that increased eye progenitor production and acceleration of eye progenitor incorporation into the eye do not require eye absence.
Figure 7. Eye absence is not required for eye progenitor amplification.
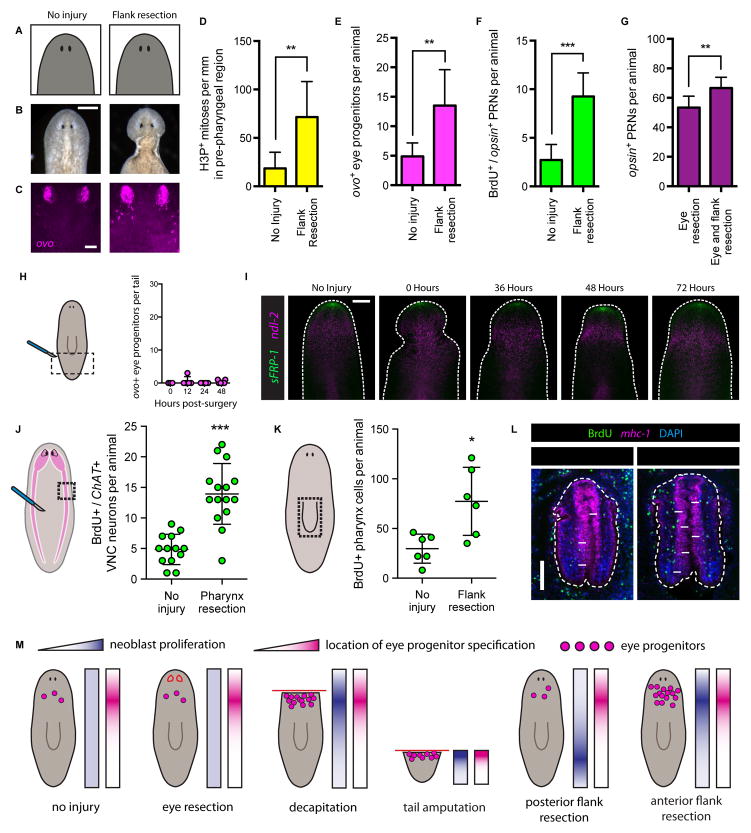
(A–C) Uninjured animals (left) and animals that underwent pre-pharyngeal lateral flank resection (right). (A) Cartoon depictions. (B) Live images. Scale bar, 200 μm. (C) FISH for ovo. Scale bar, 50 μm.
(D–G) Pre-pharyngeal flank resection increases eye tissue production. (D) H3P+ mitotic cells per mm in pre-pharyngeal region, 48 hours post surgery. (E) ovo+ eye progenitors per animal four days post surgery. (F) BrdU+/opsin+ PRNs per animal, BrdU pulse-fixation interval Day 1 – Day 7. (G) PRNs per animal one week after eye resection alone or eye and flank resection. Data are represented as mean ± SD. Statistical significance assessed using Student’s t-tests (**p<0.01, ***p<0.001). n≥7 animals per condition. See also Figure S6.
(H) ovo+ eye progenitors per tail 0, 12, 24 and 48 hours post-flank resection. Cartoon depicts surgery (red lines) and area quantified (dotted black lines). Posterior flank resection does not amplify ovo+ eye progenitors in the tail. Dots represent values from individual animals. n≥3 animals per time point.
(I) FISH for sFRP-1 and ndl-2 in anterior of uninjured and anterior flank-resected animals 0, 36, 48 and 72 hours post-surgery. Dashed white line denotes animal boundary. Arrowheads indicate posterior boundary of flank resections. sFRP-1 expression was not detected at the posterior boundary of flank resection. ndl-2 was not posteriorly expanded after flank resection. n≥5 animals per condition. Scale bar, 200 μm.
(J–K) Cartoons indicate location of tissue resection (red lines), and quantification areas (dotted black lines). For (J), pink area represents brain and ventral nerve cords (VNCs). Graphs display number of BrdU+/ChAT+ VNC neurons (J) or BrdU+ cells in pharynx as assessed by mhc-1 staining (K) per animal following no injury or indicated surgery. BrdU pulse-fixation interval Day 1 to Day 7 (J) and Day -1 to Day 4 (K). Data represented as mean ± SD. Dots represent quantified values from individual animals. Statistical significance assessed by Student’s t-test (*p<0.05, ***p<0.001). See also Figure S7.
(L) IF for BrdU with FISH for mhc-1 and DAPI labeling in pharynges of animals following no injury or flank resection. Dashed white lines outline pharynx. Arrows indicate examples of BrdU+ cells inside pharynx. Scale bar, 100 μm.
(M) Summary of conditions leading to eye progenitor amplification. Red lines indicate surgical procedures, pink dots represent eye progenitors. For each surgery, blue gradient rectangles represent regional neoblast proliferation (dark is increased), pink gradient rectangles represent location of eye progenitor specification. Eye progenitors are amplified when neoblast proliferation coincides with location of eye progenitor specification. Eye absence was not sufficient or necessary for eye progenitor amplification.
Identical flank resection injuries in the tail, where eye progenitors are not normally specified, increased neoblast proliferation locally, while not inducing proliferation in the pre-pharyngeal region (Figures S6B–S6D). Accordingly, eye progenitors were not amplified after flank resection in the tail (Figures 7H and S6E). Eye progenitor amplification is therefore not a general consequence of flank resection injury at any location. To rule out the possibility that pre-pharyngeal flank resection was simply interpreted as a decapitation injury, with the tissue posterior to the flank wounds mounting a head regeneration response, we analyzed the expression of anterior positional markers sFRP-1 and ndl-2 following flank resection (Figure 7I). We did not observe sFRP-1 expression at the posterior boundary of the flank wound by 36 hours post-surgery. Flank resection also did not induce expansion of the ndl-2 expression domain, a gene that is expressed in the muscle of the pre-pharyngeal region (Scimone et al., 2016). The absence of rescaling of anterior markers in flank-resected animals suggests that flank resection is not interpreted as a decapitation injury. This is further supported by the fact that eye progenitors were not amplified in the tail after posterior flank resection (Figure 7H). Together, our data indicate that large injuries that induce proliferation in the location of eye progenitor specification result in eye progenitor amplification, regardless of the presence or absence of the eye.
Anterior flank resection accelerates eye regeneration
It is known that wounds resulting in significant tissue loss induce a process called morphallaxis, in which overabundant tissues shrink to a size appropriate for the new size of the animal. Therefore, flank resection cannot lead to robust overgrowth of intact eyes. We therefore tested the impact of eye progenitor amplification caused by flank resection in the context of regeneration. Animals underwent eye resection alone or eye resection combined with flank resection. One week later, animals that underwent both eye and flank resection had more PRNs than did animals that underwent eye resection alone (Figure 7G). We conclude that flank resection accelerated the rate of eye regeneration.
Increased progenitor incorporation into the ventral nerve cords and pharynx does not require specific tissue removal
The pharynx is the other most accessible planarian organ for complete and specific surgical resection (Adler et al., 2014). Whereas pharynx removal increases pharynx progenitor production, in contrast to the eyes, the pharynx is a much larger organ (Figure S7A) (Cebrià and Newmark, 2007) and its removal results in significant neoblast proliferation in the location of pharynx progenitor specification (Figures S7B–S7E) (Adler et al., 2014). Because pharynx removal elevates local neoblast proliferation, we predicted that it would also lead to the amplification of non-pharynx progenitors that are specified in a nearby region, but that correspond to uninjured tissues. Indeed, pharynx resection led to increased incorporation of BrdU-labeled progenitors into the uninjured ventral nerve cords (VNCs) anterior to the pharynx, and also increased the incidence of newly differentiated SMEDWI+/ChAT+ VNC neurons (Figures 7J, S7F and S7G). This result demonstrates that the response to tissue-specific pharynx resection is not restricted to pharynx progenitors. It also demonstrates that similar to the case of the eye, removal of the VNCs is not required for increased VNC tissue production.
We also wondered whether large injuries that do not remove the pharynx, but that induce proliferation in the region of pharynx progenitor specification would lead to increased pharynx progenitor incorporation. Para-pharyngeal flank resection increased proliferation near the pharynx, where pharynx progenitor specification occurs (Figures S7H–S7K). This surgery increased incorporation of BrdU-labeled neoblast-derived cells into the intact pharynx, including more newly incorporated BrdU-labeled ChAT+ neurons in the distal tip of the pharynx (Figures 7K, 7L and S7L). These observations support a generalizable model for progenitor amplification in which large wounds that cause sustained proliferation amplify nearby progenitor types regardless of the presence or absence of their target tissue.
Discussion
How animals detect absent tissues and specifically regenerate them from diverse, unpredictable injuries is one of the great mysteries of biology. We used the planarian eye to dissect the mechanistic basis of tissue-specific regeneration in a model organ. We found that eye removal failed to specifically alter eye cell production, indicating that the eyes do not suppress their own formation by neoblasts (Figure 7M). Instead, constant progenitor production and incorporation, together with a tissue-specific decrease in the rate of cell death per eye, led to specific regeneration (Figure 5A). This passive mode of tissue-specific regeneration obviates the need for a complex tissue-specific sensing strategy by neoblasts for multiple tissues.
A similar process could allow neoblasts to fuel tissue-specific regeneration from an unlimited set of small injuries, in principle for many cell types, without sensing the identity of missing tissues. Experiments indicate that robust homeostatic tissue production exists for multiple other cell types, including the epidermis and various neural populations (Cowles et al., 2013; Newmark and Sánchez Alvarado, 2000; van Wolfswinkel et al., 2014). Furthermore, specialized neoblasts – like those for the eyes – have been identified for many planarian tissues, such as the protonephridia, epidermis, intestine, pharynx, and various neuron classes (Adler et al., 2014; Cowles et al., 2013; Currie and Pearson, 2013; Marz et al., 2013; Scimone et al., 2014; Scimone et al., 2011; van Wolfswinkel et al., 2014). Future development of tools for the specific ablation of additional tissues and quantification of their progenitors will allow assessment of the generality of our model. However, the simplicity of our passive model for tissue-specific regeneration suggests that it may explain diverse regenerative contexts in planarians and other organisms as well, particularly in cases where tissues display high turnover rates.
Our data support a model in which a decreased cell death rate emerges as a passive consequence of regenerating eyes containing fewer and younger PRNs. A regenerating eye has fewer cells available to undergo death than a homeostatic eye, which would allow net growth even if PRN cell death were purely stochastic. Nonetheless, the ratio of PRNs in regenerating versus uninjured eyes was increased following irradiation, indicating a decrease in the rate of cell loss beyond what would be expected if death were a purely stochastic process. This could be mediated by the fact that regenerating eyes, by definition, contain exclusively young PRNs. We also considered whether an active size-sensing mechanism contributed to the decreased rate of death. In contrast to regenerating eyes, partially resected eyes exhibited cell loss similar to uninjured eyes, inconsistent with active suppression of death occurring in smaller eyes in this context. Partially resected eyes contained a mixture of young and old cells, consistent with PRN age contributing to death in homeostatic and partially resected eyes, but not regenerating eyes. In principle cell death regulation could have an active role in constraining the maximum relative size of eyes and other organs, but this is not a necessary prediction. It will therefore be of interest to continue to investigate mechanisms of cell death regulation to further understand how tissue proportions are reached.
We found that eye progenitor amplification could be accelerated when large injuries coincided with the location of eye progenitor specification (Figure 7M). Because eye progenitor specification occurs in a location separate from the eyes themselves, we were able to decouple this cause of eye progenitor amplification from neoblasts sensing and responding directly to the presence or absence of the eyes. Flank resections that did not remove the eyes, but that removed tissue and increased neoblast proliferation in the location where eye progenitors are specified, induced eye progenitor amplification and increased incorporation into the eyes. We extended this finding to additional tissues, demonstrating that increased progenitor incorporation into the pharynx or ventral nerve cords does not require the removal of these tissues. We cannot exclude that tissue-specific negative feedback regulation from the pharynx or ventral nerve cords might also contribute to neoblast-derived progenitor number regulation. However, our findings support a model that explains how progenitor amplification for the eyes, pharynx, and ventral nerve cords can occur without requiring removal of those tissues.
How might a large wound in the location of progenitor specification trigger progenitor amplification? Neoblasts exhibit a general response to any wound involving substantial missing tissue that includes sustained proliferation and accumulation at the wound site (Wenemoser and Reddien, 2010). One simple scenario is that in a region where positional information is appropriate for the specification of a particular progenitor type (e.g. the eye), neoblast proliferation and accumulation simply creates more opportunities for differentiation/cell fate decisions to occur. Our finding that eye progenitor production in the pre-pharyngeal region closely reflects the degree of general neoblast proliferation is consistent with this hypothesis. Although stem cell progeny are reported to regulate stem cells in various contexts (Hsu and Fuchs, 2012), we observed that anterior wounds that did not remove the eyes or eye progenitors also increased eye progenitor numbers, suggesting that negative feedback by eye progenitors themselves also does not explain eye progenitor behavior. Furthermore, the pharynx itself does not contain VNC progenitors, yet its removal increased incorporation of progenitors into the VNCs. It will be important in future work to determine how large wounds and positional information communicate with and impact neoblast biology to give rise to progenitor amplification. However, our work indicates that progenitor amplification is not simply a result of neoblasts interpreting and responding to the exact identity of missing tissue. We propose that a target-blind mode of progenitor amplification could explain how progenitors are amplified, with neoblasts being regulated by coarse positional information and general wounding signals, rather than the presence or absence of the specific target tissues to be regenerated.
Our findings suggest that the specificity of tissue production during regeneration is inherently imprecise. Increased production of cells in uninjured tissue regions have been observed by BrdU incorporation experiments elsewhere, for example in the planarian gut, where neoblasts differentiate and incorporate into both regenerating and pre-existing intestinal branches after injury (Forsthoefel et al., 2011). In this context, incorporation into non-regenerating tissues may play a role in intestinal branch remodeling. In the axolotl brain, resection of the dorsal pallium increased production of neurons in more rostral, uninjured regions (Amamoto et al., 2016). These findings highlight the importance of considering non-tissue-specific mechanisms for explaining how animals sense and respond to injury.
Conclusion
We found that constant eye progenitor production and less cell death allow planarian eyes to passively regenerate without the need for a tissue-specific sensing strategy by neoblasts. Eye progenitors are amplified when large injuries induce proliferation in a location where eye progenitors are specified, but this process is not influenced by the presence or absence of the eye itself. We conclude that the eye does not regulate production of its own progenitors during eye regeneration. Our work identifies a mode of regeneration in which progenitor specification is target blind – not directly regulated by the presence or absence of the specific target tissue to be regenerated.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-digoxigenin-POD, Fab fragments | Roche | Ref# 11 207 733 910; RRID: AB_514500 |
| anti-fluorescein-POD, Fab fragments | Roche | Ref# 11 426 346 910; RRID: AB_840257 |
| anti-DNP-HRP conjugate | Perkin-Elmer | Cat# FP1129; RRID: AB_2629439 |
| mouse anti-BrdU | Becton Dickinson | Cat# 347580; RRID: AB_10015219 |
| goat anti-mouse-HRP | Invitrogen | Cat# G-21040; RRID: AB_2536527 |
| rabbit monoclonal anti-phospho-Histone H3 (Ser10) | Millipore | Cat# 04-817; RRID: AB_1163440 |
| goat anti-rabbit-HRP | Invitrogen | Cat# G-21234; RRID: AB_2536530 |
| Critical Commercial Assays | ||
| ApopTag Red In Situ Apoptosis Detection Kit | Millipore | Cat# S7165 |
| Experimental Models: Organisms/Strains | ||
| Schmidtea mediterranea, clonal strain CIW4, asexual | Laboratory of Peter Reddien | Sánchez Alvarado et al., 2002 |
| Oligonucleotides | ||
| Sequences used for all FISH probes and dsRNA provided in Table S2 | ||
| Software and Algorithms | ||
| Image J (FIJI) | Schindelin et al., 2012 | https://fiji.sc |
| ZEN digital imaging software | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by, the Lead Contact, Dr. Peter Reddien (reddien@wi.mit.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Asexual S. mediterranea clonal strain CIW4 animals (Sánchez Alvarado et al., 2002), starved 1–2 weeks, were used for all experiments.
METHOD DETAILS
Fluorescence In Situ Hybridization
Animals were killed in 5% NAC in 1x PBS before fixation in 4% formaldehyde in PBSTx (1x PBS containing 0.1% Triton X-100), then stored in methanol at −20°C until subsequent steps. Animals were bleached in 1x SSC solution containing 5% deionized formamide and 1.2% hydrogen peroxide for 1.5 hours while exposed to light. Animals were treated with 2 μg/ml proteinase K in PBSTx with 0.1% SDS, then hybridized with RNA probes diluted 1:800 in a solution of 50% formamide, 5x SSC, 1 mg/ml yeast RNA, 1% Tween-20 and 5% dextran sulfate at 56°C overnight. Animals were blocked for 1–2 hours prior to labeling overnight at 4°C with anti-DIG-POD (1:1500, Roche), anti-FITC-POD (1:2000, Roche), or anti-DNP-HRP (1:100, Perkin-Elmer) in blocking solutions of PBSTx containing 5% heat inactivated horse serum and 5% 10x casein solution (Sigma) for anti-DIG-POD, 10% 10x casein solution for anti-FITC-POD, and 5% horse serum and 5% western blocking reagent (Roche) for anti-DNP-HRP. For tyramide development, animals were placed for 10 minutes in borate buffer (0.1M boric acid, 2M NaCl, pH 8.5), followed by 10 minutes in borate buffer containing rhodamine (1:1000) or fluorescein (1:1500) tyramide and 0.0003% hydrogen peroxide. Prior to antibody labeling for a second probe, peroxidase inactivation was performed in 1% sodium azide overnight at 4°C. Animals were stained in a solution of 1μg/ml DAPI (Sigma) prior to mounting on slides. FISH protocol was adapted from previous work (King and Newmark, 2013; Pearson et al., 2009; Scimone et al., 2016).
BrdU Immunofluorescence
Prior to BrdU immunostaining, FISH was performed as described above, with the following exceptions: bleaching was performed overnight in methanol containing 6% hydrogen peroxide, and tyramide development was performed by placing animals in PBSTi (PBSTx containing 10 mM imidazole) for 30 minutes, then PBSTi containing rhodamine tyramide for 30 minutes, then PBSTi containing tyramide and 0.0002% hydrogen peroxide for 45 minutes. After FISH steps, animals were placed in 2N HCl with 0.5% Triton X-100 for 45 minutes, followed by 0.1M sodium borate for 3 minutes and rinsed in PBSTx. Animals were placed in blocking solution (1x PBS containing 0.3% Triton-X 100, 5mM thymidine, 0.6% BSA, 5% western blocking reagent (Roche)) for 1–2 hours, then labeled with mouse anti-BrdU (1:300, Becton Dickinson) in blocking solution overnight at 4°C. After PBSTx washes and blocking as described above, samples were labeled with goat anti-mouse-HRP secondary antibody (1:200, Invitrogen) in block overnight at 4°C. Samples were then developed with fluorescein tyramide in PBSTi containing hydrogen peroxide as described above, and stained with DAPI prior to mounting. BrdU immunofluorescence protocol was adapted from previous work (Newmark and Sánchez Alvarado, 2000; van Wolfswinkel et al., 2014).
H3P Immunofluorescence
Animals were killed in 2% HCl and placed on ice for 30 seconds, then transferred to Carnoy’s fixative (60% ethanol, 30% chloroform, 10% glacial acetic acid) for 5 minutes at room temperature then 2 hours on ice. Bleaching was performed as for BrdU immunofluorescence, followed by treatment with 2 μg/ml proteinase K in PBSTx with 0.1% SDS. Animals were blocked 1–2 hours in PBSTx containing 10% horse serum, then labeled with anti-H3P (1:100, Millipore) in block overnight at 4°C. After PBSTx washes and blocking as described above, samples were labeled with goat anti-rabbit-HRP secondary antibody (1:100, Invitrogen) in block overnight at 4°C. Samples were developed with rhodamine tyramide in PBSTi containing hydrogen peroxide as described for BrdU immunofluorescence, and stained with DAPI prior to mounting. H3P immunofluorescence protocol was adapted from previous work (Newmark and Sánchez Alvarado, 2000; Wenemoser and Reddien, 2010).
TUNEL
All FISH steps were performed as described above using opsin probe developed with fluorescein tyramide prior to TUNEL. TUNEL was performed using reagents from ApopTag Red In Situ Apoptosis Detection Kit (Millipore, #S7165). Animals were transferred to droplets on eight-well patterned microscope slides (Tekdon, Slide ID# 8-82) in PBSTx and a micropipette was used to replace PBSTx with 30 μl ApopTag equilibration buffer. Slides were incubated at room temperature for 30 minutes. Equilibration buffer was replaced with 30 μl 3 parts ApopTag TdT enzyme mix, 7 parts ApopTag reaction buffer, and slide was sealed and incubated in a dark humid chamber overnight at 37°C. Stop/wash buffer was used to transfer animals back to in situ baskets, which were incubated 5 minutes at 37°C. Animals were transferred to room temperature, washed thoroughly with PBSTx, and incubated one hour in a blocking solution of PBSTx containing 5% horse serum and 5% western blocking reagent (Roche). Animals were again transferred to droplets on eight-well patterned microscope slides, blocking solution was replaced with 30 μl of 1 part blocking solution (described above), 1 part ApopTag anti-digoxigenin rhodamine conjugate, and slides were incubated in dark sealed humid chamber at 4°C overnight. Animals were washed in PBSTx and counterstained in a solution of 1μg/ml DAPI (Sigma) in 1x PBS containing 0.1% Triton-X 100 (PBSTx) before mounting. TUNEL protocol was adapted from previous work (Pellettieri et al., 2010).
Image Acquisition and Quantification
Live images were acquired using a Zeiss Discovery V8 stereomicroscope with an AxioCam HRc camera. Fluorescence image acquisition was performed using a Zeiss LSM 700 confocal microscope. Image J software (Fiji) (Schindelin et al., 2012) or ZEN digital imaging software (Zeiss) was used for processing and quantification of all images. ovo+ eye progenitor quantification was performed on maximum intensity projections (MIPs) of optical sections using blind manual counting or a semi-automated computer protocol as described in Figure S1. ovo+, opsin+, TUNEL+/opsin+, BrdU+/opsin+, BrdU+/ChAT+, SMEDWI-1+/opsin+, SMEDWI-1+/ChAT+, BrdU+ pharynx, and H3P+ cells were quantified blind in files with randomized numerical names by examining optical sections of overlayed fluorescence channels in pre-defined regions of animals as indicated in figures. H3P counting was performed on MIPs for Figures 7D, S6C, S6D, and S7K. For PRN counting, individual eyes were cropped and left eye images were flipped horizontally so that all images appeared as right eyes (or vice versa) for blind quantification, with no indication of experimental condition.
For automated eye progenitor identification (Figure S1), the following steps were performed identically for all files in Fiji: optical section stacks were converted to 8-bit, and maximum intensity projections were generated. The processing tool “Find Edges” was applied. Threshold was applied, with minimum and maximum set at 100 and 255, respectively. The “Analyze Particles” tool was used to highlight eye progenitors and add them to the ROI manager for counting.
BrdU Delivery
BrdU (Sigma #B5002) was administered by soaking for two hours, or by injecting into the pre-pharyngeal region (Figures 7K and S7L), a solution of 1x Montjuic salts containing 25mg/ml BrdU and 3% DMSO. Following administration, animals were rinsed thoroughly with 1x Montjuic salts, then transitioned to 5g/l Instant Ocean until fixation.
Irradiation
Animals were irradiated using a dual Gammacell-40 137cesium source to deliver 6,000 rads.
RNAi
dsRNA was synthesized by in vitro transcription (Promega) from PCR-generated templates with flanking T7 promoters, ethanol precipitated, resuspended in water and annealed, and diluted in liver for delivery by feeding (Petersen and Reddien, 2008; Rouhana et al., 2013). Animals were fed 10, 8, 6, 4, and 2 days prior to eye resection with ovo or Caenorhabditis elegans unc-22 (control) (Benian et al., 1989) dsRNA.
Surgical Procedures
Animals were placed on moist filter paper on a cold block in order to limit movement, and a microsurgery blade was used to remove desired tissues. Pharynx resection was performed by surgical extraction through a small longitudinal dorsal incision. Chemical amputation by exposure to sodium azide was avoided because of its metabolic effects that cause global suppression of mitotic activity (Adler et al., 2014).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed in GraphPad Prism software. Statistical tests, significance, data points, error bars, and other information relevant to figures are described and explained in corresponding legends. Exact animal numbers for all experiments are defined in Table S1.
Supplementary Material
Highlights.
Eye regeneration occurs without regulation of planarian stem cells by the eye
Decreased cell death in regenerating eyes facilitates tissue-specific regeneration
Large head injuries without eye removal increased eye progenitor formation
Eye absence is not sufficient or necessary for increased eye progenitor production
Acknowledgments
We thank D. Wagner and J. van Wolfswinkel for BrdU, FISH, IF and TUNEL advice, K. Martin for technical assistance, and K.D. Atabay and members of the Reddien lab for comments and discussion. We acknowledge NIH (R01GM080639) support. S.A.L. was supported by a National Defense Science & Engineering Graduate (NDSEG) Fellowship. P.W.R. is an Investigator of the Howard Hughes Medical Institute and an associate member of the Broad Institute of Harvard and MIT.
Footnotes
Supplemental Information includes seven figures and two tables and can be found with this article online.
Author contributions
S.A.L. and P.W.R. conceived, designed and interpreted the experiments and wrote the manuscript. S.W.L. made preliminary observations on eye resection and eye progenitor amplification. S.A.L. performed the experiments, acquired and analyzed images and data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler CE, Sánchez Alvarado A. Types or states? Cellular dynamics and regenerative potential. Trends Cell Biol. 2015;25:687–696. doi: 10.1016/j.tcb.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler CE, Seidel CW, McKinney SA, Sánchez Alvarado A. Selective amputation of the pharynx identifies a FoxA-dependent regeneration program in planaria. eLife. 2014;3:e02238. doi: 10.7554/eLife.02238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amamoto R, Huerta VG, Takahashi E, Dai G, Grant AK, Fu Z, Arlotta P. Adult axolotls can regenerate original neuronal diversity in response to brain injury. eLife. 2016;5:e13998. doi: 10.7554/eLife.13998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguñà J. Mitosis in the intact and regenerating planarian Dugesia mediterranea n. sp II Mitotic studies during regeneration, and a possible mechanism of blastema formation. J Exp Zool. 1976;195:65–80. [Google Scholar]
- Benian GM, Kiff JE, Neckelmann N, Moerman DG, Waterston RH. Sequence of an unusually large protein implicated in regulation of myosin activity in C. elegans. Nature. 1989;342:45–50. doi: 10.1038/342045a0. [DOI] [PubMed] [Google Scholar]
- Cebrià F, Newmark PA. Morphogenesis defects are associated with abnormal nervous system regeneration following roboA RNAi in planarians. Development. 2007;134:833–837. doi: 10.1242/dev.02794. [DOI] [PubMed] [Google Scholar]
- Cowles MW, Brown DD, Nisperos SV, Stanley BN, Pearson BJ, Zayas RM. Genome-wide analysis of the bHLH gene family in planarians identifies factors required for adult neurogenesis and neuronal regeneration. Development. 2013;140:4691–4702. doi: 10.1242/dev.098616. [DOI] [PubMed] [Google Scholar]
- Currie KW, Pearson BJ. Transcription factors lhx1/5-1 and pitx are required for the maintenance and regeneration of serotonergic neurons in planarians. Development. 2013;140:3577–3588. doi: 10.1242/dev.098590. [DOI] [PubMed] [Google Scholar]
- Dubois F. Contribution á l ’ètude de la migration des cellules de règènèration chez les Planaires dulcicoles. Bull Biol Fr Belg. 1949;83:213–283. [Google Scholar]
- Forsthoefel DJ, Park AE, Newmark PA. Stem cell-based growth, regeneration, and remodeling of the planarian intestine. Dev Biol. 2011;356:445–459. doi: 10.1016/j.ydbio.2011.05.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Peters AH, Newmark PA. A Bruno-like gene is required for stem cell maintenance in planarians. Dev Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, Sánchez Alvarado A. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol. 2010;347:24–39. doi: 10.1016/j.ydbio.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Fuchs E. A family business: stem cell progeny join the niche to regulate homeostasis. Nat Rev Mol Cell Biol. 2012;13:103–114. doi: 10.1038/nrm3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RS, Newmark PA. In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev Biol. 2013;13:8. doi: 10.1186/1471-213X-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan SW, Reddien PW. dlx and sp6-9 control optic cup regeneration in a prototypic eye. PLoS Genet. 2011;7:e1002226. doi: 10.1371/journal.pgen.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan SW, Reddien PW. Transcriptome analysis of the planarian eye identifies ovo as a specific regulator of eye regeneration. Cell Rep. 2012;2:294–307. doi: 10.1016/j.celrep.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangel M, Bonsall MB, Aboobaker A. Feedback control in planarian stem cell systems. BMC Syst Biol. 2016;10:17. doi: 10.1186/s12918-016-0261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz M, Seebeck F, Bartscherer K. A Pitx transcription factor controls the establishment and maintenance of the serotonergic lineage in planarians. Development. 2013;140:4499–4509. doi: 10.1242/dev.100081. [DOI] [PubMed] [Google Scholar]
- Newmark P, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Inoue T, Yoshimoto K, Taniguchi T, Kitamura Y, Agata K. Regeneration of dopaminergic neurons after 6-hydroxydopamine-induced lesion in planarian brain. J Neurochem. 2011;119:1217–1231. doi: 10.1111/j.1471-4159.2011.07518.x. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dynam. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sánchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Dev Biol. 2010;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Smed-βcatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. A wound-induced Wnt expression program controls planarian regeneration polarity. Proc Natl Acad Sci USA. 2009;106:17061–17066. doi: 10.1073/pnas.0906823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet. 2010;11:710–722. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW. Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet. 2011;27:277–285. doi: 10.1016/j.tig.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Ann Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Rouhana L, Weiss JA, Forsthoefel DJ, Lee H, King RS, Inoue T, Shibata N, Agata K, Newmark PA. RNA interference by feeding in vitro-synthesized double-stranded RNA to planarians: methodology and dynamics. Dev Dynam. 2013;242:718–730. doi: 10.1002/dvdy.23950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A. Regeneration in the metazoans: why does it happen? Bioessays. 2000;22:578–590. doi: 10.1002/(SICI)1521-1878(200006)22:6<578::AID-BIES11>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci USA. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA, Robb SM, Juste R. The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development. 2002;129:5659–5665. doi: 10.1242/dev.00167. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Cote LE, Rogers T, Reddien PW. Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. eLife. 2016;5:e12845. doi: 10.7554/eLife.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Kravarik KM, Lapan SW, Reddien PW. Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Reports. 2014;3:339–352. doi: 10.1016/j.stemcr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Meisel J, Reddien PW. The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea. Development. 2010;137:1231–1241. doi: 10.1242/dev.042051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Srivastava M, Bell GW, Reddien PW. A regulatory program for excretory system regeneration in planarians. Development. 2011;138:4387–4398. doi: 10.1242/dev.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Dev Cell. 2011;21:172–185. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Wagner DE, Reddien PW. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell. 2014;15:326–339. doi: 10.1016/j.stem.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Ho JJ, Reddien PW. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell. 2012;10:299–311. doi: 10.1016/j.stem.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol. 2010;344:979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchley JN, Mayer M, Wagner DE, Owen JH, Reddien PW. Muscle cells provide instructions for planarian regeneration. Cell Rep. 2013;4:633–641. doi: 10.1016/j.celrep.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.